Abstract
Summary: Plant long non-coding RNA database (PLncDB) attempts to provide the following functions related to long non-coding RNAs (lncRNAs): (i) Genomic information for a large number of lncRNAs collected from various resources; (ii) an online genome browser for plant lncRNAs based on a platform similar to that of the UCSC Genome Browser; (iii) Integration of transcriptome datasets derived from various samples including different tissues, developmental stages, mutants and stress treatments; and (iv) A list of epigenetic modification datasets and small RNA datasets. Currently, our PLncDB provides a comprehensive genomic view of Arabidopsis lncRNAs for the plant research community. This database will be regularly updated with new plant genome when available so as to greatly facilitate future investigations on plant lncRNAs.
Availability: PLncDB is freely accessible at http://chualab.rockefeller.edu/gbrowse2/homepage.html and all results can be downloaded for free at the website.
Contact: chua@rockefeller.edu
1 INTRODUCTION
Non-coding RNAs (ncRNAs) are a family of RNAs that do not encode proteins. On the basis of their length and genomic locations, ncRNAs can be further classified as (i) small ncRNAs including miRNAs and small interfering RNAs (siRNAs); (ii) natural antisense transcripts (NATs) (Wang et al., 2005, 2006; Zhang et al., 2012); (iii) long intronic non-coding RNAs (incRNAs); and (iv) long intergenic non-coding RNAs (lincRNAs) (Guttman et al., 2009; Liu et al., 2012). RNAs in the last three categories are at least 200 nt or longer and they are referred to as long non-coding RNAs (lncRNAs).
Genomes of human (Gupta et al., 2010; Khalil et al., 2009), mouse (Dinger et al., 2008) and fly (Tupy et al., 2005) have been shown to encode lncRNAs that play important roles in cell differentiation, immune response, imprinting, tumor genesis and other important biological processes (Dinger et al., 2008; Gupta et al., 2010; Khalil et al., 2009; Liao et al., 2011a; Wilusz et al., 2009). Besides, genetic mutations of human lncRNAs have been shown to be associated with diseases and pathophysiological conditions (Cabianca et al., 2012; Gupta et al., 2010; Hu et al., 2011).
For plants, genome-wide search for ncRNAs has been previously conducted in Arabidopsis thaliana (MacIntosh et al., 2001; Marker et al., 2002; Rymarquis et al., 2008; Song et al., 2009), Medicago truncatula (Wen et al., 2007), Zea mays (Boerner and McGinnis, 2012) and Tritucum aestivum (Xin et al., 2011). The recent genome-wide study based on around 200 Arabidopsis tilling array datasets and RNA sequencing (RNA-seq) has identified thousands of lncRNAs in Arabidopsis (Liu et al., 2012). These lncRNAs show tissue-specific expression, and a large number of them are responsive to abiotic stresses (Liu et al., 2012). However, the function of these lncRNAs remains largely unexplored. Genomic loci of many lncRNAs are associated with histone modifications and DNA methylations suggesting an epigenetic regulation of these loci (Guttman et al., 2009; Liu et al., 2012). In addition, biogenesis of a subgroup of lncRNAs is co-regulated by CBP20, CBP80 and SERRATE (Liu et al., 2012). Some sense and antisense double-stranded RNAs involving lncRNA partners are processed by the RNA interference machinery into siRNAs (Zhang et al., 2012).
Although thousands of lncRNAs have been identified in Arabidopsis and other plants and their expression has been profiled on a genome-wide basis, these RNAs have not been fully recorded and annotated in public databases. As far as we know, there are only seven databases and one server related to currently available lncRNAs: TAIR (Swarbreck et al., 2008), PlantNATsDB (Chen et al., 2012), lncRNAdb (Amaral et al., 2011), NRED (Dinger et al., 2009), ncRNAimprint (Zhang et al., 2010), NONCODE (Bu et al., 2012) and ncFANs (Liao et al., 2011b). Among them, only PlantNATsDB (Chen et al., 2012) is designed to query about NATs pairs; however, this database just lists all NATs pair and does not provide a genome view. The other six databases are not specifically designed for plant lncRNAs (Table 1). Therefore, a database that contains comprehensive information related to lncRNAs, such as genomic information, expression profiles, siRNA information and associated epigenetic markers is warranted. Here, we attempt to develop an online database for plant lncRNAs, named PLncDB (Plant long non-coding RNA database), with the aim to provide comprehensive information for plant lncRNAs. Table 1 compares information content between our database and those of others.
Table 1.
Comparison between PLncDB with related databases
| Database | lncRNA | Expression | Source | Organism | Description |
|---|---|---|---|---|---|
| TAIR10 | 478 | × | × | Arabidopsis | No expression data |
| lncRNAdb | 5/176 | × | × | All | Only validated lncRNAs |
| NRED | 0 (plant) | √ | Design arrays | Human, mouse | Very few lncRNAs |
| ncRNAi-mprint | 26/7094 | × | × | 9 mammalian | Only imprinted lncRNAs |
| NONCO-DE | 73 372 | × | × | Human, mouse | From literature |
| PlantNATsDB | 2 138 498 | × | × | Plant | Predicted NATs |
| PLncDB | 16 227 | √ | Tiling, lincRNA arrays RNA-seq | Arabidopsis | Mapped lncRNAs, Expression, Epigenetic data |
2 AIMS OF DATABASE
Recent studies in mammalian genomes have shown that lncRNAs are generally characterized by four interesting features: (i) eukaryotic genome codes a few thousand lincRNAs (Cabili et al., 2011; Dinger et al., 2008; Guttman et al., 2009); (ii) lncRNA genes are expressed in a temporal and/or spatial specific manner (Dinger et al., 2008; Managadze et al., 2011); (iii) genomic loci encoding lncRNAs are associated with epigenetic markers (Guttman et al., 2009; Khalil et al., 2009); (iv) sense and antisense transcripts double-stranded structure may be processed into siRNAs (Zhang et al., 2012).
Based on the characteristics of lncRNAs, our PlncDB aims to provide the following four essential functions: (i) a collection and integration of lncRNAs from different data resources; (ii) lncRNA expression levels in various samples including different tissues, developmental stages, mutants and stress treatments; (iii) epigenetic modifications (e.g. DNA methylations and histone modifications) on lncRNA-encoding loci and their flanking genomic regions; and (iv) a collection of siRNA sequencing dataset across the whole genome (Table 2).
Table 2.
Detail information about PLncDB
| Dataset | Number | Description |
|---|---|---|
| lncRNA (RepTAS) | ||
| Flower/root/leaf | 4915 | lincRNA array |
| lincRNA | 3718 | |
| Pri-miRNAs | 173 | |
| Protein-coding gene | 90 | |
| DNA methylation | ||
| Met1/DDC | Tilling array | Ryan (Gerhard et al., 2004) |
| Gene | ||
| Protein coding gene | 33 323 | TAIR 10 |
| Small RNA | 134 478 | siRNA sequence |
| Histone modification | ||
| Dataset1 (WT and VIP3) | Sookyung (Oh et al., 2008) | |
| H3K27me3/2 | Tilling array | |
| H3K36me2/H3K4me3 | Tilling array | |
| Dataset2 (WT and Met1) | Xiaoyu Z (Zhang et al., 2009) | |
| H3K4me1/2/3 | Tilling array | |
| Tiling array | ||
| Phosphate (Shoot/root) 10 d/13 d | Genome | Tiling array |
| ABA/drought/cold/salt 2 h/10 h | Genome | Tiling (Matsui et al., 2008) |
3 DATABASE ACCESS
We constructed a genome browser database using the open source GBrowse library (Stein et al., 2002) to integrate and visualize these different sources PLncDB. In the case of Arabidopsis, we have also provided an updated version from TAIR10 with respect to genomic context, alignment information, protein coding gene annotation and known ncRNAs. As for lncRNA expression information, we adopted a new file format BigWig (Kent et al., 2010)
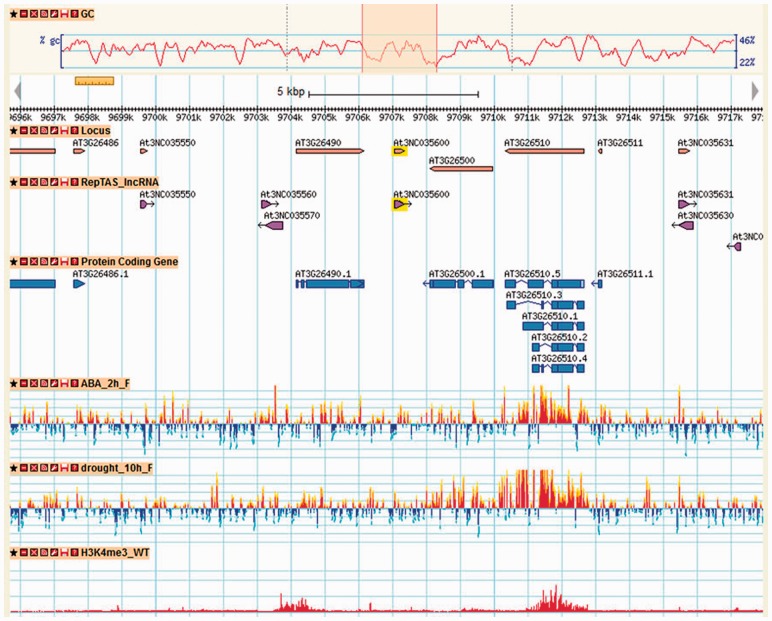
to expedite the querying. The database can be accessed or queried in various ways. Just by clicking on a specific lncRNA, one can visualize related mutant/stress information (Fig. 1). Specific searches can be performed using the name/keywords of gene/protein and/or location on the chromosome. At the same time the entire database is available for download in different format on the website.
Fig. 1.
Snapshot of PLncDB
4 FUNCTION OF THE DATABASE
4.1 An online database to deposit, browse and download information relating to a large number of lncRNAs
We collected a total of 16 227 Arabidopsis lncRNAs from various resources published in the past decade (Liu et al., 2012). These lncRNAs were identified based on different versions of genome sequences and were annotated separately using different criteria. For our Reproducibility-based Tiling array Analysis Strategy (RepTAS) method, 13 466 transcript units (TU) were identified (Liu et al., 2012). To provide uniformed and comprehensive information for Arabidopsis lncRNAs, by comparing the genomic loci of TUs with exons, pseudogenes, repeat sequences and transposable elements annotated in TAIR10, we finally reclassified the remaining TUs into the following six categories: (i) TU encoding NATs, (ii) Repeats-Containing TUs, (iii) Gene-Associated TU, (iv) TUs encoding transcripts with long open reading frames suggesting novel protein-coding genes, also named TUs of Unknown Coding Potential (Cabili et al., 2011); (v) TUs for lincRNAs; (vi) Other Intergenic TUs. Recently, using a RepTAS, we identified 6480 genes encoding lincRNAs (Liu et al., 2012).
4.2 An online genome browser to show lncRNA expression of various transcriptome data
An interesting feature of lncRNAs is their significant tissue-specific expression pattern compared with mRNAs (Cabili et al., 2011; Dinger et al., 2008). In our previous study, we profiled transcriptome of Arabidopsis lncRNAs using multiple platforms including high-throughput RNA-seq, tiling-arrays and a custom designed long oligonucleotide microarray (Liu et al., 2012). Dynamic expression patterns of thousands of lncRNA were found in a number of tissues, developmental time-points, biotic/abiotic stress conditions and Arabidopsis mutants deficient in several RNA-interacting proteins. It is reasonable to hypothesize that the expression specificity of lncRNA may imply biological functions. Therefore, an online genome browser with a large set of transcriptome data will be useful to biologists to further investigate functional roles of the lincRNAs.
In this study, we collected datasets of three transcriptome detection platforms and analyzed their data quality. The raw datasets of the selective samples were normalized and re-analyzed using a uniformed analysis protocol. We then integrated the processed signal intensities into the genome browser (Fig. 1). The current version of PLncDB is version 1.
4.3 An Association of lncRNA-encoding genomic loci with epigenetic markers
Expression of thousands of lncRNAs is associated with epigenetic regulation. Yet, little is known about epigenetic regulations of lncRNA expression in plants. Two widely used strategies have been used to profile epigenetic regulation in a genome-wide view: (i) detection of epigenetic modification sites or modifier binding sites using immunoprecipitation followed by high-throughput detection such as Chromatin Immunoprecipitation on chip (ChIP-chip) and Chip-sequencing (ChIP-seq). (ii) Profiling of transcriptome changes in mutants deficient in epigenetic modifiers. In our study, we analyzed ChIP-chip data of the following histone modifications (H3K27me3, H3K4me3, H3K36me3 and H3K9me3).
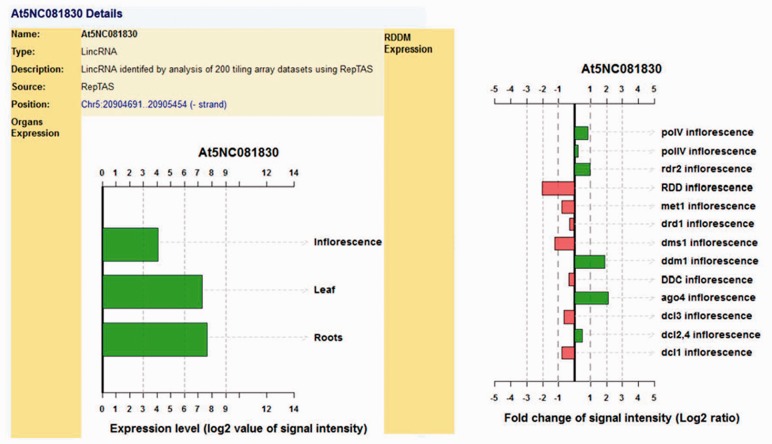
Besides, we also profiled expression changes of lncRNAs in a number of RdRM-related mutants (RDD, DCL1/2/3/4, AGO4, RDR2 and DMS1) (Fig. 2). These results have been integrated into the genome browser of PLncDB for public access (Figs 1 and 2).
Fig. 2.
An example: detailed expression for At5NC081830
4.4 Sense and antisense double strands are precursor of siRNA
Sense and antisense transcripts may form double-stranded RNAs that are subsequently processed by the RNA interference machinery into siRNAs (Zhang et al., 2012). A few so-called nat-siRNAs have been reported in plants, mammals, Drosophila and yeasts. However, many questions remain regarding the features and biogenesis of nat-siRNAs (Luo et al., 2009; Wang et al., 2006; Zhang et al., 2012). For this reason, we also included our previous small RNAs sequence dataset in this database (Wang et al., 2011).
5 FUTURE DIRECTION
In future, we also plan to include in our database other lncRNA datasets, like those for intronic non-coding RNAs (incRNAs) and Natural Antisense RNAs (NATs). Furthermore, information on lncRNAs of other plant species, e.g. rice, corn, etc, will be also included as the data become available.
ACKNOWLEDGEMENTS
We thank all people’s contribution on this work.
Funding: This work was supported by Singapore Ministry of Education Tier-2 grant MOE2009-T2-2-004 to L.S.W. and NIH GM44640 and the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ906910) Rural Development Administration, Republic of Korea to N.-H.C.
Conflict of Interest: none declared.
REFERENCES
- Amaral PP, et al. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner S, McGinnis KM. Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS One. 2012;7:e43047. doi: 10.1371/journal.pone.0043047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu D, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–D215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, et al. PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 2012;40:D1187–D1193. doi: 10.1093/nar/gkr823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, et al. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DS, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, et al. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011a;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, et al. ncFANs: a web server for functional annotation of long non-coding RNAs. Nucleic Acids Res. 2011b;39:W118–W124. doi: 10.1093/nar/gkr432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo QJ, et al. Evidence for antisense transcription associated with microRNA target mRNAs in Arabidopsis. PLoS Genet. 2009;5:e1000457. doi: 10.1371/journal.pgen.1000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, et al. Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol. 2001;127:765–776. [PMC free article] [PubMed] [Google Scholar]
- Managadze D, et al. Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol. Evol. 2011;3:1390–1404. doi: 10.1093/gbe/evr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker C, et al. Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr Biol. 2002;12:2002–2013. doi: 10.1016/s0960-9822(02)01304-0. [DOI] [PubMed] [Google Scholar]
- Matsui A, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Oh S, et al. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, et al. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008;13:329–334. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Song D, et al. Computational prediction of novel non-coding RNAs in Arabidopsis thaliana. BMC Bioinformatics. 2009;10(Suppl. 1):S36. doi: 10.1186/1471-2105-10-S1-S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupy JL, et al. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, et al. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 2006;7:R92. doi: 10.1186/gb-2006-7-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 2011;67:292–304. doi: 10.1111/j.1365-313X.2011.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, et al. In Silico identification and characterization of mRNA-like noncoding transcripts in Medicago truncatula. In Silico Biol. 2007;7:485–505. [PubMed] [Google Scholar]
- Wilusz JE, et al. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, et al. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. ncRNAimprint: a comprehensive database of mammalian imprinted noncoding RNAs. RNA. 2010;16:1889–1901. doi: 10.1261/rna.2226910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012;13:R20. doi: 10.1186/gb-2012-13-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]