Abstract
Degradation of mRNAs plays an essential role in modulation of gene expression and in quality control of mRNA biogenesis. Nearly all major mRNA decay pathways characterized thus far in eukaryotes are initiated by deadenylation, i.e., shortening of the mRNA 3′ poly(A) tail. Deadenylation is often a rate-limiting step for mRNA degradation and translational silencing, making it an important control point for both processes. In this review, we discuss the fundamental principles that govern mRNA deadenylation in eukaryotes. We use several major mRNA decay pathways in mammalian cells to illustrate mechanisms and regulation of deadenylation-dependent mRNA decay, including decay directed by AU-rich elements (AREs) in the 3′ untranslated region (UTR), the rapid decay mediated by destabilizing elements in protein-coding regions, the surveillance mechanism that detects and degrades nonsense-containing mRNA (i.e., NMD), the decay directed by miRNAs, and the default decay pathway for stable messages. Mammalian mRNA deadenylation involves two consecutive phases mediated by the PAN2-PAN3 and the CCR4-CAF1 complexes, respectively. Decapping takes place after deadenylation and may serve as a backup mechanism to trigger mRNA decay if initial deadenylation is compromised. In addition, we discuss how deadenylation impacts the dynamics of RNA processing bodies (P-bodies), where non-translatable mRNAs can be degraded or stored. Possible models for mechanisms of various deadenylation-dependent mRNA decay pathways are also discussed.
Keywords: deadenylation, mRNA turnover, poly(A), microRNA, ARE, NMD, P-body, deadenylase, translation, PABP
Messenger RNA (mRNA) mediates the flow of genetic information from the nucleus to the cytoplasm, where it serves as a template for translation by ribosomes into protein product. Once in the cytoplasm, the fate of an mRNA is greatly influenced by how well it is translated and where it is localized or stored subcellularly [1–6]. Eventually, all mRNAs are targeted for degradation. Thus, regulation of mRNA turnover is important for controlling the abundance of cellular transcripts and thus the levels of protein expression [4, 7, 8]. All mRNAs contain a 7-methyl guanosine cap (m7G-cap) structure at their 5′ termini, which in conjunction with the cap binding protein complex renders the mRNA resistant to 5′ to 3′ exonucleases [9]. At the 3′ termini of all mRNAs, with the exception of histone mRNA, a poly(A) tail (i.e., a stretch of adenylate residues) along with the poly(A) binding protein (PABP) protects the mRNA from ribonuclease attack at the 3′ end [10]. Moreover, the 5′ m7G-cap/cap-binding complex and the 3′ poly(A)/PABP complex can interact with each other to form a closed loop that enhances translation initiation and provides an effective means to protect mRNA ends from nuclease attack [11, 12]. The length of a poly(A) tail is a critical determinant of mRNA stability and translation efficiency [13–15]. Following transport of mRNAs to the cytoplasm, poly(A) tails undergo shortening at different rates to approximately 10–60 nucleotides (nt), a process termed deadenylation [16, 17]. Deadenylation impacts mRNA by reducing its translatability and/or inducing its degradation. Recent identification and characterization of at least eight distinct deadenylases in metazoa highlights the diverse biological functions of deadenylases, and thus of deadenylation, in mRNA regulation [18–20]. Moreover, deadenylation has been demonstrated to be the major step triggering mRNA decay in eukaryotic cells (see below). It is not surprising that regulating deadenylation emerges as a key means of controlling eukaryotic gene expression.
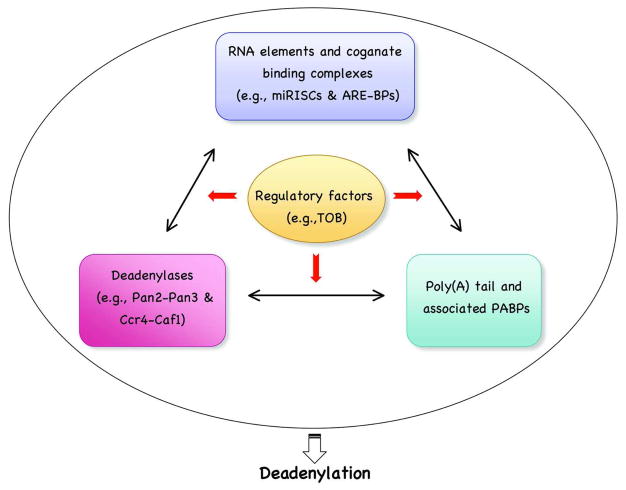
In this review, we first discuss the fundamental principles that govern mRNA deadenylation in eukaryotes. We focus on what has been learned about the mechanistic steps in yeast and mammalian cells. We then use examples, including mRNA decay mediated by adenine/uridine (AU)-rich elements (AREs), nonsense-mediated decay (NMD), microRNA (miRNA)-mediated decay (miRMD), and decay mediated by the determinant in the c-fos protein coding region, to illustrate mechanisms and regulation of deadenylation-dependent mRNA decay in mammalian cells. We also discuss how deadenylation impacts the dynamics of RNA processing bodies (P-bodies), the recently identified cytoplasmic foci that are enriched in non-translatable mRNA-protein complexes (mRNPs) and may be the trash bin for at least some mRNAs [21, 22]. We do not discuss the removal of oligo(A) from RNA intermediates associated with nuclear RNA decay [23], or deadenylation that occurs in the cytoplasm of oocytes and early embryos where shortening of the 3′ poly(A) tails of maternal mRNAs is used as a major means to silence translation of the transcripts [1, 24]. We also do not discuss deadenylases per se, as they are considered by other reviews (see Cross-references). We discuss new findings that highlight the importance of regulating deadenylation in controlling eukaryotic gene expression, particularly in mammalian somatic cells. We propose that the deadenylation rate of any given mRNA in a specific cellular environment is a summation of interactions among several key players for deadenylation, including deadenylases, the poly(A)-PABP complex, mRNA element-binding complexes, and factors that modulate these interactions (Figure 1).
Figure 1.
The deadenylation rate of an mRNA in mammalian cells is determined by interplays among several key players.
I. Development of an approach to study mRNA decay kinetics
Nearly four decades ago, experiments on the RNase susceptibility of different RNAs led to the discovery of a poly(A) tail at the 3′ end of eukaryotic mRNAs [25, 26]. It is now clear that all eukaryotic mRNAs, with the exception of histone mRNA, contain a 3′ poly(A) tail that associates with multiple copies of PABP. The mature mRNAs that have just arrived in the cytoplasm are fairly homogenous in the size of their poly(A) tails, ranging from 200 to 250 residues in mammalian cells and from 70 to 80 residues in budding yeast, Saccharomyces cerevisiae [11, 16]. Owing to the extreme lability of decay intermediates in vivo and the lack of proper methodologies to rigorously monitor deadenylation and decay kinetics, the exact role of the 3′ poly(A) tail in mRNA turnover were nebulous for nearly two decades since its discovery in the early 70s [16]. Existing methodologies were inadequate to establish the precise precursor-product relationships between an mRNA and its decay intermediates, making it hard to figure out how mRNA degradation is triggered in the cytoplasm and what mechanistic steps are involved.
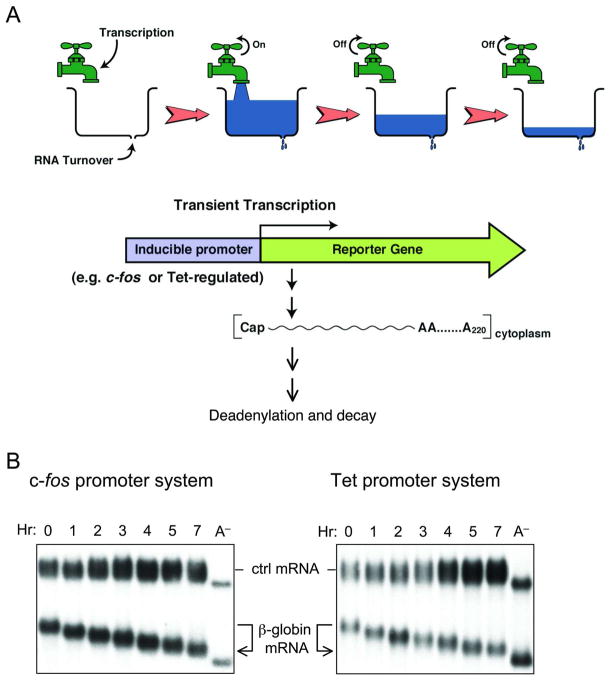
To decipher the mechanism(s) that initiate decay of eukaryotic mRNA in vivo, one has to be able to chase the mature form of an mRNA that has just arrived in the cytoplasm from the nucleus, and thus contains a full-length poly(A) tail, into its decay intermediates and final products. Importantly, the chase has to be done in the absence of further transcription of the mRNA of interest so that changes in the 3′ poly(A) length and the entire mRNA size during the time-course can be accurately monitored. These considerations led to the development of the transcriptional pulsing approach (Figure 2A), which employs the c-fos or the Tet-regulated promoter to drive a highly robust but transient transcription of a reporter gene in mammalian cells [27–31].
Figure 2.
The transcriptional pulsing approach to determine mRNA deadenylation and decay kinetics. A. Schematic diagram illustrating how an inducible promoter can be used to achieve a short burst of mRNA production for kinetic studies. B. Northern blots showing default deadenylation exhibited by the stable β-globin mRNA. Transient transcription is driven by either the c-fos promoter (left) or the Tet-promoter (right). Times indicate hours (Hr) after transcriptional pulsing of β-globin gene. A− : poly(A)− mRNA as a size marker; ctrl : control mRNA constitutively expressed used as an internal control.
The c-fos promoter can be induced quickly and transiently in response to serum or growth factor addition [32, 33]. Following serum induction, transcription from the c-fos promoter only lasts about 30 minutes (min), thereby providing a reliable and simple way of achieving a transient burst of transcription. The resulting mRNA population synthesized during the 30 min time-window is fairly homogenous in length. In a typical time-course experiment, RNA samples are collected at different time points after serum induction and are analyzed by Northern blot analysis (Figure 2B, left). This approach has led to the finding that both stable transcripts (e.g.,β-globin mRNA) and unstable messages (e.g., c-fos mRNA) undergo deadenylation that precedes decay of the RNA body in mammalian cells [28, 30].
The c-fos system is restricted to analysis of mRNA decay in cells undergoing the G0 to G1 transition triggered by serum induction of growth-arrested cells. In the late 90’s, taking advantage of the tight control of transcription made possible by the Tet-off promoter and tTA trans-activator system in mammalian cells [34], a new transcriptional pulsing approach was established to complement the c-fos system [27, 31]. In the new system, a transient transcription from a Tet-off promoter driven reporter gene can be accomplished by including or excluding tetracycline in the culture medium. This new system makes it possible to conduct time-course experiments in mammalian cells (e.g., Figure 2B, right) under any physiological or cellular state and thus study the regulation of mRNA turnover by a signaling pathway. Combined with small interfering RNA (siRNA)-mediated gene knockdown and overexpression of dominant-negative mutants of different factors, this new transcriptional pulsing approach has led to the finding that deadenylation is biphasic, involving consecutive actions of Pan2-Pan3 and Ccr4-Caf1 deadenylase complexes in mammalian cells (see section III below) [35].
A similar transcriptional pulsing approach was developed in budding yeast carrying a polymerase II temperature-sensitive mutation [36]. Transient transcription of a reporter gene was under the regulation of the GAL1 promoter. Cells were cultured in medium containing galactose; transcription was induced at 30 °C for 10–15 min and then rapidly repressed by shifting the cells to 36 °C. Combining this transcriptional pulsing approach with both genetic and biochemical approaches, a general turnover pathway for both stable and unstable transcripts in budding yeast was revealed. In this pathway, shortening of the 3′ poly(A) tail of an mRNA to oligo(A) leads to removal of the cap structure (or decapping) at the 5′ end followed by 5′ to 3′ exonucloelytic digestion of the RNA body [9, 36, 37].
II. Deadenylation: the first major step triggering mRNA decay
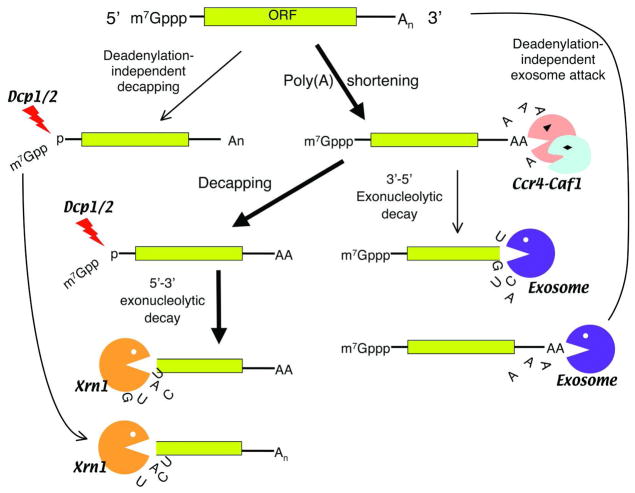
In theory, mRNA degradation can be initiated by deadenylation from the 3′ end, decapping at the 5′ end, or endonucleolytic cleavage within the message. A newly created end would then be unprotected from subsequent exonuclease attack. However, studies using the transcriptional pulsing approaches showed that mRNA decay in eukaryotes involves multiple steps and is more complicated than simply creating free ends followed by exonucleolytic digestion (Figure 3) [8].
Figure 3.
Schematic diagram showing eukarytoic mRNA decay pathways in the cytoplasm. Thick arrows indicate the major pathway for cytoplasmic mRNA degradation triggered by deadenylation. Note that the decay pathway triggered by an endonucleolytic cleavage (e.g., siRNA-mediated decay) is not depicted here.
Nearly all major mRNA decay pathways yet characterized in eukaryotes are initiated by deadenylation, with the predominant poly(A) nuclease being the Ccr4-Caf1 complex [35, 38–40]. Deadenylation primarily leads to decapping by the Dcp1-Dcp2 complex at the 5′ end with subsequent 5′to 3′ exonucleolytic digestion of the RNA body by Xrn1 [9, 41–44]. Alternatively, when the 5′ to 3′ decay pathway is compromised, after deadenylation the mRNA body can be degraded from the 3′ end by a large complex called an exosome (Figure 3) [45–48]. Deadenylation-independent pathways of mRNA degradation also exist. For example, siRNA-mediated decay is accomplished through an endonucleolytic cleavage of the siRNA-targets [49–51], and in Drosophila, nonsense or premature termination codons (PTC) trigger an endonucleolytic cleavage followed by exonuclease digestion of the exposed ends [52]. A similar pathway may also contribute to human NMD as SMG6, a factor involved in NMD, was reported to promote endonucleolytic cleavage of nonsense mRNA in human cells [79]. Other deadenylation-independent decay pathways were observed mainly in yeast. For instance, when an aberrant yeast mRNA loses its stop codon and becomes a non-stop mRNA, the exosome is able to completely degrade the mRNA from the 3′ end of the poly(A) tail without involving any known deadenylases [53, 54]. Also, a decay pathway for mRNAs with ribosomal pausing (“no-go” decay pathway) triggered by endonucleolytic cleavage was observed in yeast [55]. Additionally, in yeast, nonsense codons usually trigger rapid Dcp1-Dcp2-mediated decapping followed by 5′ to 3′ Xrn1 digestion of the RNA body before the poly(A) tail is significantly removed [56]. Interestingly, when nonsense codons are present in the mid-region of an open-reading frame (ORF) in yeast, the corresponding mRNAs are triggered for rapid decay through deadenylation [57].
It remains unclear whether deadenylation-independent decapping operates in mammalian cells. All major modes of mammalian mRNA decay observed thus far are triggered by deadenylation, including decay directed by AREs in the 3′ untranslated region (UTR), the rapid decay mediated by destabilizing elements in protein-coding regions, the surveillance mechanism that detects and degrades nonsense-containing mRNA (i.e., NMD), the decay directed by miRNAs, and the default decay pathway for stable messages (Figure 4) (see section IV below).
Figure 4.
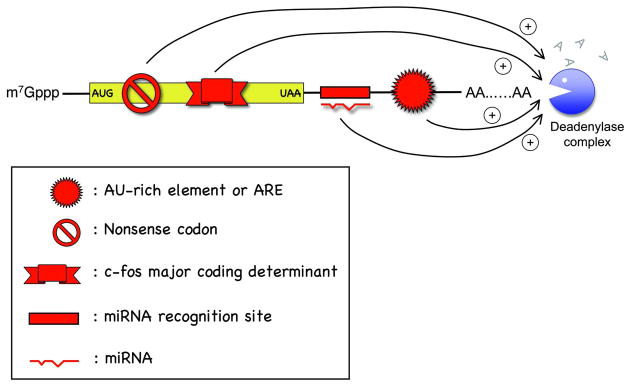
Schematic diagram illustrating cis-acting elements or mutations in mammalian mRNA that promote accelerated deadenylation.
III. Mammalian deadenylation involves two consecutive steps
Among various deadenylases, Ccr4-Caf1 and Pan2-Pan3 are highly conserved throughout evolution from yeast to mammals [7, 18–20]. In yeast, mRNA deadenylation is mainly mediated by the Ccr4p-Caf1p complex [39, 40, 58], while the Pan2p-Pan3p complex is thought to trim the 3′ poly(A) to approximately 70–80 nt in the nucleus during the mRNA maturation process [59]. When the Ccr4p-Caf1p deadenylating function is compromised, Pan2p-Pan3p accounts for some residual deadenylase activity in the cytoplasm [39]. In contrast, knocking out Pan2p-Pan3p function has little effect on deadenylation or decay. This appears to be the case in Drosophila as well [60].
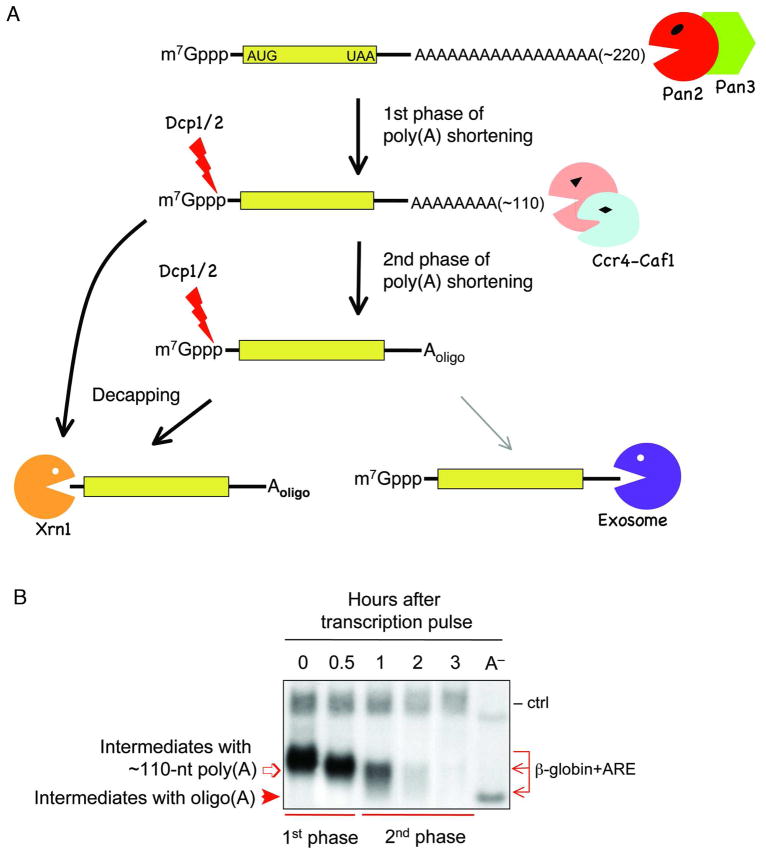
Studies using the Tet-promoter driven transcriptional pulsing approach discovered that mammalian deadenylation exhibits biphasic kinetics (Figure 5A) [35]. In the first phase, mRNAs undergo slow but fairly synchronous deadenylation that shortens the 3′ poly(A) tail to approximately 110 nt with no detectable decay of the RNA body (Figure 5B). The observed kinetics are consistent with a distributive enzymatic digestion of the poly(A) tail. An enzyme catalyzing distributive deadenylation dissociates from a poly(A) tail after hydrolyzing only a few nucleotides and then switches to another poly(A) tail, whereas a deadenylase catalyzing processive digestion remains associated with and nearly completely hydrolyze a poly(A) tail prior to switching to another poly(A) tail [61]. In the second phase of deadenylation, mRNAs become less synchronous, with a poly(A) size distribution ranging from 110 nt to 10 nt. During this phase, decay of the RNA body is also observed (Figure 5B). The second phase kinetics are consistent with processive enzymatic digestion of the poly(A) tail to oligo(A), leading to decay of the RNA body.
Figure 5.
A: Schematic diagram showing the consecutive phases of deadenylation and mechanistic steps following deadenylation in mammalian cells. B: Northern blot showing that biphasic deadenylation precedes decay of the RNA body. Transient transcription of β-globin carrying the c-fos ARE is driven by the Tet-promoter. Times indicate hours after transcriptional pulsing of the gene. A− : poly(A)− mRNA as a size marker; ctrl : control mRNA constitutively expressed used as an internal control.
The mechanistic steps and enzymes involved in the biphasic kinetics of deadenylation were defined by studies that monitored mRNA deadenylation and decay using the transcriptional pulsing approach after individual poly(A) nucleases, Dcp2 decapping enzyme, or a combination of these factors were inactivated by over-expression of dominant-negative mutants or by siRNA-mediated gene knockdown [35]. When Pan2 activity was compromised, the first phase of deadenylation was slowed down. However, there was no significant change in mRNA decay, suggesting that Ccr4-Caf1 can eventually take over deadenylation when Pan2 activity is impeded. In contrast, when either Ccr4 or Caf1 activity was compromised, an mRNA intermediate with a ~110 nt poly(A) tail accumulated following the first phase of deadenylation. Moreover, knockdown of Dcp2 decapping activity resulted in accumulation of two distinct mRNA intermediates with ~110 nt or 10–20 nt poly(A) tails, but not accumulation of mRNAs with a full-length poly(A) tail. These results indicate that decapping is not a major means of triggering mRNA decay in mammalian cells. Instead, mammalian decapping occurs after the first phase and/or the second phase of deadenylation (Figure 5A).
Simultaneously compromising both Ccr4-Caf1 and Dcp2 activities essentially halts various mRNA decays with concomitant accumulation of a stable intermediate containing 110 nt of poly(A) tail [35, 38, 62], lending further support to the idea that the major route for mRNA decay in mammalian cells is triggered by deadenylation followed by decapping and 5′ to 3′ exonuleolytic digestion of the RNA body. Thus, while there is a difference between the mechanisms for mRNA degradation in yeast and in mammals, the major mRNA decay pathway is highly conserved throughout evolution. Since deadenylation is a reversible process, it is plausible that in eukaryotes, deadenylation serves as an important checkpoint before an mRNA is committed to elimination during embryogenesis and cell growth and differentiation and thus is an important step for regulation of gene expression.
IV. Examples of mRNA decay triggered by deadenylation
A recurring theme over the past two decades is that RNA destabilizing elements or RNA destabilizing mutations exert their effects mainly by inducing accelerated deadenylation leading to rapid mRNA decay in mammalian cells. Consistent with this notion, computational modeling of eukaryotic mRNA turnover indicates that changes in levels of mRNA are highly leveraged to the rate of deadenylation [63]. In this section, we discuss five different examples to illustrate this theme.
A. Slow deadenylation of stable mRNAs: the default state
The 3′poly(A) tail undergoes shortening as soon as a mature mRNA, whether stable or labile, arrives in the cytoplasm from the nucleus [16, 17]. In other words, there is a “default” deadenylation, which occurs regardless of whether a transcript contains a destabilizing element, a stabilizing element, or neither. Over the past two decades, β-globin mRNA has been used as a representative of “stable” messages whose slow deadenylation and decay was considered to reflect the default activity of the major cytoplasmic mRNA decay pathway tested in various mammalian cells (e.g., refs [28, 31, 64, 65]). Introducing a strong RNA hairpin to the 5′ UTR of β-globin mRNA specifically blocks its translation but does not affect its deadenylation [66, 67], suggesting that the default deadenylation is irrelevant to the translation status of mRNAs. However, since deadenylation of an mRNA can be regulated both on global level (i.e., the basal level of slow default deadenylation of virtually all mRNAs) and on an mRNA-specific level [20], translation may play a critical role in regulating deadenylation of certain mRNAs carrying specific elements (see examples B and C below).
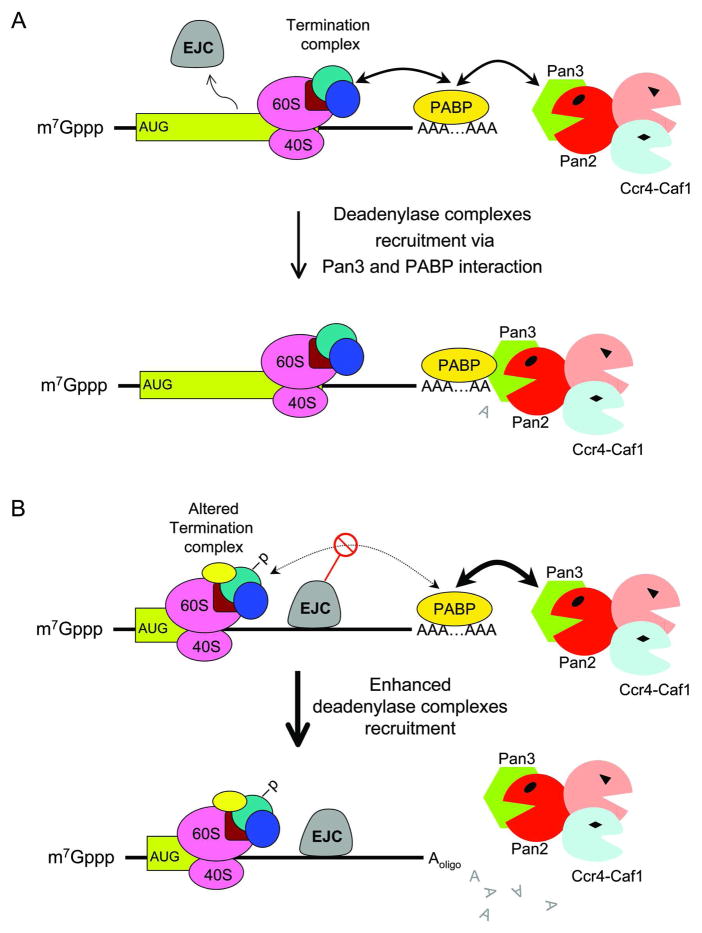
What is the mechanism underlying default deadenylation in the cytoplasm? Kinetic evidence obtained from the transcriptional pulsing approach shows that deadenylation of the stable β-globin mRNA is a result of consecutive action of Pan2-Pan3 and Ccr4-Caf1 deadenylases [35]. Previous studies indicated that Pan3 is a genuine PABP-interacting protein that confers to Pan2 a PABP-dependent deadenylase activity [68–70]. Additionally, Pan2-Pan3 can form a super complex with Ccr4-Caf1, which provides an opportunity to coordinate the two phases of deadenylation [38]. Therefore, one may envisage that the Pan2-Pan3/Ccr4-Caf1 deadenylase super-complex can be randomly recruited to the 3′ poly(A)-PABP complex through the interaction between Pan3 and PABP (Figure 6A). Since the first phase of deadenylation kinetics is consistent with a distributive and not a processive enzymatic action [35], deadenylation by Pan2-Pan3 involves constant recruitment and release of the 3′ poly(A) tail, reflecting the slow default deadenylation of the cell.
Figure 6.
Schematic diagram showing a model for default deadenylation (A) and for acclerated deadenylation by nonsense codon (B) in mammalian cells. -P: modification of termination complex, e.g., phosphorylation. EJC: exon-exon junction complex.
B. Mammalian nonsense-mediated decay
Most normal eukaryotic mRNAs contain a stop codon at the end of the coding region and are not subject to NMD [71]. In contrast, mutant mRNAs that have an in-frame stop codon upstream of the normal stop codon are recognized as premature by the NMD machinery, leading to mRNA destabilization. Although the NMD pathway can be found in Saccharomyces cerevisiae [72], Drosophila melanogaster [73], Caenorhabditis elegans [74], mammals [75], and plants [76], the immediate consequences of premature termination codons (PTC) recognition leading to mRNA decay appear to be different in eukaryotes. In yeast, a major consequence of PTC recognition is decapping [56], whereas in flies, PTC recognition leads to endonucleolytic cleavage of the mRNA in the vicinity of the aberrant stop codon [52]. In other species, including mammals, PTC recognition leads to accelerated deadenylation [77, 78].
The discovery that SMG6, a key factor involved in NMD, induces endocleavage of a PTC-containing mRNA led to a hypothesis that mammalian NMD may be initiated by SMG6-mediated endonucleolytic cleavage [79], which is reminiscent of NMD in Drosophila. However, there is no direct evidence to support this hypothesis thus far. Kinetic studies (e.g., using the transcriptional pulsing approach combined with gene knockdown of SMG6) have not been performed to directly show that SMG6-mediated endocleavage induces rapid decay or precedes accelerated deadenylation of the PTC-containing transcript. In contrast, kinetics studies have demonstrated that mammalian NMD is triggered by deadenylation [35, 38, 78, 80]. The PTC-containing β-globin mRNA undergoes biphasic deadenylation at accelerated rates followed by a rapid decay of the mRNA body. Decapping of the PTC-containing transcript does not occur until the end of the first or second phase of deadenylation [35]. Compromising deadenylating activity greatly impairs decay of the PTC-containing transcript, whereas compromising decapping activity has only a marginal effect [35]. Moreover, when translation of the PTC-containing β–globin mRNA was blocked by introducing a strong hairpin in the 5 UTR, the corresponding transcript exhibited slow deadenylation kinetics similar to those of the stable β-globin transcript [78], indicating that NMD is tightly coupled to translation [81].
Given that NMD is strictly coupled to active translation, accumulating evidence indicates that premature termination at a nonsense codon is physically and functionally different from termination at a native stop codon. For instance, in mammalian cells, a large exon junction protein complex (EJC) deposited upstream of the exon-exon junctions during RNA splicing is widely considered to be a marker that triggers NMD [82]. A normal transcript that contains a stop codon in the last exon [71] is not recognized by the NMD machinery because EJC or NMD marker proteins are displaced by the translating ribosome and/or associated factors (Figure 6A). During normal translation termination, the termination complex containing the UPF1-eRF1-eRF3 trimer interacts well with 3′ poly(A)-PABP (Figure 6A) [83, 84]. This interaction may be disrupted in a PTC-containing transcript due to the presence of EJC (Figure 6B), which causes alteration (including phosphorylation of UPF1) of the termination complex [85, 86]. Since both Pan3 and eRF3 are PABP-interacting proteins, the absence of an effective interaction between eRF3 and PABP may increase the interaction between Pan3 and PABP, thereby enhancing the recruitment of the Pan2-Pan3 and Ccr4-Caf1 super deadenylase complex to the 3′ poly(A)-PABP (Figure 6B). This possibility is also consistent with observations that a tethered EJC complex or an elongated 3′ UTR destabilized the transcript [87–90], as the tethered EJC complex or lengthened 3′ UTR may interfere with the proper “communication” between the termination complex and the 3′ poly(A)-PABP [83]. In addition, alteration of the termination complex or phosphorylation of UPF1 may cause destabilization of the 5′-cap/3′-poly(A) closed-loop in a PTC-containing transcript, making the 3′ poly(A) tail susceptible to deadenylase attack and leading to accelerated deadenylation.
C. Decay mediated by the c-fos coding region determinant
The mRNA decay mediated by the major coding-region determinant (mCRD) of the c-fos proto-oncogene transcript is another example illustrating a novel mechanism that requires translation or ribosome transit of the ORF to trigger accelerated deadenylation of the transcript.
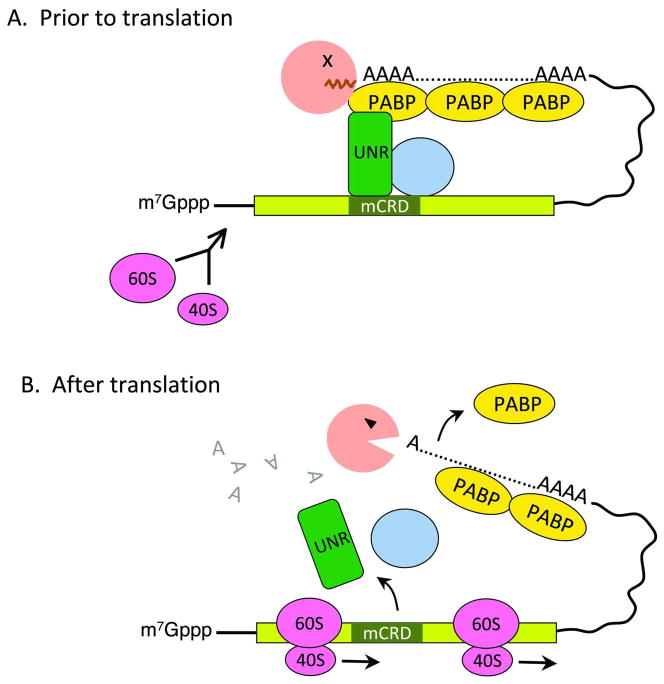
The c-fos mCRD is located in the central portion of the protein-coding region of the transcript [91]. A minimal spacer sequence between the mCRD and the poly(A) tail is required to allow the formation of a unique protein complex that brings the mCRD and the poly(A) tail together (Figure 7A) [92]. A key member of this “bridging” complex is a cold-shock domain-containing RNA-binding protein, UNR. Knocking down UNR via RNAi resulted in retardation of deadenylation of an mCRD-containing mRNA [93]. UNR binds directly and simultaneously to both the mCRD sequence and PABP [93], thus establishing a physical interaction between the poly(A)/PABP complex and the mCRD/UNR complex. Interference with this interaction diminishes the mCRD-destabilizing function in vivo. The mCRD/UNR complex also associates with the CCR4 deadenylase. Intriguingly, blocking translation initiation (e.g., by introducing a strong hairpin structure into the 5′ UTR) of an mRNA whose decay is mediated by the mCRD effectively impeded its deadenylation, leading to full stabilization of the message [93]. These findings support a model in which the mCRD/UNR complex serves as a “landing/assembly” platform for formation of a deadenylation/decay mRNP complex involving the 3′ poly(A)-PABP complex and Ccr4 deadenylase (Figure 7A). The deadenylation/decay complex is kept in a dormant state prior to translation. Ribosome transit is required to remodel the complex and somehow activate Ccr4 nuclease, thereby triggering rapid deadenylation and subsequent decay of the RNA body (Figure 7B).
Figure 7.
A model for translationally coupled deadenylation mediated by the c-fos major coding region determinant of instability (mCRD). Prior to translation (A), Unr, other auxiliary factors (shown here as a golden yellow circle), and Ccr4 form a bridging complex that brings the 3′ poly(A) tail and the mCRD together. Ccr4 access to the poly(A) tail is blocked. Following translation initiation (B), ribosome transit knocks off the bridging complex and somehow activates Ccr4 for accelerated deadenylation.
The translationally coupled decay directed by the c-fos mCRD illustrates a suicidal mechanism by which translation of a transcript immediately triggers the death of the transcript through rapid deadenylation. Through this pathway, a transient production of a protein can be effectively achieved. This novel mechanism also illustrates the importance of mRNP remodeling in regulating mRNA turnover. It will be interesting to see whether other protein coding region determinants of instability, such as the one found in the c-myc transcript [94–97], may also use this mechanism to mediate rapid mRNA decay.
D. Rapid mRNA decay mediated by AU-rich elements
AREs are the most commonly found destabilizing elements in unstable mRNAs. They have been shown to destabilize mRNAs in species ranging from yeast and Drosophila, to mammals [64, 98]. Interestingly, AREs do not share any significant sequence homology aside from two elusive features, the high AU content and in most cases the presence of various copies of AUUUA motifs [99–101]. Nearly three decades ago, studies using the c-fos promoter-driven transcriptional pulsing system showed that AREs induce rapid deadenylation followed by decay of the RNA body, suggesting that ARE-mediated decay is triggered by deadenylation [28, 30]. However, the participating deadenylases were unknown and the cause-effect relationship between deadenylation and decay was not firmly established. In the past decade, some ARE-binding proteins (ARE-BPs) were found to associate with the exosome complex that degrades the RNA body exonucleolytically from 3′ to 5′ or with the decapping complex that removes the 5′ cap to facilitate decay of the RNA body from the 5′ end [48, 102, 103]. Discussions of the mechanism of ARE-mediated decay were often focused on whether the transcripts undergo 5′ to 3′ or 3′ to 5′ decay or on the role of decapping. The exact step that triggers ARE-mediated mRNA decay, and thus the role of deadenylation in this critical event, was frequently ignored. Moreover, even though knocking down the decapping enzyme and exosome components via RNA interference (RNAi) affected ARE-mediated decay, it was not directly tested (e.g., by kinetic studies) whether decapping or the exosome’s action actually preceded deadenylation or triggered mRNA decay in vivo.
Recent identification and characterization of deadenylases involved in general deadenylation in mammalian cells has helped address the triggering event for ARE-mediated decay. Kinetic studies using the Tet-off promoter driven transcriptional pulsing system combined with approaches compromising the function of deadenylases showed that disrupting Caf1 function severely impedes deadenylation induced by the c-fos ARE, leading to nearly full stabilization of the corresponding ARE-containing transcript as an intermediate with a poly(A) tail of about 110 nt [38]. This result indicates that when Caf1 function is compromised, the second phase but not the first phase of deadenylation is blocked. On the other hand, disrupting decapping activity has little effect on the instability of ARE-containing transcripts. Collectively, these results support the model that ARE-mediated mRNA decay is triggered by deadenylation mediated by the consecutive action of Pan2-Pan3 and Ccr4-Caf1.
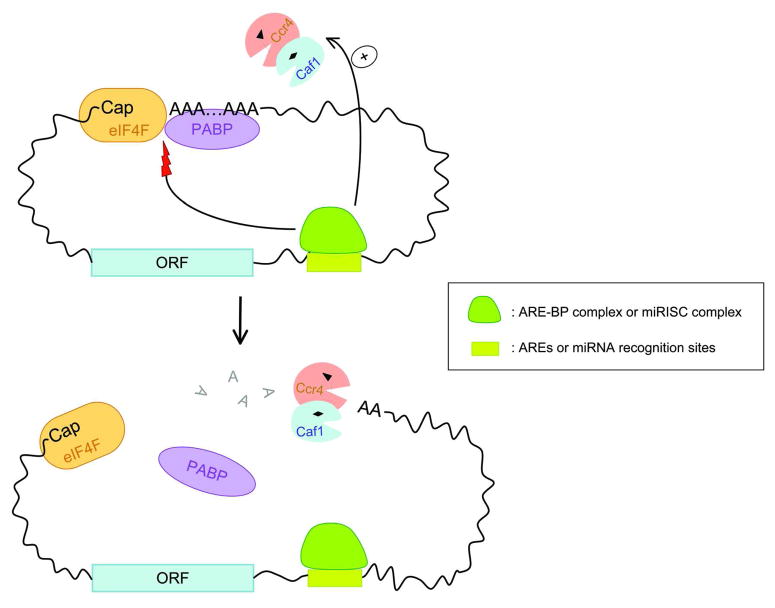
How AREs and their cognate ARE-BPs act to promote rapid deadenylation remains unclear. Since ARE/ARE-BP complexes have not yet been reported to directly interact with either the Pan2-Pan3 or Ccr4-Caf1 complex, one possibility is that the binding of ARE-BPs to AREs may interfere with the interaction between the 5′ cap/eIF4F complex and the 3′ poly(A)/PABP complex, thus preventing the formation of a closed-loop structure (Figure 8). This could in turn make the 3′ poly(A) tail susceptible to deadenylase attack, leading to accelerated deadenylation. Alternatively, Pan2-Pan3 and Ccr4-Caf1 may be recruited to the vicinity of the 3′ poly(A) tails of ARE-containing transcripts through a transient interaction, not readily detected by co-immunoprecipitation assay, or through an “adapter” that binds both the ARE/ARE-BP complex and deadenylases. These possibilities are not mutually exclusive, as AREs may evolve to gain other “backup” ways to ensure rapid decay of their mRNA targets.
Figure 8.
Schematic diagram showing possible modes of accelerated deadenylation promoted by AREs or miRNA binding sites in mammalian cells, which involve either direct recruitment of Ccr4-Caf1 and/or disruption of eIF4F-PABP interaction to open up the closed loop formed between the 5′ Cap and 3′ poly(A) tail. Note that these two mechanisms are not mutually exclusive.
E. MicroRNA-mediated mRNA decay
MicroRNAs (miRNAs) are 20- to 22-nt long noncoding RNAs regulating gene expression post-transcriptionally by base-pairing to target mRNAs [104–108]. In general, the miRNA association results in translational repression, often accompanied by degradation of the target mRNA. Following the findings that miRNAs cause decay of their mRNA targets, a general picture of miRNA-mediated decay (miRMD) has gradually emerged in the past few years. Studies in zebrafish embryos, Drosophila cells and human cells first showed that mRNAs that were targeted by miRNAs for degradation have undergone prior deadenylation [109–111]. Studies in Drosophila cells showed that miRNAs promote deadenylation and decapping and their action involves GW182, Argonaute (Ago)1, the Ccr4-NOT complex, and the Dcp1-Dcp2 complex [112]. It was unclear from these studies whether miRMD is triggered by deadenylation, decapping, or both nor were the participating decay enzymes and factors identified in mammalian cells. More recently, combining the transcriptional pulsing approach with approaches for compromising specific endogenous decay enzymes, kinetic studies revealed that the miRNA-induced silencing complex (miRISC) routes its mRNA target to the major decay pathway in mammalian cells [62], i.e., mRNA decay triggered by biphasic deadenylation involving Pan2-Pan3 and Ccr4-Caf1 deadenylase complexes, followed by Dcp1-Dcp2 complex-directed decapping (Figure 5). Mammalian miRISCs contain Ago and TNRC6 (also known as GW182) proteins. When tethered to mRNAs, all four human Ago proteins and TNRC6C are each able to recapitulate the two deadenylation steps [62]. These findings indicate that promotion of biphasic deadenylation to trigger mRNA decay is an intrinsic property of miRNAs in mammalian cells.
Several recent studies provide some clues as to how miRISC induces rapid deadenylation. For example, both Caf1 and PABP were shown to be required for miRISC-mediated deadenylation in mouse acites cell extracts and were able to interact with GW182 in vitro [113]. In addition, tethering Drosophila GW182 to an mRNA can bypass the requirement of Ago proteins for promoting mRNA decay, but not the converse [112]. One possibility is that the interaction of GW182 in RISC with PABP may destabilize the 3′ poly(A)-PABP complex or open up the closed-loop formed between the 5′ cap-binding complex and the 3′ poly(A)-PABP complex, leading to accelerated deadenylation (Figure 8). Given that Caf1 interacts with GW182 in an in vitro system, another possibility is that GW182 in RISC directly recruits Caf1 to a miRNA-targeted transcript to elicit rapid deadenylation. However, it should be noted that in several other studies an interaction between RISC components and Caf1 cannot be detected [62, 114–117], suggesting a weak, indirect or transient interaction between GW182 and Caf1. These observations lay a foundation for determining how miRISC promotes deadenylation in vivo.
V. Deadenylation is a prerequisite for RNA processing body assembly
RNA processing bodies (P-bodies) are specific cytoplasmic foci that contain proteins known to function in mRNA metabolism [21, 22, 118]. These foci are also referred to as GW bodies because they carry GW182 proteins [112, 115, 119–122]. The function of P-bodies is not yet fully understood, but it is clear that the mRNAs in P-bodies can be stored, degraded, or reenter the translating pool of mRNAs. Because P-bodies contain the Dcp1-Dcp2 decapping enzyme and the 5′-3′ exonuclease XRN1 [123, 124] but lack the exosome complex (which contains 3′-5′ exonucleases), it is likely that mRNAs are degraded via the 5′ to 3′ decay pathway in P-bodies. More recently, the components of the two major poly(A) nuclease complexes, including Pan2, Pan3, Ccr4 and Caf1, were found in mammalian P-bodies [38, 125]. This finding links all major mRNA decay factors except the exosome components to P-bodies. Interestingly, P-bodies lack ribosomal components, most translation initiation factors, and PABP, indicating that ribosomes, PABP, and translation initiation factors must dissociate from mRNPs before or immediately after they enter or aggregate to form P-bodies.
In addition to general mRNA decay factors, many key factors required for ARE-mediated decay, NMD, and miRMD as well as the corresponding mRNA substrates of these major decay pathways are also found in P-bodies (reviewed in [21, 22, 118, 126]). Since deadenylation is the major step that initiates mRNA decay in all of these pathways, it is not surprising that microscopically visible P-bodies disappear concomitantly when deadenylation is blocked [38]. Moreover, when deadenylation is restored, P-bodies also reappear in the cytoplasm [38]. The key evidence supporting that deadenylation is a prerequisite for P-body formation and not the opposite came from the observation that knockdown of Pan3, a key component of P-bodies, impairs P-body formation but has little effect on the deadenylation and decay of ARE-containing transcripts and miRNA targeted mRNAs [38]. Thus, although the non-translatable mRNPs may be prepared and/or assembled into P-bodies in different ways, deadenylation is always a necessary and perhaps the most upstream step that enables required mRNP remodeling for P-body assembly. The observation that Pan3 is physically required for P-body formation but its knockdown has little effect on deadenylation argues for an additional role of Pan3 in P-body formation. This is further substantiated by the finding that Pan3 greatly enhances the localization of other P-body components to P-bodies [38]. Thus, deadenylation and participating deadenylases are not simply required for preparing mRNA substrates; in fact, they play an indispensable role both structurally and functionally in P-body formation and regulation.
Conclusion
Deadenylation triggers mRNA decay in nearly all known major decay pathways. The recent observations that miRISC promotes rapid deadenylation to trigger target mRNA decay and that deadenylation is prerequisite for P-body formation further underscore the importance of regulating deadenylation in controlling gene expression. Given the presence of a variety of cis-acting RNA elements in mRNAs and the abundance of cognate trans-acting factors that promote rapid deadenylation, deadenylation-based regulation is apparently more common than previously realized.
How is deadenylation regulated mechanistically? Based on the findings and the examples discussed in this review, it appears that deadenylation is regulated at an mRNA-specific level, depending on the RNA regulatory elements that reside in a given transcript. One model proposes that RNA elements and the associated cognate protein complexes recruit deadenylase complexes directly to the target mRNAs and induce accelerated deadenylation (Figure 8). Another model posits that RNA elements and their cognate binding protein complexes destabilize the closed-loop formed between the 5′ cap and 3′ poly(A) tail of target mRNAs, thereby making the 3′ poly(A) tail more susceptible to attack by deadenylase complexes (Figure 8). While these two means of regulation are not mutually exclusive, several related issues remain to be addressed. For example, do the RNA-specific binding complexes interfere with the interaction between eIF4G and PABP or do they destabilize the interaction between PABP and poly(A) tail? Under what mechanism does this kind of interference occur? Moreover, an mRNA may carry more than one destabilizing element (e.g., the c-fos mRNA contains multiple RNA destabilizing elements both in the ORF and 3′ UTR) [28, 29]. The fact that deadenylation represents a major way to trigger mRNA decay mediated by different elements suggests that the actions of multiple decay elements of a transcript converge to exert a synergistic effect to promote highly accelerated deadenylation.
In addition to regulation at the mRNA-specific level, deadenylation can also be regulated on a global level (i.e., the basal level of slow default deadenylation of all poly(A)+ mRNAs), such as that achieved by modulating Pan2-Pan3 or Ccr4-Caf1 deadenylase activity per se. Global regulation can also be achieved by controlling the expression of factors that mediate the interaction between mRNAs and deadenylase complexes [127]. For example, the TOB proteins directly recruit the Ccr4-Caf1 complex to the 3′ poly(A)-PABP complex in a transcript-independent manner via their ability to simultaneously interact with PABP and Caf1 [128, 129]. Interestingly, TOB factors are a family of antiproliferative proteins and have been implicated in cell differentiation, development, and apoptosis [130]. As this mode of global regulation of deadenylation is likely to impact the cellular mRNA expression profile and thus alter cellular homeostasis, it will be interesting to find out whether it may assist cellular transitions during cell growth and differentiation.
The finding that mammalian deadenylation is a two-step process may have an important implication for regulating mRNA turnover. The Pan2-Pan3 complex, which is responsible for the first step (i.e., shortening the poly(A) tail to 110 nt), is evolutionarily conserved; however, its function in deadenylation appears to be dispensable. Therefore, what is the functional significance of the first phase of deadenylation? Could this phase represent a “maturation” step or a remodeling step that better prepares mRNAs for their functions in the cytoplasm? Also, since Pan2-Pan3 and Ccr4-Caf1 deadenylases are all found in P-bodies, do both steps of deadenylation occur inside P-bodies? As PABP is not found in mammalian P-bodies, how do PABPs dissociate from the 3′ poly(A) tail prior to an mRNP entering P-bodies? This is a nontrivial issue because PABP exhibits a very high binding affinity for the poly(A) tail (at nanomolar range). Apparently, the regulation of deadenylation, a key event controlling mRNA turnover, is more complicated than previously realized. For the coming years, we will witness the emergence of more factors that modulate deadenylation through their interactions, directly or indirectly, with the deadenylation machinery and gain mew insight into the biological consequences.
References
- 1.Wickens M, et al. Translational control in developmental decisions. In: Mathews M, editor. Translational Control. 2. Cold Spring Harbor Press; 2000. [Google Scholar]
- 2.Gebauer F, Hentze MW. Molecular Mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 4.Garneau NL, et al. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 5.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 6.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends in Cell Biology. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Meyer S, et al. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 8.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Strut Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 9.Muhlrad D, et al. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-->3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 10.Mangus DA, et al. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:233. doi: 10.1186/gb-2003-4-7-223. http://genomebiology.com/2003/2004/2007/2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobson A. Poly(A) metabolism and translation: The colsed-loop Model. In: Hershey JWB, et al., editors. Tanslational Control. Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 12.Sachs AB. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N, et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 13.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annual Review Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 14.Wickens M, et al. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 15.Wilusz CW, et al. The cap-to-tail guide to mRNA turnover. Nature Reviews Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 16.Baker EJ. Control of poly(A) length. In: Belasco JG, Brawerman G, editors. Control of messenger RNA stability. Academic Press; 1993. [Google Scholar]
- 17.Brawerman G. The role of the poly(A) sequence in mammalian messenger RNA. Crit Rev Biochem. 1981;10:1–38. doi: 10.3109/10409238109114634. [DOI] [PubMed] [Google Scholar]
- 18.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucl Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupressoir A, et al. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 21.Eulalio A, et al. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 22.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Houseley J, Tollervey D. The Many Pathways of RNA Degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Huang YS, Richter JD. Regulation of local mRNA translation. Curr Opin Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Lim L, Canellakis ES. Adenine-rich polymer associated with rabbit reticulocyte messenger RNA. Nature. 1970;227:710–712. doi: 10.1038/227710a0. [DOI] [PubMed] [Google Scholar]
- 26.Darnell JD, et al. An Adenylic Acid-Rich Sequence in Messenger RNA of HeLa Cells and Its Possible Relationship to Reiterated Sites in DNA. Proc Natl Acad Sci USA. 1971;68:1321–1325. doi: 10.1073/pnas.68.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loflin PT, et al. Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods. 1999;17:11–20. doi: 10.1006/meth.1998.0702. [DOI] [PubMed] [Google Scholar]
- 28.Shyu A-B, et al. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–232. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 29.Shyu A-B, et al. The c-fos mRNA is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 30.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 31.Xu N, et al. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucl Acids Res. 1998;26:558–565. doi: 10.1093/nar/26.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenberg ME, Ziff E. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature (London) 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 33.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 34.Gossen M, et al. Control of gene activity in higher eukaryotic cells by prokaryotic regulatory elements. TIBS. 1993;18:471–475. doi: 10.1016/0968-0004(93)90009-c. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita A, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 36.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes & Development. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 37.Decker CJ, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends in Biochemical Sciences. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 38.Zheng D, et al. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker M, et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, et al. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 42.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;74:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 43.Cougot N, et al. Cap-tabolism. Trends in Biochemical Sciences. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Kiledjian M. Decapping the message: a beginning or an end. Biochem Soc Trans. 2006;34:35–38. doi: 10.1042/BST20060035. [DOI] [PubMed] [Google Scholar]
- 45.Allmang C, et al. The yeast exosome and human PM–Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs JS, et al. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell P, et al. The Exosome: A Conserved Eukaryotic RNA Processing Complex Containing Multiple 3′-->5′ Exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 48.Chen CY, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein E, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 50.Hammond SM, et al. Argonaute2, a Link Between Genetic and Biochemical Analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, et al. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 52.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 53.Frischmeyer PA, et al. An mRNA Surveillance Mechanism That Eliminates Transcripts Lacking Termination Codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 54.van Hoof A, et al. Exosome-Mediated Recognition and Degradation of mRNAs Lacking a Termination Codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 55.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 57.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′->5′ degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 58.Tucker M, et al. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mangus DA, et al. Positive and Negative Regulation of Poly(A) Nuclease. Mol Cell Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Temme C, et al. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu N, et al. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: Key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen CYA, et al. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:553–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 64.Chen AC-Y, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 65.Loflin PT, et al. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan XC, et al. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CY, et al. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchida N, et al. Identification of a Human Cytoplasmic Poly(A) Nuclease Complex Stimulated by Poly(A)-binding Protein. J Biol Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- 69.Boeck R, et al. The Yeast Pan2 Protein Is Required for Poly(A)-binding Protein-stimulated Poly(A)-nuclease Activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 70.Brown CE, et al. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends In Biochemical Sciences. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 72.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. PNAS. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brogna S. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA. 1999;5:562–573. doi: 10.1017/s1355838299981359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hodgkin J, et al. A New Kind of Informational Suppression in the Nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maquat LE, et al. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27:543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 76.van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. The Plant Journal. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- 77.Cao D, Parker R. Computational Modeling and Experimental Analysis of Nonsense-Mediated Decay in Yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 78.Chen CYA, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23:4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eberle AB, et al. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 80.Couttet P, Grange T. Premature termination codons enhance mRNA decapping in human cells. Nucl Acids Res. 2004;32:488–494. doi: 10.1093/nar/gkh218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thermann R, et al. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Hir H, et al. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 83.Ivanov PV, et al. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shyu AB, et al. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Current Opinion in Cell Biology. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 87.Gehring K, et al. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 88.Behm-Ansmant I, et al. A conserved role for cytoplasmic poly(A)-binding protein 1(PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buhler M, et al. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- 90.Lykke-Andersen J, et al. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 91.Schiavi SC, et al. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J Biol Chem. 1994;269:3441–3448. [PubMed] [Google Scholar]
- 92.Grosset C, et al. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 93.Chang T-C, et al. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernstein PL, et al. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes & Development. 1992;6:642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- 95.Herrick DJ, Ross J. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol Cell Biol. 1994;14:2119–2128. doi: 10.1128/mcb.14.3.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lemm I, Ross J. Regulation of c-myc mRNA Decay by Translational Pausing in a Coding Region Instability Determinant. Mol Cell Biol. 2002;22:3959–3969. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wisdom R, Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein sythesis inhibitors. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 98.Barreau C, et al. AU-rich elements and associated factors: are there unifying principles? Nucl Acids Res. 2006;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen CY, et al. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen CY, Shyu AB. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lagnado CA, et al. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stoecklin G, et al. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fenger-Gron M, et al. Mltiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 104.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 105.Bushati N, Cohen SM. microRNA Functions. Annual Review of Cell and Developmental Biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 106.Filipowicz W, et al. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 107.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 108.Nissan T, Parker R. Computational analysis of miRNA-mediated repression of translation: Implications for models of translation initiation inhibition. RNA. 2008;14:1480–1491. doi: 10.1261/rna.1072808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 110.Wu L, et al. MicroRNAs direct rapid deadenylation of mRNA. PNAS. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Behm-Ansmant I, et al. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp on Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 112.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fabian MR, et al. Mammalian miRNA RISC Recruits CAF1 and PABP to Affect PABP-Dependent Deadenylation. Molecular Cell. 2009 doi: 10.1016/j.molcel.2009.08.004. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baillat D, Shiekhattar R. Functional Dissection of the Human TNRC6 (GW182-Related) Family of Proteins. Mol Cell Biol. 2009;29:4144–4155. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meister G, et al. Identificiation of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 116.Landthaler M, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang L, et al. Systematic Identification of C.†elegans miRISC Proteins, miRNAs, and mRNA Targets by Their Interactions with GW182 Proteins AIN-1 and AIN-2. Molecular Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 119.Eystathioy T, et al. A Phosphorylated Cytoplasmic Autoantigen, GW182, Associates with a Unique Population of Human mRNAs within Novel Cytoplasmic Speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jakymiw A, et al. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;8:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 121.Liu J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rehwinkel JAN, et al. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sheth U, Parker R. Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ingelfinger D, et al. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 125.Cougot N, et al. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Franks TM, Lykke-Andersen J. The Control of mRNA Decapping and P-Body Formation. Molecular Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mauxion F, et al. BTG/TOB factors impact deadenylases. Trends in Biochemical Sciences. 2009;34:640–647. doi: 10.1016/j.tibs.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ezzeddine N, et al. Human TOB, an Antiproliferative Transcription Factor, Is a Poly(A)-Binding Protein-Dependent Positive Regulator of Cytoplasmic mRNA Deadenylation. Mol Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takashi Miyasaka MMKITSHFSTJIIKSTY. Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Science. 2008;99:755–761. doi: 10.1111/j.1349-7006.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Matsuda S, et al. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Letters. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]