Abstract
F-box proteins and DCAF proteins are the substrate binding subunits of SCF (Skp1-Cul1-F-box protein) and CRL4 (Cul4-RING protein Ligase) ubiquitin ligase complexes, respectively. Using affinity purification and mass spectrometry, we determined that the F-box protein FBXO11 interacts with CDT2, a DCAF protein that controls cell cycle progression, and recruits CDT2 to the SCFFBXO11 complex to promote its proteasomal degradation. In contrast to most SCF substrates, which exhibit phosphodegrondependent binding to F-box proteins, CDK-mediated phosphorylation of Thr464 present in the CDT2 degron inhibits recognition by FBXO11. Finally, our results show that the functional interaction between FBXO11 and CDT2 is evolutionary conserved from worms to humans and plays an important role in regulating the timing of cell cycle exit.
INTRODUCTION
Unidirectional progression through the cell cycle depends on the specific, rapid, and temporally-controlled proteolysis of key cellular regulators by the ubiquitin-proteasome system (UPS). E3 ubiquitin ligases confer substrate specificity to the UPS. Among the eukaryotic E3s, Cullin-RING Ligases (CRLs) constitute the largest family of multi-subunit ubiquitin ligases (Petroski and Deshaies, 2005). The archetypes of the CRL family are the CRL1/SCF (Skp1-Cul1-F-box protein) E3s, which utilize different F-box proteins (69 in humans) as receptors that bind substrates. Significantly, multiple F-box proteins are mutated or display altered expression in a variety of diseases, including cancer (Frescas and Pagano, 2008; Lipkowitz and Weissman, 2011; Skaar et al., 2009a).
FBXO11 is conserved from nematodes to mammals and both human FBXO11 and its worm ortholog (DRE-1) form functional SCF ubiquitin ligases (Fielenbach et al., 2007). DRE-1 deletion causes larva lethality, whereas DRE-1 mutation induces precocious terminal differentiation of epidermal stem cells and altered temporal patterning of gonadal outgrowths, indicating an important role for DRE-1 in controlling cell fate determination (Fielenbach et al., 2007).
In mice, homozygous mutation of Fbxo11 results in cleft palate defects, facial clefting, and perinatal lethality. Moreover, haploinsufficient mutant alleles cause otitis media, a disorder that affects approximately 15 % of children (Hardisty-Hughes et al., 2006). Accordingly, genetic studies show a correlation between particular SNP variants of FBXO11 and the development of chronic otitis media (Segade et al., 2006). Finally, FBXO11 inactivating mutations contribute to the pathogenesis of diffuse large B-cell lymphoma (DLBCL) through BCL6 stabilization, a B-cell specific oncoprotein (Duan et al., 2012). FBXO11 mutations are also present in other human cancers, such as colon, lung, ovary, and head and neck tumors (Kan et al., 2010; Cancer Genome Atlas Research Network, 2011; Stransky et al., 2011; Yoshida et al., 2011; Lohr et al., 2012). These data suggest that FBXO11 may function as a tumor suppressor, whose loss of function contributes to the pathogenesis of DLBCL (via BCL6accumulation) and other cancers (through the stabilization of unidentified pro-oncogenic substrates).
In an effort to elucidate FBXO11 functions, we have identified CDT2 as a novel interactor of FBXO11. CDT2 belongs to the family of WD40 repeat-containing DCAF proteins that work as substrate receptors for CRL4 ubiquitin ligases. CDT2 is conserved from nematodes to humans and plays fundamental roles in the regulation of the S-phase of the cell cycle by controlling the degradation of SET8, CDT1, and p21 under normal and stress conditions (Abbas and Dutta, 2011; Abbas et al., 2010; Abbas et al., 2008; Centore et al., 2010; Havens and Walter, 2011; Higa et al., 2006; Jorgensen et al., 2011; Kim et al., 2008; Oda et al., 2010).
In this study, we demonstrate that SCFFBXO11 targets CDT2 for proteasomal degradation, elucidating a critical and conserved control mechanism for the timing of cell cycle exit.
RESULTS
To identify SCFFBXO11 substrates, FLAG-HA-tagged FBXO11 was transiently expressed in HEK-293T cells. To block the degradation of SCFFBXO11 substrates and increase their co-purification with FBXO11, cells were either co-transfected with CUL1(1–385), a dominant negative CUL1 mutant, or treated for four hours with the proteasome inhibitor MG132. Purifications of FLAG-HA-FBXO10, a paralog of FBXO11, were used as a control. FBXO11 and FBXO10 complexes were immunopurified for analysis by Multidimensional Protein Identification Technology (MudPIT) (Florens and Washburn, 2006). Peptides corresponding to CDT2 were specifically identified in FBXO11 immunoprecipitates from cells in which either CUL1(1–385) was co-expressed or MG132 was added (Table S1).
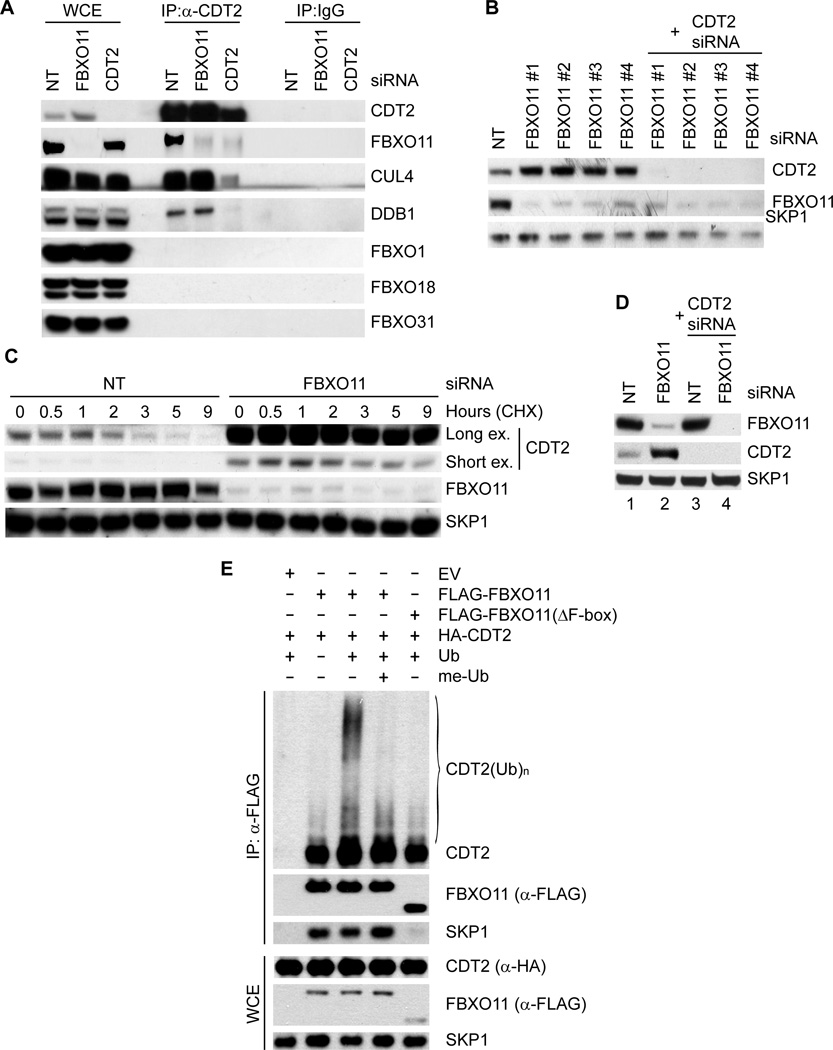
To investigate whether the binding between CDT2 and FBXO11 is specific, FLAG-tagged CDT2 was expressed in HeLa cells and immunoprecipitated with an anti-FLAG resin. We found that FLAG-tagged CDT2 was able to co-immunoprecipitate endogenous FBXO11 and, as expected, endogenous CUL4 and DDB1, but it was unable to co-immunoprecipitate any of the nine additional F-box proteins tested (Fig. S1A). Furthermore, endogenous CDT2 co-immunoprecipitated endogenous FBXO11, except when CDT2 expression was silenced by siRNA (Fig. 1A), proving that FBXO11 was co-immunoprecipitated via CDT2. In contrast, when FBXO11 was depleted, CDT2 still co-immunoprecipitated CUL4 and DDB1, but not FBXO11 (Fig. 1A).
Fig. 1. FBXO11 targets CDT2 for ubiquitylation and degradation.
(A) Endogenous FBXO11 and endogenous CDT2 associate in HeLa cells. HeLa cells were transfected with either siRNAs to the indicated mRNAs or a non-targeting siRNA (NT). Lysates were immunoprecipitated with either an affinity purified polyclonal antibody against FBXO11, an affinity purified polyclonal antibody to CDT2, or an affinity purified rabbit IgG and analyzed by immunoblotting, as indicated.
(B) HeLa cells were transfected with either siRNAs to the indicated mRNAs (four different FBXO11 siRNAs) or a non-targeting siRNA (NT). Cells were collected 48 hours after transfection, lysed, and processed for immunoblotting with antibodies to the indicated proteins.
(C) Silencing of FBXO11 results in the stabilization of CDT2. HeLa cells were transfected with either an siRNA to FBXO11 or a non-targeting siRNA (NT), before treatment with cycloheximide (CHX) for the indicated times. Protein extracts were then immunoblotted for the indicated proteins. Long and short exposures (ex.) are shown for CDT2.
(D) The experiment was performed as in (B).
(E) HEK-293T cells were transfected with HA-tagged CDT2, FLAG-tagged FBXO11, FLAG-tagged FBXO11(ΔFbox), and/or an empty vector (EV) as indicated. After immunopurification with anti-FLAG resin, in vitro ubiquitylation of CDT2 was performed in the presence or absence of E1, E2s, and Ubiquitin (Ub). Where indicated, methylated ubiquitin (me-Ub) was added. Samples were analyzed by immunoblotting with an anti-CDT2 antibody. The bracket on the right marks a ladder of bands >100 kDa corresponding to ubiquitylated CDT2. Immunoblots of whole cell extracts (WCE) are shown at the bottom. (see also Figure S1 and Table S1).
Significantly, the FBXO11-CDT2 interaction is conserved through evolution as CDT-2 (the C. elegans ortholog of CDT2) expressed in HeLa cells was able to immunoprecipitate both endogenous human FBXO11 and exogenous C. elegans DRE-1 (Fig. S1B–C).
To test whether FBXO11 might regulate the degradation of CDT2, we used four different siRNAs to reduce its expression in HeLa and U-2OS cells. Depletion of FBXO11 by all four siRNAs induced an increase in both the steady state levels and stability of CDT2 (Fig. 1B–C and Fig. S1D). In contrast, silencing of CDT2 did not induce changes in the abundance of FBXO11 (Fig. 1D). These experiments indicate that, whereas SCFFBXO11 controls the stability of CDT2, CRL4CDT2 does not control FBXO11 abundance.
We also observed that expression of FBXO11 induced the appearance of high molecular weight species of CDT2, which are likely ubiquitylated forms of CDT2 (Fig. S1E). Moreover, we reconstituted the ubiquitylation of CDT2 by FBXO11 in vitro. Immunopurified FBXO11, but not FBXO11(ΔF-box), a mutant that is unable form an active SCF complex, promoted the in vitro ubiquitylation of CDT2 when ubiquitin was present in the reaction (Fig. 1E). Methylated ubiquitin inhibited the formation of the highest molecular weight forms of CDT2, demonstrating that the high molecular weight forms of CDT2 are indeed polyubiquitylated. These results support the hypothesis that FBXO11 directly controls the ubiquitin-mediated degradation of CDT2.
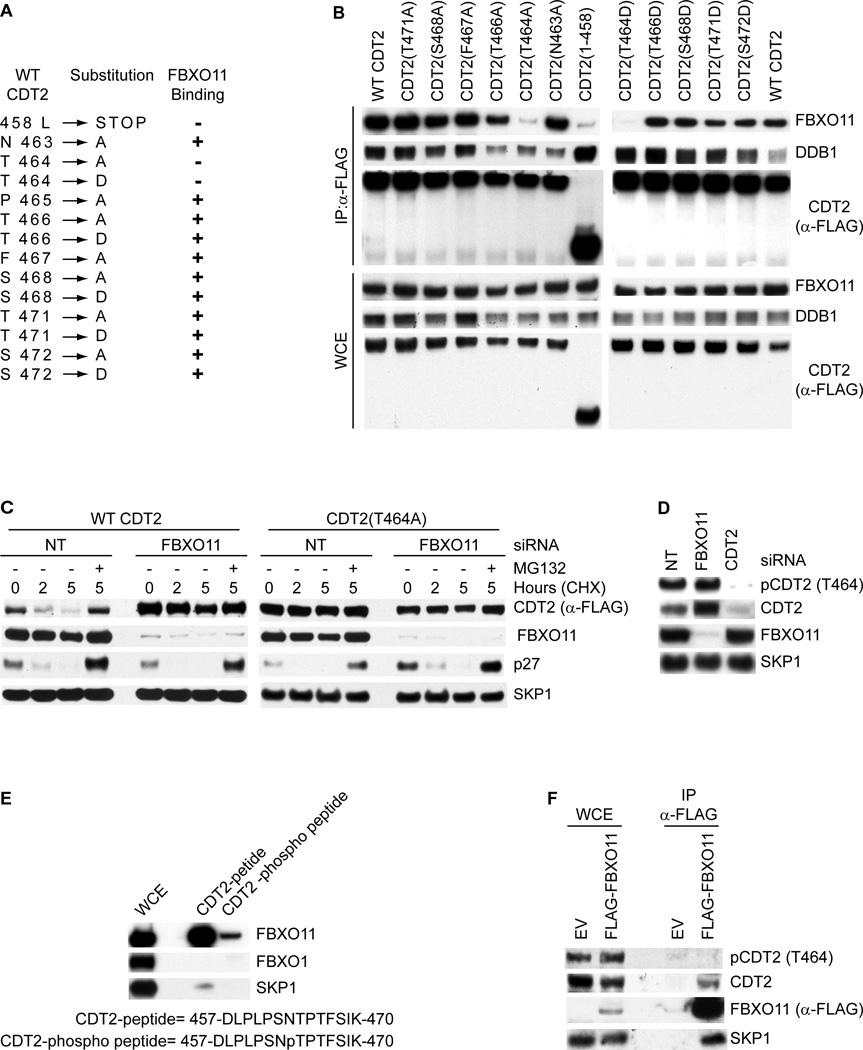
To map the FBXO11 binding motif in CDT2, we generated CDT2 deletion mutants and found that the FBXO11 binding motif of human CDT2 is present in a region between amino acids 458 and 471 (Fig. S2A–C). Then, to further narrow down the FBXO11 interacting region we mutagenized individual residues present in this 14 amino acid region of CDT2 to either Asp or Ala (Fig. 2A). The only amino acid necessary for stable association between CDT2 and FBXO11 was Thr464, and its mutation to either Ala or Asp strongly decreased the ability of CDT2 to co-immunoprecipitate endogenous FBXO11 (Fig. 2B). Significantly, Thr464 is conserved through evolution (Fig. S2D), and a CDT2(Thr464Ala) mutant displayed a significantly longer half-life than wild type CDT2 (Fig. 2C and Fig. S2E). Moreover, in contrast to wild type CDT2, CDT2(Thr464Ala) was not stabilized by FBXO11 silencing or MG132 (Fig. 2C and Fig. S2E).
Fig. 2. CDT2 is phosphorylated in vivo on Thr464, thereby inhibiting CDT2 binding to FBXO11.
(A) List of CDT2 mutants tested for binding to FBXO11. CDT2 mutants that interacted with endogenous FBXO11 are designated with the symbol (+).
(B) HeLa cells were transfected with either FLAG-tagged CDT2 or the indicated FLAG-tagged CDT2 mutants. Whole cell extracts (WCE) were immunoprecipitated (IP) with anti-FLAG resin, and immunocomplexes were immunoblotted as indicated.
(C) HeLa cells were infected with either an empty virus (EV), a retrovirus expressing FLAG-HA-tagged wild type CDT2 or FLAG-HA-tagged CDT2(T464A). Cells were then transfected with either an siRNA to FBXO11 or a non-targeting siRNA (NT), before treatment with cycloheximide (CHX) for the indicated times. Protein extracts were then immunoblotted for the indicated proteins.
(D) HeLa cells were transfected with either siRNAs to the indicated mRNAs or a non-targeting siRNA (NT). Cells were collected 48 hours after transfection, lysed, and processed for immunoblotting with antibodies to the indicated proteins.
(E) HeLa cell extracts were incubated with beads coupled to the indicated peptide or phospho-peptide. Beads were extensively washed, and bound proteins were immunoblotted as indicated.
(F) HeLa cells were transfected with FLAG-tagged FBXO11 or an empty vector (EV). Whole cell extracts were immunoprecipitated (IP) with anti-FLAG resin and probed with antibodies to the indicated proteins.
(see also Figure S2)
Most often, phosphorylation of substrates is necessary for their binding to F-box proteins (Cardozo and Pagano, 2004; Skaar et al., 2009a; Skaar et al., 2009b). Therefore, we examined whether Thr464 phosphorylation of CDT2 plays a role in FBXO11 binding. To this end, we generated a phospho-specific antibody against a peptide containing phospho-threonine at position 464. This antibody recognized wild type CDT2, but not three different CDT2 mutants in which Thr464 was substituted to Asp, Glu, or Ala (Fig. S2F). In addition, λ-phosphatase treatment of immunopurified CDT2 or CDT2 silencing abolished recognition of CDT2 by the phospho-specific antibody (Fig. S2F–G, Fig. 2D). These results show that CDT2 is phosphorylated in vivo on Thr464.
Interestingly, we also observed that the CDT2(Pro465Ala) mutant was not phosphorylated in cultured cells, but it was still able to bind FBXO11 (Fig. S3A), suggesting that this phosphorylation is not essential for CDT2 binding to FBXO11. To further investigate whether phosphorylation affects the CDT2-FBXO11 interaction, we used immobilized, synthetic peptides containing the FBXO11 binding sequence (aa 457–470 in human). While non-phosphorylated peptide efficiently bound FBXO11 (but not FBXO1), a corresponding peptide containing phosphorylated Thr464 displayed a strongly reduced ability to bind FBXO11 (Fig. 2E), indicating that Thr464 phosphorylation inhibits the interaction between the two proteins. Accordingly, CDT2 phosphorylated on Thr464 was not co-immunoprecipitated with FBXO11 (Fig. 2F). Thus, the presence of unphosphorylated Thr464 appears crucial for the binding of CDT2 to FBXO11, explaining why mutation of Thr464 to either Ala (which eliminates Thr464) or Asp (which presumably mimics Thr464 phosphorylation) reduced the in vivo binding to FBXO11. Notably, although mutation of Pro465 inhibits Thr464 phosphorylation, this mutation did not affect the CDT2-FBXO11 interaction.
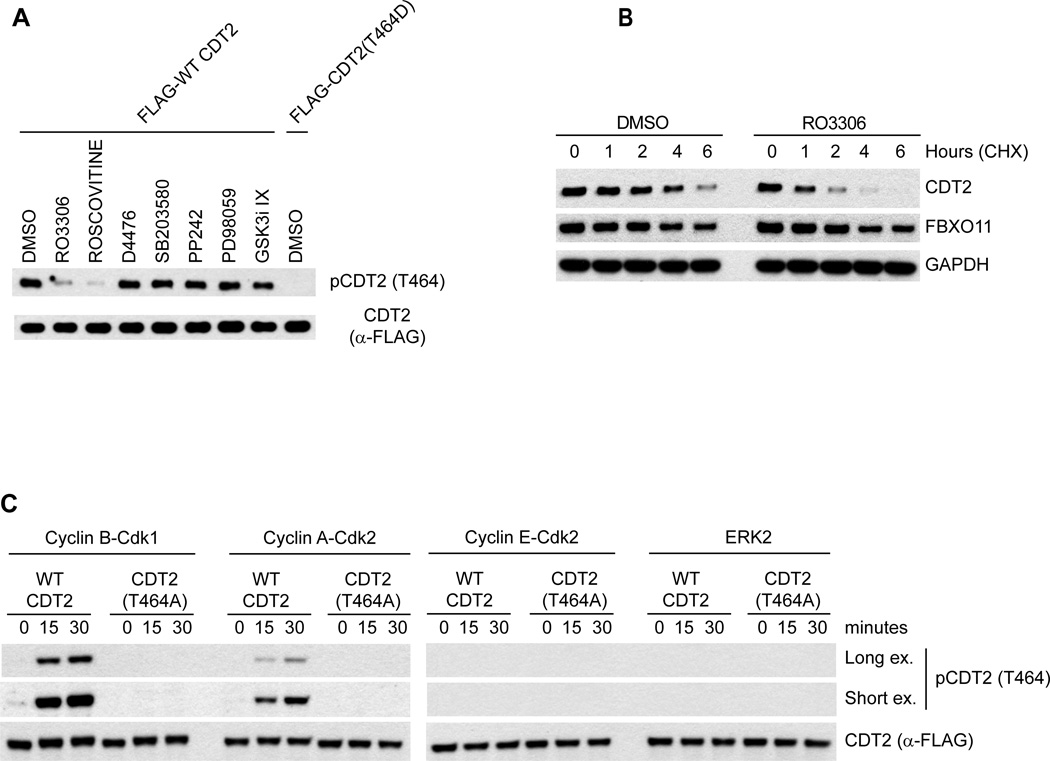
The fact that mutation of Pro465 prevents Thr464 phosphorylation suggested that a proline-directed kinase phosphorylates Thr464. As a first step in the identification of this kinase, we treated cultured cells with a panel of kinase inhibitors. We found that two different CDK inhibitors (RO-3306 and Roscovitine) reduced CDT2 phosphorylation on Thr464, whereas SB203580 (a p38 inhibitor), D4476 (a CKI inhibitor), SB203580 (a p38 inhibitor), pp242 (a mTOR inhibitor), PD98059 (an ERK inhibitor), GSK iX (a GSK inhibitor), U0126 (a MEK inhibitor), DMAT (a CKII inhibitor), and LY293646 (a DNA-PK inhibitor) had no effect (Fig. 3A and data not shown). Importantly, RO-3306 significantly reduced the half-life of CDT2 (Fig. 3B) and increased the co-immunoprecipitation of endogenous FBXO11 with FLAG-CDT2 (Fig. S3B), in agreement with the idea that phosphorylation of Thr464 stabilizes the protein. Moreover, both Cyclin B-Cdk1 and Cyclin A-Cdk2, but not Cyclin E-Cdk2, phosphorylated CDT2 on Thr464 in vitro (Fig. 3C), even though all three Pro-directed kinases were able to phosphorylate p27 on Thr187 (Fig. S3C). Finally, ERK2 (another proline-directed kinase) and Plk1 (another cell cycle regulatory kinase), failed to phosphorylate CDT2 in vitro (Figs. 3C and S3D). These data suggest that CDK-dependent phosphorylation of CDT2 blocks its interaction with FBXO11 and promote CDT2 stability.
Fig. 3. CDT2 is phosphorylated by CDKs.
(A) HeLa cells were treated with the indicated kinase inhibitors. Cells were collected four hours post-treatment, lysed, and immunoblotted as indicated.
(B) HeLa cells were treated with either DMSO or RO-3306. One hour later, cycloheximide (CHX) was added for the indicated times. Cells were collected, lysed, and immunoblotted as indicated.
(C) In vitro-translated CDT2 was incubated at 30°C with the indicated kinases. After the indicated times, the reactions were stopped, and the samples were immunoblotted as indicated.
(see also Figure S3)
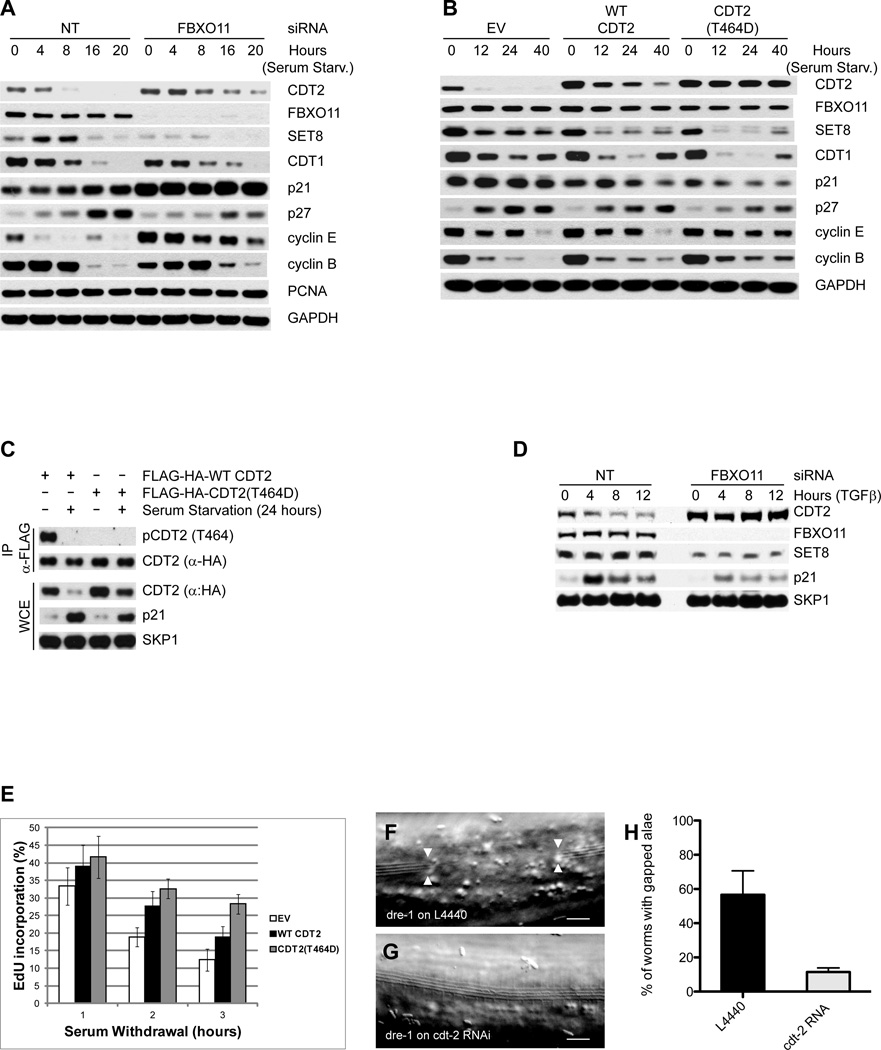
We then sought to find the physiological conditions required for CDT2 degradation, focusing our attention on conditions that inhibit CDK activity. We found that, in two different immortalized cell lines (RPE1-hTERT and HaCaT), CDT2 levels dramatically decreased after cell cycle withdraw obtained by serum starvation (Fig. 4A and Fig. S4A). This decrease was dependent on FBXO11, as shown by CDT2 stabilization following FBXO11 silencing (Fig. 4A and Fig. S4A) or inhibition of the proteasome with MG132 (not shown). To further investigate the degradation of CDT2, we serum starved RPE1-hTERT and HaCaT cells expressing either an empty virus, FLAG-tagged wild-type CDT2, or FLAG-tagged CDT2(T464D). Wild-type CDT2 (both endogenous and exogenous) was degraded after serum withdraw, while CDT2(T464D) remained stable (Fig. 4B and Fig. S4B), in agreement with its inability to bind FBXO11 (Fig. 2B). Significantly, CDT2 was phosphorylated on Thr464 in the presence of serum, but not in the absence of serum (Fig. 4C), supporting the notion that CDT2 degradation is blocked by Thr464 phosphorylation.
Fig. 4. FBXO11-dependent CDT2 degradation regulates the timing of cell cycle exit.
(A) RPE1-hTERT cells were transfected with either an siRNA to FBXO11 or a non-targeting siRNA (NT) and serum starved for the indicated times. Protein extracts were immunoblotted for the indicated proteins.
(B) RPE1-hTERT cells were infected with an empty virus (EV), a retrovirus expressing FLAG-HA-tagged wild type CDT2, or FLAG-HA-tagged CDT2(T464D). Cells were serum starved for the indicated times. Protein extracts were immunoblotted for the indicated proteins.
(C) HaCaT cells infected as in (B) were serum starved for 24 hours. Whole cell lysates were then immunoprecipitated with anti-FLAG resin and immunoblotted as indicated.
(D) MDA-MB-231 cells were transfected with either an siRNA to FBXO11 or a non-targeting siRNA (NT) and treated with TGFβ for the indicated times. Protein extracts were immunoblotted for the indicated proteins.
(E) The experiment was performed as in (B), except that cells were incubated with EdU for the last 20 minutes before collection. The percentage of EdU positive cells was determined by FACS analysis in triplicate experiments. The graph shows the percentage of cells incorporating EdU normalized to the percentage of cells incorporating EdU prior to serum starvation, which was set as 100%. Data are represented as mean of three independent experiments +/− SEM.
(F–G) Adult alae gaps of L4 molt dre-1(dh99) mutants are observed with an empty vector control (F, arrowheads). These gaps rarely occur when dre-1(dh99) worms are grown on cdt-2 RNAi (H). Scale bars, 10 µm.
(H) Percentage of dre-1(dh99) mutants with interrupted adult alae at the L4 molt when grown on L4440 or cdt-2 RNAi, respectively. These data are from four experiments with ≥ 20 worms each. Error bars indicate the standard deviation. **p≤0.001.
(see also Figure S4)
In order to confirm our results using a different stimulus that inhibits CDK activity and induces cell cycle arrest, we used TGF-β, whose downstream target SMAD2 has been proven to genetically interact with FBXO11 in a mouse model (Tateossian et al., 2009). We observed that CDT2 levels decreased in two TGF-β–responsive cell lines (HaCaT and MDA-MB-231), but not in the TGF-β–unresponsive T47-D cell line (Fig. 4D and Fig. S4C), and this decrease was dependent on FBXO11 (Fig. 4D and Fig. S4D). Moreover, wild-type CDT2 was degraded in HaCaT cells upon TGF-β treatment, while CDT2(T464D) was not (Fig. S4E). Finally, TGF-β treatment induced a decrease in the phosphorylation of CDT2 on Thr464 (Fig. S4F). Thus, cell cycle exit induced by serum starvation or TGF-β affects the phosphorylation and SCFFBXO11-dependent degradation of CDT2.
Inhibition of CDT2 degradation by FBXO11 depletion or expression of the stable CDT2(T464D) mutant in cells deprived of serum or treated with TGFβ resulted in a consistent decrease in SET8 levels; however, the effect on other CDT2 substrates (CDT1 and p21) was less consistent (Fig. 4A–B, Fig. S4A–B). Moreover, CDT2 stabilization resulted in a consistent delay in cell cycle exit, as indicated by the percentage of cells in S phase (Fig. 4E and Fig. S4G–I), slower kinetics of p27 accumulation (p27 is marker of G0/G1), and the delayed disappearance of S-phase and mitotic cyclins (cyclin E and cyclin B, respectively) (Fig. 4A–B and Fig. S4A–B). Wild type CDT2 produced intermediate effects since, although degraded in response to serum withdraw, it was expressed at levels above those of the endogenous protein (Fig. 4B). Taken together, these results indicate that FBXO11-dependent degradation of CDT2 plays a role in controlling the timing of cell cycle exit.
To validate our phenotypic observations in vivo, we used C. elegans as a genetic model to examine the FBXO11-CDT2 interaction. C. elegans epidermal stem cells, called seam cells, undergo various larval stage-specific patterns of proliferation and asymmetric cell division. Early in L4, they cease dividing, exit the cell cycle, and fuse into a long syncytium along the lateral midline. Finally, they differentiate and synthesize an adult-specific cuticular structure called adult alae. dre-1 mutants show various alterations in seam cell fates, including precocious seam fusion and gaps in the adult alae. The latter have been reported in various retarded heterochronic mutants like let-7 (Slack et al., 2000), which fail to exit the cell cycle. CKI-1 contributes to cell cycle exit and is upregulated in arresting cells. We observed cki-1::gfp expression in all seam cells by mid-L4 of wild type worms (n≥15), after seam cells exit the cell cycle and synthesize the adult alae (Fig. S4J,K). In contrast, cki-1::gfp was not expressed in the region of the alae gaps in dre-1 mutants (n≥15), even though cells flanking the gap showed expression. These findings suggest that the alae gaps in dre-1 mutants arise from a failure to exit the cell cycle and differentiate properly.
If CDT-2 were a DRE-1 substrate, then cdt-2 knockdown would be predicted to suppress dre-1 mutant defects. Depleting cdt-2 levels using RNAi strikingly reduced the appearance of gaps in the adult alae of dre-1(dh99) mutants from 56% to 13% (Fig. 4F–H). Interestingly, the precocious seam cell fusion of dre-1(dh99) mutants and a synthetic gonadal migration defect observed in dre-1(dh99);daf-12(rh61rh411) double mutants were not affected by cdt-2 depletion (Fig. S4L–S). These results reveal that dre-1 and cdt-2 specifically interact for some developmental processes (adult alae formation) but not others (seam fusion and gonadal outgrowth).
DISCUSSION
It is believed that substrate recognition subunits of CRL ubiquitin ligases are degraded via an auto-ubiquitylation process that is promoted by the lack of substrate availability (Lee et al., 2011; Yen and Elledge, 2008). An exception to this rule is the degradation of the substrate binding subunits of SCFSKP2, namely, SKP2, which is ubiquitylated and degraded via the APC/CCDH1 ubiquitin ligase (Bashir et al., 2004; Wei et al., 2004). We now show that the degradation of a CRL4 substrate receptor (CDT2) is regulated by a SCF ubiquitin ligase (SCFFBXO11). In proliferating cells, this event is inhibited by the CDK-dependent phosphorylation of CDT2 on Thr464, which is present in the FBXO11 binding domain of CDT2. Thus, in contrast to other F-box proteins, which bind substrates only when they are phosphorylated in their degrons, FBXO11 behaves in the opposite way, dissociating from the substrate when the degron is phosphorylated. This identifies a novel mechanism for regulation of substrate recognition by an F-box protein.
Significantly, we found that CDT2 phosphorylation on Thr464 and CDT2 stability decrease when cells exit the cell cycle. The decrease in CDT2 protein levels is mediated by SCFFBXO11, and inhibition of this event produces a delay in cell cycle exit. In serumstarved cells containing high levels of CDT2, we observed a consistent decrease in SET8 levels, but decreases in the levels of the other two CDT2 substrates (i.e., CDT1 and p21) were less consistent. The reason for this differential effect is currently not known.
In worms, cdt-2 depletion suppresses the appearance of gaps in the adult alae of dre-1 mutants. Conceivably, these gaps could occur as a consequence of delayed cell cycle exit of seam cells, which then fail to execute adult-specific cellular differentiation programs on schedule (Slack et al., 2000). This hypothesis is supported by the lack of cki-1::gfp expression, a marker for arresting cells, in the epidermal tissues underlying the alae gaps. The physical interaction between CDT-2 and DRE-1, and the genetic interactions observed in C. elegans are consistent with the results in human cells and support a model in which CDT-2 is a substrate of SCFDRE-1. However, the observation that cdt-2 depletion cannot rescue the precocious seam cell fusion even or the synthetic gonadal migration present in the dre-1 suggests that accumulation of distinct SCFDRE-1 substrates is responsible for the diverse heterochronic phenotypes of dre-1 mutants.
In summary, our findings indicate that the FBXO11/DRE-1-dependent degradation of CDT2/CDT-2 is part of a highly conserved cellular program that controls the timing of cell cycle exit and differentiation. In the future, it will be important to determine the specific cellular context under which mammalian CDT2 is degraded to control developmental and cell differentiation.
EXPERIMENTAL PROCEDURES
Cell Lines, Serum Starvation, and Drug Treatments
HeLa, RPE1-hTERT, HEK293T, and HaCaT, cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS). Cells were starved for the indicated time periods in 0.1% serum. The following drugs were used: MG132 (Peptides International; 10 mM), cycloheximide (Sigma; 100 mg/ml), recombinant TGF-β (Cell Signaling, 5 ng/ml), RO3306 (Tocris, 20 µM), Roscovitine (Cell Signaling, 10 µM), pp242 (Sigma, 2.5 µM), PD98059 (Cell Signaling, 20 µM), D4476 (Calbiochem, 50 µM), GSK3i IX (Calbiochem, 5 µM), DMAT (Sigma, 200 nM), LY293646 (Millipore, 1 µM), UO126 (Cell Signaling, 30 µM), SB203580 (Calbiochem, 10 µM).
Antibodies
The following Rabbit polyclonal antibodies were used: FBXO11 (Novus Biological), CDT2 (Novus Biological), p21 (Santa Cruz Biotechnology), SET8 (Cell Signaling), phospho SMAD2 (S465/467) (Cell Signaling), FBXO1 (C-20; Santa Cruz Biotechnology), FBXO5 (Invitrogen), FBXO31 (Bethyl), FBXL1 (Invitrogen), FBXW1 (Cell Signaling), FBXW7 (Bethyl) anti CUL4 (Rockland/VWR), anti-FLAG (Sigma), phospho p27 (T187) (Invitrogen), and MYC (Bethyl Laboratories). Antibodies against FBXO18, FBXW9, FBXL11, cyclin B, and SKP1 were generated and characterized in the Pagano laboratory. The rabbit polyclonal phosphospecific antibody pCDT2 (T464) was generated using a peptide containing the sequence DLPLPSNpTPTFSIK. The following mouse monoclonal antibodies rabbit were used: DDB1 (Invitrogen), PLK1 (Invitrogen), anti-FLAG M2 (Sigma), p27 (BD Biosciences) and anti-HA (Covance).
Biochemical Methods
Extract preparation, immunoprecipitation, and western blotting were performed as previously described (Duan et al., 2012; Duan et al., 2011). Cell lysis, immunopurification, and MS/MS analysis have been previously described (D'Angiolella et al., 2012). In Vitro Phosphorylation of CDT2 and p27 were performed as previously described for other CDK substrates (D'Angiolella et al., 2012). CDT2 ubiquitylation was performed in 30 µl reaction buffer containing 0.1 µM E1, 0.25 µM Ubc3, 0.25 µM Ubc5, 1.5 µg/µl ubiquitin, and 1 µl of in vitro translated CDT2. Full length FBXO11 or a deletion mutant lacking its N-terminus that contains the F-box domain FBXO11(ΔFbox) were immunopurified with anti-FLAG M2 affinity gel from transfected HEK293T cells and eluted by competition with FLAG peptide. Samples were then incubated at 30°C for the indicated times and analyzed by SDS-PAGE and immunoblotting.
siRNAs and shRNAs
siRNA duplexes were transfected into subconfluent RPE1-hTERT, HeLa and HaCaT cells using HiPerfect reagent (QIAGEN) according to the manufacturer’s instructions. The Non Targeting (NT), CDT2 and FBXO11 siRNAs were purchased from Dharmacon (On target Plus D-001810-10-05, L-020543-00-0005 and L-012428-00-0005, respectively.). The retroviral vectors expressing shRNA control or against FBXO11 was a gift from Dr. Carroll (NYU Medical Center).
Mammalian cell Infection
Retrovirus-mediated gene transfer was performed as previously described (D'Angiolella et al., 2010).
C. elegans RNAi
The strains used in this study were: N2 (Bristol), AA426 dre-1(dh99), VT825 dpy20(e1282);maIs113(cki-1::gfp). RNAi was performed using the feeding protocol as described (Kamath et al., 2001). Worms were placed on RNAi for one generation before phenotypic analysis. RNAi efficiency was validated by the observation of cdt-2 knockdown related phenotypes (Kim et al., 2008), as well as qPCR (data not shown). All experiments were conducted at 20°C.
Supplementary Material
Highlights.
FBXO11 targets CDT2, a CRL4 substrate receptor, for proteasomal degradation
CDK-mediated phosphorylation of CDT2 degron inhibits recognition by FBXO11
FBXO11-mediated degradation of CDT2 controls the timing of cell cycle exit
FBXO11-CDT2 functional interaction is evolutionary conserved from worms to humans
Acknowledgements
The authors thank W. Carroll for reagents, Scott Millman for his help with flow cytometry, and J. R. Skaar for critical reading of the manuscript. MP is grateful to T.M. Thor for continuous support. This work was funded by grants from the National Institutes of Health (R01-GM057587, R37-CA076584, R21-CA161108, and R03 TW009040) to MP, the Max Planck Gesellschaft, SFB635 of the DFG to AA, Susan G. Komen for the Cure to S.D and Agencia Nacional de Promoción Científica y Tecnológica-Argentina (ANPCyT) (PICT-2011-1231) to M.R. AS, LF, and MPW are supported by the Stowers Institute for Medical Research. MP is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors of this work has a financial interest related to this work.
Contributor Information
Mario Rossi, Email: mrossi@ibioba-mpsp-conicet.gov.ar.
Michele Pagano, Email: michele.pagano@nyumc.org.
References
- Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle. 2011;10:241–249. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-Mediated Degradation of Ribonucleotide Reductase M2 Controls Genome Integrity and DNA Repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Saraf A, Vijayakumar S, Florens L, Washburn M, Dynlacht B, Pagano M. SCFCyclin Fcontrols centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Guardavaccaro D, Neubert K, Chan T, Li D, Feng Q, Hutter H, Pagano M, Antebi A. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisty-Hughes RE, Tateossian H, Morse SA, Romero MR, Middleton A, Tymowska-Lalanne Z, Hunter AJ, Cheeseman M, Brown SD. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet. 2006;15:3273–3279. doi: 10.1093/hmg/ddl403. [DOI] [PubMed] [Google Scholar]
- Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011;25:1568–1582. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol. 2011;192:43–54. doi: 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Sweredoski MJ, Graham RL, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006460. M110 006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Segade F, Daly KA, Allred D, Hicks PJ, Cox M, Brown M, Hardisty-Hughes RE, Brown SD, Rich SS, Bowden DW. Association of the FBXO11 gene with chronic otitis media with effusion and recurrent otitis media: the Minnesota COME/ROM Family Study. Arch Otolaryngol Head Neck Surg. 2006;132:729–733. doi: 10.1001/archotol.132.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, D'Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009a;137:1358. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. SnapShot: F box proteins I. Cell. 2009b;137:1160. doi: 10.1016/j.cell.2009.05.039. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateossian H, Hardisty-Hughes RE, Morse S, Romero MR, Hilton H, Dean C, Brown SD. Regulation of TGF-beta signalling by Fbxo11, the gene mutated in the Jeff otitis media mouse mutant. Pathogenetics. 2009;2:5. doi: 10.1186/1755-8417-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.