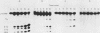Abstract
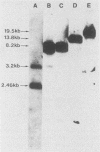
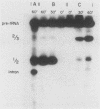
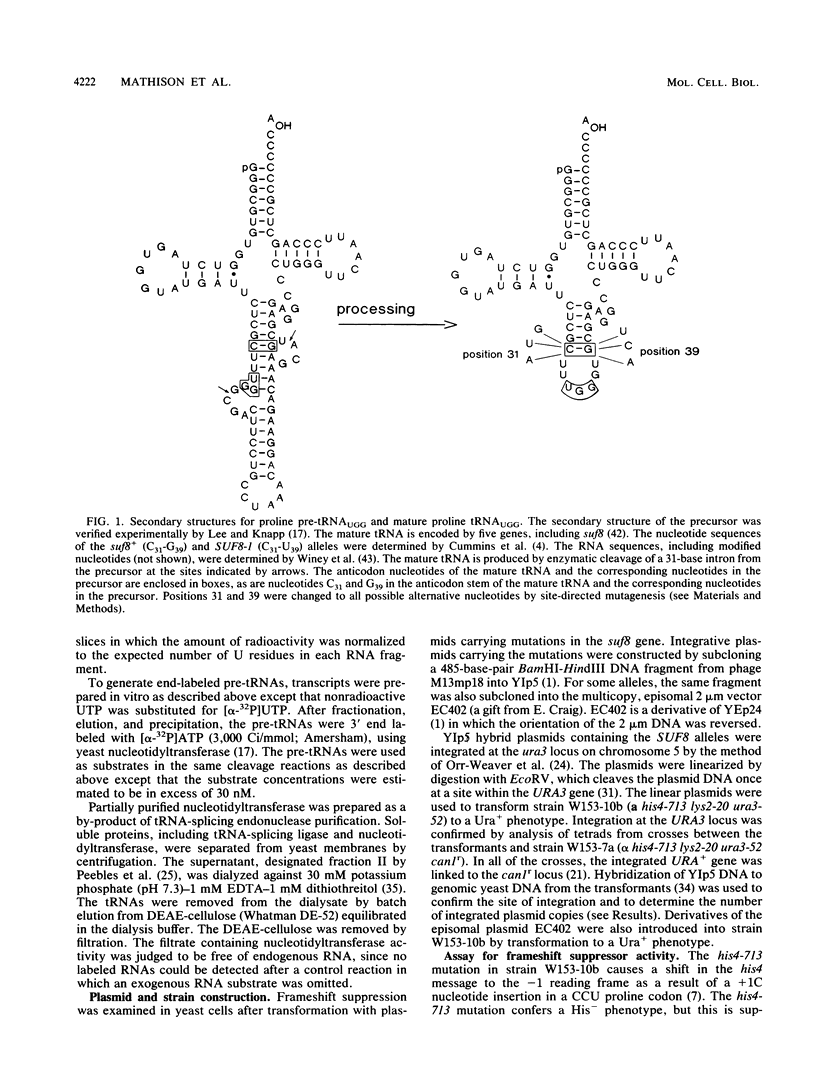
To evaluate the role of exon domains in tRNA splicing, the anti-codon stem of proline pre-tRNAUGG from Saccharomyces cerevisiae was altered by site-directed mutagenesis of the suf8 gene. Sixteen alleles were constructed that encode mutant pre-tRNAs containing all possible base combinations in the last base pair of the anticodon stem adjacent to the anticodon loop (positions 31 and 39). The altered pre-tRNAs were screened by using an in vitro endonucleolytic cleavage assay to determine whether perturbations in secondary structure affect the intron excision reaction. The pre-tRNAs were cleaved efficiently whenever secondary structure in the anticodon stem was maintained through standard base pairing or G.U interactions. However, most of the pre-tRNAs with disrupted secondary structure were poor substrates for intron excision. We also determined the extent to which the suf8 alleles produce functional products in vivo. Each allele was integrated in one to three copies into a yeast chromosome or introduced on a high-copy-number plasmid by transformation. The formation of a functional product was assayed by the ability of each allele to suppress the +1 frameshift mutation his4-713 through four-base codon reading, as shown previously for the SUF8-1 suppressor allele. We found that alleles containing any standard base pair or G.U pair at position 31/39 in the anticodon stem failed to suppress his4-713. We could not assess in vivo splicing with these alleles because the tRNA products, even if they are made, would be expected to read a normal triplet rather than a quadruplet codon. However, all of the alleles that contained a disrupted base pair at position 31/ 39 in the anticodon stem altered the structure of the tRNA in a manner that caused frameshift suppression. Suppression indicated that splicing must have occurred to some extent in vivo even though most of the suppression alleles produced pre-tRNAs that were cleaved with low efficiency or not at all in vitro. These results have important implications for the interpretation of in vitro cleavage assays in general and for the potential use of suppressors to select mutations that affects tRNA splicing.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clark M. W., Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987 Oct;105(4):1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Jacobsen K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984 Oct;38(3):841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- Cummins C. M., Gaber R. F., Culbertson M. R., Mann R., Fink G. R. Frameshift suppression in Saccharomyces cerevisiae. III. Isolation and genetic properties of group III suppressors. Genetics. 1980 Aug;95(4):855–879. doi: 10.1093/genetics/95.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987 Dec 11;238(4833):1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. Suppressible four-base glycine and proline codons in yeast. Science. 1981 Apr 24;212(4493):455–457. doi: 10.1126/science.7010605. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Frameshift suppression in Saccharomyces cerevisiae. IV. New suppressors among spontaneous co-revertants of the Group II his4-206 and leu 2-3 frameshift mutations. Genetics. 1982 Jul-Aug;101(3-4):345–367. doi: 10.1093/genetics/101.3-4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C. L. Assembly of a tRNA splicing complex: evidence for concerted excision and joining steps in splicing in vitro. Mol Cell Biol. 1986 Feb;6(2):635–644. doi: 10.1128/mcb.6.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983 Feb;32(2):537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Greer C. L., Söll D., Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):76–84. doi: 10.1128/mcb.7.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G., Beckmann J. S., Johnson P. F., Fuhrman S. A., Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978 Jun;14(2):221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., De Robertis E. M. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol. 1981 Jan 15;145(2):405–420. doi: 10.1016/0022-2836(81)90212-6. [DOI] [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983 Feb;32(2):525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Ogden R. C., Knapp G., Abelson J. Splicing of yeast tRNA precursors: a two-stage reaction. Cell. 1979 Sep;18(1):27–35. doi: 10.1016/0092-8674(79)90350-7. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Broach J. R., Wensink P. C., Hereford L. M., Fink G. R., Botstein D. Isolation and analysis of recombinant DNA molecules containing yeast DNA. Gene. 1978 Sep;4(1):37–49. doi: 10.1016/0378-1119(78)90013-6. [DOI] [PubMed] [Google Scholar]
- Phizicky E. M., Schwartz R. C., Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986 Feb 25;261(6):2978–2986. [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988 Nov 18;55(4):719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. DNA rearrangements associated with a transposable element in yeast. Cell. 1980 Aug;21(1):239–249. doi: 10.1016/0092-8674(80)90131-2. [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sternbach H., von der Haar F., Schlimme E., Gaertner E., Cramer F. Isolation and properties of tRNA nucleotidyl transferase from yeast. Eur J Biochem. 1971 Sep 24;22(2):166–172. doi: 10.1111/j.1432-1033.1971.tb01528.x. [DOI] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Tobian J. A., Drinkard L., Zasloff M. tRNA nuclear transport: defining the critical regions of human tRNAimet by point mutagenesis. Cell. 1985 Dec;43(2 Pt 1):415–422. doi: 10.1016/0092-8674(85)90171-0. [DOI] [PubMed] [Google Scholar]
- Willis I., Hottinger H., Pearson D., Chisholm V., Leupold U., Söll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984 Jul;3(7):1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Culbertson M. R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988 Apr;118(4):609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Edelman I., Culbertson M. R. A synthetic intron in a naturally intronless yeast pre-tRNA is spliced efficiently in vivo. Mol Cell Biol. 1989 Jan;9(1):329–331. doi: 10.1128/mcb.9.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mathison L., Soref C. M., Culbertson M. R. Distribution of introns in frameshift-suppressor proline-tRNA genes of Saccharomyces cerevisiae. Gene. 1989 Mar 15;76(1):89–97. doi: 10.1016/0378-1119(89)90011-5. [DOI] [PubMed] [Google Scholar]
- Winey M., Mendenhall M. D., Cummins C. M., Culbertson M. R., Knapp G. Splicing of a yeast proline tRNA containing a novel suppressor mutation in the anticodon stem. J Mol Biol. 1986 Nov 5;192(1):49–63. doi: 10.1016/0022-2836(86)90463-8. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- van Tol H., Gross H. J., Beier H. Non-enzymatic excision of pre-tRNA introns? EMBO J. 1989 Jan;8(1):293–300. doi: 10.1002/j.1460-2075.1989.tb03376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]