Abstract
The eukaryotic cytoskeleton is a vulnerable target of many microbial pathogens during the course of infection. Rearrangements of host cytoskeleton benefit microbes in various stages of their infection cycle such as invasion, motility, and persistence. Bacterial pathogens deliver a number of effector proteins into host cells for modulating the dynamics of actin and microtubule cytoskeleton. Alteration of the actin cytoskeleton is generally achieved by bacterial effectors that target the small GTPases of the host. Modulation of microtubule dynamics involves direct interaction of effector proteins with the subunits of microtubules or recruiting cellular proteins that affect microtubule dynamics. This review will discuss effector proteins from animal and human bacterial pathogens that either destabilize or stabilize host micro-tubules to advance the infectious process. A compilation of these research findings will provide an overview of known and unknown strategies used by various bacterial effectors to modulate the host microtubule dynamics. The present review will undoubtedly help direct future research to determine the mechanisms of action of many bacterial effector proteins and contribute to understanding the survival strategies of diverse adherent and invasive bacterial pathogens.
Keywords: bacteria, bacterial effector proteins, cytoskeleton, microtubules
Introduction
The cytoskeleton is a cytoplasmic scaffold that determines cell shape, enables cell movement, and plays an essential role in intracellular organelle transport and cell division. The cytoskeleton network consists of three types of protein filaments: actin filaments, intermediate filaments, and microtubules. The eukaryotic cytoskeleton is targeted by a variety of bacterial and viral pathogens during the course of infection, and dynamic changes of the cytoskeleton influence the interaction of microbial pathogens with the host cells. Consequently, successful microbial pathogens modulate cytoskeleton dynamics to facilitate adherence to the cells, invasion, intra- and intercellular trafficking, and to prevent intracellular killing (1–6). Microbial pathogens deliver a number of effector proteins to the host cells to rearrange the cytoskeleton to benefit the infection process. These effector proteins essentially target small GTPases to modulate the dynamics of the actin cytoskeleton of host cells (3, 7–11). Rearrangement of the actin cytoskeleton by pathogenic microorganisms has been extensively reviewed elsewhere (3, 8, 10, 12–19). This review will focus on modulation of host microtubule dynamics by pathogenic bacteria.
Microtubules are essential components of the eukaryotic cytoskeleton composed of heterodimers of α- and β-tubulin. Tubulin dimers polymerize to form a microtubule that consists of 13 linear protofilaments assembled around a hollow core (20, 21). Microtubules are polar structures with a fast-growing plus end and a slow-growing minus end, and this polarity determines the direction of movement along micro-tubules (22, 23). Important to cell function, microtubules are dynamic structures that undergo continual assembly and disassembly within the cell (24, 25). Many bacterial pathogens modulate this microtubule dynamics by employing virulence proteins to promote infection (Table 1). This review will discuss various bacterial effectors that destabilize or stabilize host microtubule networks.
Table 1.
Bacterial effector proteins that modulate microtubule dynamics.
| Pathogen | Effector proteins | References |
|---|---|---|
| Microtubule destabilizers | ||
| Shigella flexneri | VirA | (29–31) |
| Enteropathogenic | EspG and EspG2 | (36–38) |
| Escherichia coli | ||
| Citrobacter rodentium | EspG | (44) |
| Chlamydia | CopN | (49–51) |
| Edwardsiella tarda | EseG | (53) |
| Listeria monocytogenes | ActA | (60, 67, 72) |
| Microtubule stabilizers | ||
| Salmonella enterica | SifA, SseF, SseG | (54, 77) |
| Brucella spp. | TcpB/Bpt1 | (83–85) |
| Clostridium difficile | CDT | (89) |
| Clostridium botulinum | C2 | (89) |
| Clostridium perfringens | Iota toxin | (89) |
| Streptococcus pneumonia | Pneumolysin | (95) |
Destabilization of host microtubule cytoskeleton by bacterial pathogens
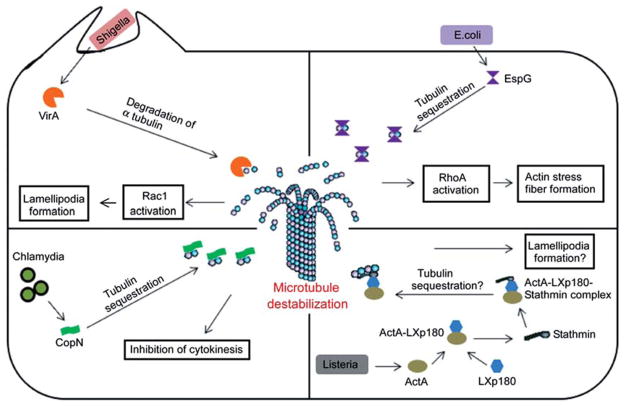
Destabilization of host microtubules is a common strategy adopted by various bacterial pathogens (Figure 1). Microtubule destabilization benefits these pathogens in many ways, including the free movement of pathogens through the cytoplasm and modulation of actin cytoskeleton through the activation of small GTPases. Rearrangement of the actin cytoskeleton facilitates formation of membrane ruffles and pseudopodia that promotes bacterial invasion and movement. Examples of major bacterial effectors that destabilize micro-tubule networks are discussed below.
Figure 1.
Bacterial effector proteins that destabilize host microtubules. The mechanism of action of effector proteins that has been demonstrated or hypothesized is illustrated. The possible benefits of host microtubule destruction for the pathogen have also been depicted in the figure.
VirA of Shigella
Shigella flexneri, the causative agent of bacterial dysentery, harbors an important virulence gene, VirA, that encodes a 45-kDa protein (26). Shigella delivers VirA into the host cells using its type III secretion system (T3SS), and intracellular VirA modulates the cytoskeleton dynamics to facilitate bacterial entry and intracellular movement (27, 28). Shigella spp. deficient in the VirA gene are defective in intracellular movement and present an attenuated phenotype in a mouse model of infection (29). Studies have established that the VirA effector protein modulates host microtubule dynamics by acting as a destabilization factor (29). Shigella destroys the microtubule network in the infected cells and creates a tunnel through which the bacteria move smoothly. Infection studies using mutant bacteria indicated that these properties are attributed to the VirA protein of Shigella.
VirA interacts with the subunits of the microtubule through an N-terminal domain that is located between amino acid residues 224 and 315 (28). VirA can efficiently inhibit microtubule polymerization and induce depolymerization of assembled microtubules in a dose-dependent manner (28). However, the mechanism whereby VirA induces microtubule destabilization remained controversial. More recently, VirA was reported as a cysteine protease that specifically targets the α-subunit of the tubulin heterodimer (29). VirA is capable of degrading purified human α-tubulin, and the protease activity is sensitive to the protease inhibitors leupeptin and cystatin C. Mutation studies identified the catalytic cysteine residue at the N-terminus of VirA (C34), and its mutation to serine (VirAC34S) affects the activity of the protein (29). VirAC34S does not exhibit micro-tubule disruption in COS-7 cells, and Shigella expressing the respective mutant presented a defective intracellular movement (29). However, experimental data from two independent structural and functional studies contradicted the identified VirA mechanism of action (30, 31). The elucidated crystal structure of VirA reveals that it harbors two independently folded domains that resemble the letter ‘V’ (30, 31). VirA represents a novel protein fold and does not show any significant structural homology to papain-like cysteine proteases as indicated by previous biochemical assays. However, the N-terminal domain of VirA exhibits limited similarity to the inhibitors of cysteine proteases. The putative active site of VirA that contains the catalytic residue Cys34 appears disordered in the crystal structure. Structural analysis also pointed out that the N-terminal domain comprising the 224–315 amino acid region of VirA is very likely involved in dimer formation rather than tubulin interaction (30, 31). Nevertheless, there are conflicting reports on the existence of VirA as a monomer or dimer. In contrast to the previous report (29), others could not identify any obvious proteolytic activity or microtubule depolymerization properties by purified VirA protein (30, 31). VirA may act as a scaffold for a host papain-like cysteine protease, or recruits an unidentified microtubule-destabilizing protein to facilitate the microtubule destruction.
In addition to creating a tunnel for bacterial movement by destroying microtubule networks, VirA has the ability to induce membrane ruffles in various mammalian cells. VirA-induced membrane ruffles are reported to be mediated by the activation of the small GTPase Rac1. It has been hypothesized that microtubule regrowth after its depolymerization may activate Rac1, resulting in the development of membrane ruffles and lamellipodial protrusions that promote bacterial entry into host cells (28).
EspG of Escherichia coli
Enteropathogenic E. coli (EPEC) is a bacterial pathogen that causes gastroenteritis in humans (32). EPEC adheres to the gastrointestinal mucosa and forms attaching and effacing lesions that are characterized by localized destruction of the gastric microvillus brush border, intimate adherence of bacteria to epithelial cells, and cytoskeleton reorganization (32–34). EPEC injects a battery of effector proteins into intestinal epithelial cells through its T3SS to subvert the host cell processes to benefit the extracellular bacterium (33, 35). EPEC secretes effector proteins, EspG and EspG2, to destabilize microtubules. Importantly, EspG and EspG2 share a striking similarity (40% and 38%, respectively) to the VirA protein of Shigella, which is also a microtubule-destabilizing protein (36). There is a 62% similarity between EspG and EspG2 (36). Interestingly, E. coli EspG as well as EspG2 can restore the intracellular persistence of the Shigella VirA mutant, indicating a functional level similarity of EspG/EspG2 and VirA (36). In vitro infection studies using EspG mutant EPEC did not indicate any observable attenuated phenotype, whereas in vivo studies using a rabbit model of diarrhea presented with attenuated gut colonization by an EspG mutant (36).
Wild-type EPEC caused localized microtubule destruction beneath the site of adherence (37). Infection studies using mutant EPEC demonstrate that microtubule depletion is attributed to the EspG and EspG2 or orf3 genes. However, a single mutant of EspG did not exhibit microtubule destruction due to the functional redundancy of EspG and EspG2, whereas a double mutant was defective in microtubule depletion (37). Both EspG and EspG2 are capable of binding tubulins but not Taxol-stabilized microtubules (38). In vitro studies also demonstrate that EspG and EspG2 can efficiently inhibit microtubule polymerization as well as trigger the destabilization of polymerized microtubules (38). The exact mechanism of EspG/EspG2-mediated microtubule destruction remains unknown. Given the high similarity of VirA and EspG/EspG2, a cysteine protease activity has been proposed as the mechanism of microtubule destruction by EspG/EspG2. However, the protease activity of EspG/EspG2 has not been demonstrated. An alternative hypothesis suggests that EspG/EspG2 acts similarly to the microtubule-destabilizing protein, stathmin, which sequesters tubulins and reduces the concentration of tubulin available for microtubule assembly (38–40).
Escherichia coli is an extracellular bacterium, and how EspG/EspG2-induced microtubule destruction may benefit the pathogen remains largely unknown. EspG/EspG2-induced microtubule destruction triggers the release and activation of a microtubule-associated RhoA-specific guanine nucleotide exchange factor, GEF-H1. Activated GEF-H1 in turn activates a RhoA-ROCK signaling pathway and induces actin stress fiber formation (38). GEF-H1 associates with the epithelial tight junctions and regulates the paracellular permeability by reorganizing the actin cytoskeleton (41). Therefore, it is assumed that the microtubule destruction by EspG/EspG2 leads to actin rearrangements and increased paracellular permeability, which contributes to EPEC-induced diarrhea.
EspG of Citrobacter
Citrobacter rodentium is a murine attaching and effacing pathogen that causes mild diarrhea and colonic hyperplasia in mice (42). EspG encoded by C. rodentium shares strong homology with the EspG of EPEC (43, 44). EspG, secreted by T3SS, of C. rodentium also binds to human tubulin and induces localized microtubule destruction and stimulates actin stress fiber formation (45). Disrupting microtubules in colonocytes by EspG and potentially with EspF, the various cell membrane aquaporin water channels that normally absorb water from the gut are repositioned to the cell cytoplasm, contributing to diarrhea during bacterial infection (46).
CopN of Chlamydia
The Chlamydiae are Gram-negative, obligate intracellular pathogens that cause a range of human diseases, including genital, ocular, and respiratory infections (47). They undergo a biphasic developmental cycle involving the infectious elementary body (EB), and the replicative, non-infectious reticulate body (RB) (48). After entering their target eukaryotic cells, EBs differentiate into RBs and replicate within an endosome-derived membranous vacuole, termed ‘inclusion’ (48). Microtubules play an essential role in the intracellular lifestyle of Chlamydia. Chlamydial inclusions are trafficked along microtubules toward the minus ends and aggregate at the microtubule organizing center (MTOC) (49, 50). Disruption of microtubules by nocodazole treatment in Cos-7 cells inhibited the characteristic localization of Chlamydia inclusions. Studies have shown that inclusion body translocation depends on the minus-end-directed microtubule motor complex dyenin (49).
Even though Chlamydia exploits the microtubule network for trafficking of inclusions, one of the Chlamydia effectors, CopN, exhibits microtubule destabilization properties (51). Heterologous expression of CopN in yeast and mammalian cells affected the formation of microtubule structures and blocks cell division (52). Recently, CopN was shown to directly bind non-polymerized α and β-tubulins but not to polymerized microtubules (53). CopN can efficiently inhibit microtubule polymerization but cannot induce depolymerization due to its inability to bind to the polymerized microtubules. On the basis of these observations, it is hypothesized that CopN may act like stathmin and destabilize the microtubules by sequestrating α- and β-tubulins (53). Chlamydia resides at the MTOC, and the microtubule destabilization property of CopN may disrupt the mitotic spindles, leading to chromosomal segregation defects and inhibition of cytokinesis.
EseG of Edwardsiella tarda
Edwardsiella tarda is an enteric pathogen that causes septicemia of fish and gastroenteritis of humans (54). Edwardsiella tarda secretes an effector protein, EseG, to the host cells through a T3SS. Overexpression of EseG in HeLa cells induces dramatic microtubule destruction (55). EseG does not share any homology with other microtubule-destabilizing proteins such as VirA and EspG. However, EseG does share a conserved domain with the SseF and SseG proteins of Salmonella. A microtubule destabilization property has not been demonstrated for SseF or SseG, and these effector proteins play an essential role in the perinuclear localization of Salmonella-containing vacuoles. In fact, SseG co-localizes with microtubules and exhibit microtubule-bundling properties (56). Nevertheless, the conserved domain is reported to be essential for EseG to destabilize microtubules (55). EseG interacts with α-tubulin through a separate domain at the N-terminus of the protein (55). The actual mechanism of EseG-mediated microtubule destruction remains to be elucidated.
ActA of Listeria
Listeria monocytogenes is an intracellular pathogenic bacterium that causes the severe food-borne infection listeriosis (57). Listeria is capable of invading and replicating in a variety of mammalian cells. After internalization, Listeria prevents phagosome-lysosome fusion and escapes from the phagosome with the help of its virulence protein, LLO (58). Free bacteria then replicate in the cells and spread to neighboring cells without inducing cell lysis (58–60). Listeria encodes a number of virulence factors to facilitate its intra-cellular survival and spread (61). Studies have shown that ActA is essential for inter- and intracellular movement of Listeria. ActA induces a comet-shaped actin polymerization at the posterior pole of the bacterium that generates unidirectional propulsion force to push the bacterium through the cytoplasm. ActA mimics the C-terminal domain of Wiskott-Aldrich syndrome protein (WASP) and activates actin-related protein (Arp) 2/3 complex to facilitate the comet formation at the pole of the bacterium (62). The microtubule-binding protein dynamin-2 co-localizes with the Listeria-induced actin comets. Dynamin-2 is a GTPase protein that is ubiquitously expressed in mammals and plays key roles in various cellular processes, including fission of clathrin-coated endocytic vesicles, vesicle trafficking, centrosome cohesion, actin reorganization, and microtubule dynamics (63–67). Dynamin-2 polymerizes the entire length around the microtubules and contributes to the correct bundling of microtubules. Silencing of dynamin-2 by siRNA results in dynamic instability of microtubules and induces accumulation of acetylated and stable microtubules (68). Infection of dynamin-2-depleted HeLa cells with Listeria reduces actin comet tail formation and diminishes the speed of bacterial movement (69). These studies imply that the alteration of microtubule dynamics influences the Listeria-induced actin comet tail formation. A number of actin regulatory proteins such as Rho family GTPases are associated with microtubules, and microtubule dynamics regulate actin regulatory protein release and activation (70–72). Therefore, alterations of microtubule dynamics likely affect the reorganization of actin cytoskeleton and the formation of actin comets.
A high-throughput yeast two-hybrid screen identified a mammalian protein, LaXp180, that interacts with ActA of Listeria (73). LaXp180 interacts with a well-characterized microtubule-destabilizing protein, stathmin (74). Stathmin sequestrates tubulin dimers by forming a complex and reduces the concentration of free tubulin available for polymerization (39, 40, 75). In addition to the tubulin sequestration, stathmin also promotes microtubule catastrophe or shortening through an unidentified mechanism (75). The role of microtubules on the intracellular lifestyle of Listeria has not been investigated in detail. However, stathmin-mediated microtubule depletion by Listeria proteins may lead to actin rearrangement that promotes bacterial movement and spread. Studies have shown that the microtubule destabilization property of stathmin is inactivated by its phosphorylation, and the phosphorylated stathmin induces lamellipodia formation that is mediated by a multiprotein complex termed WAVE-2 (76, 77). Whether the ActA-LaXp180-stathmin interaction induces lamellipodia formation to promote the spread of Listeria is an area that needs to be addressed.
Stabilization of host microtubule cytoskeleton by bacterial pathogens
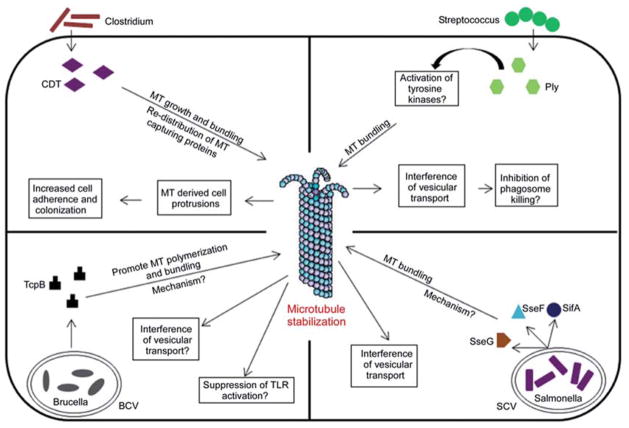
Many invasive and adherent bacterial pathogens induce stabilization of host microtubules to promote their survival and persistence (Figure 2). The mechanism by which bacterial effector proteins induce the stabilization of host microtubules and the beneficial role of stabilized microtubules for survival of certain pathogens in the host are poorly understood. Major bacterial effector proteins that stabilize host microtubules are as follows.
Figure 2.
Bacterial effector proteins that stabilize host microtubules. The known mechanism of action of effector proteins and the benefits of host microtubule stabilization for the pathogen have been depicted in the figure. MT, microtubule; BCV, Brucella-containing vacuoles; SCV, Salmonella-containing vacuole; Ply, pneumolysin; CDT, Clostridium difficile toxin.
SifA of Salmonella
Salmonellae are gastrointestinal pathogens causing diseases ranging from enteritis to typhoid fever (78). They are facultative intracellular pathogens and reside in membranous compartments termed Salmonella-containing vacuoles (SCV). Salmonella possess two T3SS to deliver its effector proteins to the host cytosol to modify the host cell processes to benefit the pathogen. The Salmonella-secreted proteins SifA, SseF, and SseG have been reported to affect micro-tubule dynamics (56). SifA is an important virulence protein of Salmonella and induces tubular networks, termed as Salmonella-induced filaments (SIFs), that extend from the SCVs (79). SIF formation is essential for pathogenicity as the SifA mutant Salmonella presents an attenuated phenotype in mice and macrophages (80, 81). Intact microtubules are required for the formation of SIFs, indicating that SIF structures are formed on the scaffolding of microtubules (56, 79). Recent studies have shown that SIFs constitute the tubular aggregates of phagosomes, and the tubular networks require the participation of host SifA kinesin-interacting protein, SKIP, and the microtubule motor, kinesin-1 (82, 83). The Salmonella effectors SseG and SseF interfere with microtubule organization and induce massive microtubule bundling (56). It is hypothesized that microtubule reorganization may reduce or block the vesicles that transport along the microtubules. Therefore, microtubule bundling may bring the vesicles in close proximity, leading to their fusion and formation of tubular networks along the microtubules (56) benefiting the Salmonella. The mechanism by which these Salmonella effectors alter the microtubule organization remains obscure.
TcpB of Brucella spp
Brucella spp. are infectious intracellular pathogens causing brucellosis of animals and humans (84). Brucella spp. encode a Toll/interleukin-like receptor domain(TIRdomain)-containing protein termed TcpB/Bpt1. TcpB harbors a phosphoinositide-binding domain at the N-terminus and a TIR domain at the C-terminus (85). TcpB inhibits host innate immune responses meditated by TLR2 and TLR4 (85–87). Recent studies have shown that TcpB targets a TLR adaptor protein, TIRAP, to inhibit TLRs by inducing the ubiquitination and degradation of TIRAP (85, 88). Overexpression of TcpB in mammalian cells resulted in dramatic cell shrinkage and rounding up, suggesting a potential interaction with microtubules of the cell. Subcellular localization studies indicate that TcpB co-localizes predominantly with the microtubules (Figure 3) (85). Microtubule localization is attributed to the TIR domain of TcpB and a point mutation at the active site of TIR domain, i.e., BB-loop, abolished the affinity for microtubules. TcpB-expressing cells display thickened and bundled microtubule networks indicative of microtubule stabilization.
Figure 3.
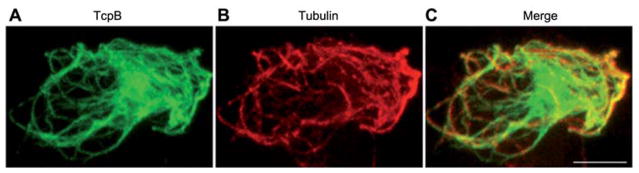
TcpB co-localizes with host microtubules. HEK-293 cells were transfected with pCMV-HA-TcpB plasmid and stained for HA-TcpB (A), tubulin (B), and merged (C). Scale bar, 5 μm.
Polymerization of microtubules in the presence of purified TcpB reveals a robust microtubule stabilization property of TcpB. TcpB acts like the microtubule-stabilizing drug paclitaxel, and dramatically enhances the nucleation and growth phases of microtubule polymerization (Figure 4) (89). In addition, TcpB can efficiently suppress the inhibition of microtubule depolymerization by nocodazole or cold. In agreement with the subcellular localization studies, a BB-loop mutant TcpB exhibits defective microtubule binding and stabilization properties (89).
Figure 4.
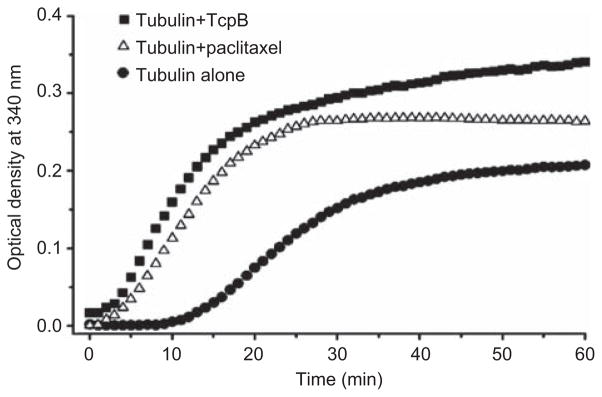
TcpB enhances the rate of microtubule polymerization like paclitaxel (Taxol). In vitro microtubule polymerization assay in the presence of purified TcpB (0.5 mg/ml) or paclitaxel (5 μM). TcpB could efficiently enhance the nucleation and growth phases of microtubule polymerization and the total amount of microtubules polymerized similar to the microtubule stabilization drug paclitaxel. Adapted from ref. (87).
The significance of TcpB-microtubule interaction and microtubule stabilization remains obscure. Potentially, TcpB induces microtubule bundling to interfere with the vesicular transport along microtubule tracks that may benefit Brucellae to prevent phagosome-lysosome fusion and subsequent phagosomal killing. However, experiments to demonstrate the secretion of TcpB by Brucella have not yet been successful. As the intact BB-loop is crucial for both microtubule stabilization as well as TLR inhibition, a correlation between these two properties has also been hypothesized. Nevertheless, the role of microtubules in the regulation of TLR signaling remains enigmatic. Studies are in progress to address the significance and mechanism of TcpB-induced microtubule stabilization.
CDT toxin of Clostridium difficile
Clostridium difficile is a major cause of chronic antibiotic-associated diarrhea and pseudomembranous colitis (90). Hypervirulent strains of C. difficile produce the binary actin-ADP ribosylating toxin, C. difficile transferase (CDT). CDT depolymerizes the actin cytoskeleton using its actin-modifying ADP-ribosyltransferase property. Expression of CDT in the human colon carcinoma cell line Caco-2 induced characteristic cell surface projections that consist of microtubules (91). Subsequent analyses indicate that in the presence of CDT, microtubules form bundles and grow along the cell cortex and project from the cell surface. The microtubules that cross the cell borders are capped with a microtubule plus-end-tracking protein, EB1, indicating that microtubule protrusions are the result of tubulin polymerization rather than sliding of microtubules. In addition, CDT causes redistribution of microtubule-capturing proteins, CLASP2 and ACF7, from the cell periphery to the cell interior (91). The capture of growing microtubules at the cell cortex by tip-associated proteins is an important process that regulates microtubule dynamics (92, 93). Interfering with this capturing process by CDT may contribute to the growth of microtubules beyond the cell borders. Induction of microtubule-derived cell protrusions increase adherence and colonization of Clostridia on epithelial cells (91). The iota toxin of Clostridium perfringes and C2 toxin of Clostridium botulinum also induce microtubule-based protrusions on the surface of epithelial cells (91).
Pneumolysin of Streptococcus pneumonia
Streptococcus pneumoniae is a major causative agent of bacterial meningitis (94). Streptococcus pneumoniae encodes pneumolysin (Ply), a member of the cholesterol-dependent cytolysins that is essential for the virulence of the bacteria (95, 96). Microtubule stabilization is observed in pneumococcal meningitis and is attributed to the pneumolysin of S. pneumoniae (97). The expression of sublytic levels of pneumolysin in SH-SY5Y neuron cells induces massive microtubule bundling and increased levels of acetylated tubulin and stabilized microtubules (97). Ply-induced microtubule bundling is partially affected by the addition of Src-kinase family inhibitors. As members of the tyrosine kinase family are known to promote tubulin polymerization and stabilize microtubules, Ply-mediated microtubule stabilization is thought to be mediated by tyrosine kinases (97–99). Ply-mediated microtubule stability and bundling inhibits organelle transport as demonstrated for defective mitochondrial transport in Chinese hamster ovary cells (97). Therefore, defective organelle transport caused by microtubule stabilization may explain the neuronal dysfunction in the course of pneumococcal meningitis. Streptococcus pneumoniae is not a classic intracellular pathogen; however, it is capable of invading and propagating in host cells (100, 101). Therefore, Ply-induced microtubule stabilization may help the intracellular survival of the bacterium by interfering with the vesicular transport and inhibiting phagosome killing.
Despite extensive research in the field of host-pathogen interaction, the mechanism of action of many of the above-discussed effector proteins remains to be elucidated. Therefore, future studies need to address various effector protein strategies that modulate the host microtubule dynamics and its contribution to the survival and persistence of bacterial pathogens in the host. Insights into the effector protein-micro-tubule interaction will undoubtedly provide an opportunity to develop innovative therapeutic strategies, such as antivirulence drugs that inhibit the specific functions of the effector protein. Future research should also focus on identifying novel virulence proteins of pathogenic microorganisms that target the organization of host microtubule networks.
Conclusion
Microtubules are easy targets of pathogenic microorganisms for hijacking the cellular processes to create a replication-permissive niche. Modulation of microtubule dynamics benefits the pathogen in various ways, including promoting intracellular motility, interfering with vesicular trafficking, and reorganizing the actin cytoskeleton. Therefore, detailed studies on microtubule-pathogen interaction will undoubtedly contribute to our understanding of pathogenicity and host adaptation of several infectious pathogens. Additionally, the virulence proteins that affect microtubule dynamics constitute a handy tool for dissecting various cellular processes that are regulated by microtubules. Similar studies will also provide valuable insight into the influence of microtubules on the dynamics of the actin cytoskeleton.
Acknowledgments
This work was supported by the National Institutes of Health (grant nos. R03AI101611 and R01 AIO73558) and the Binational Agricultural Research and Development (BARD) Fund (grant no. US-4378-11).
References
- 1.Roberts KL, Baines JD. Actin in herpesvirus infection. Viruses. 2011;3:336–46. doi: 10.3390/v3040336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida S, Sasakawa C. Exploiting host microtubule dynamics: a new aspect of bacterial invasion. Trends Microbiol. 2003;11:139–43. doi: 10.1016/s0966-842x(03)00023-4. [DOI] [PubMed] [Google Scholar]
- 3.Aktories K. Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol. 2011;9:487–98. doi: 10.1038/nrmicro2592. [DOI] [PubMed] [Google Scholar]
- 4.Goosney DL, Knoechel DG, Finlay BB. Enteropathogenic E. coli, Salmonella, and Shigella: masters of host cell cytoskeletal exploitation. Emerg Infect Dis. 1999;5:216–23. doi: 10.3201/eid0502.990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stebbins CE. Structural insights into bacterial modulation of the host cytoskeleton. Curr Opin Struct Biol. 2004;14:731–40. doi: 10.1016/j.sbi.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Stolp B, Fackler OT. How HIV takes advantage of the cytoskeleton in entry and replication. Viruses. 2011;3:293–311. doi: 10.3390/v3040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campellone KG. Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU and actin pedestal assembly. FEBS J. 2010;277:2390–402. doi: 10.1111/j.1742-4658.2010.07653.x. [DOI] [PubMed] [Google Scholar]
- 8.Aepfelbacher M, Zumbihl R, Heesemann J. Modulation of Rho GTPases and the actin cytoskeleton by YopT of Yersinia. Curr Top Microbiol Immunol. 2005;291:167–75. doi: 10.1007/3-540-27511-8_9. [DOI] [PubMed] [Google Scholar]
- 9.Boquet P. Small GTP binding proteins and bacterial virulence. Microbes Infect. 2000;2:837–43. doi: 10.1016/s1286-4579(00)90369-1. [DOI] [PubMed] [Google Scholar]
- 10.Finlay BB. Bacterial virulence strategies that utilize Rho GTPases. Curr Top Microbiol Immunol. 2005;291:1–10. doi: 10.1007/3-540-27511-8_1. [DOI] [PubMed] [Google Scholar]
- 11.Schlumberger MC, Hardt WD. Triggered phagocytosis by Salmonella: bacterial molecular mimicry of RhoGTPase activation/deactivation. Curr Top Microbiol Immunol. 2005;291:29–42. doi: 10.1007/3-540-27511-8_3. [DOI] [PubMed] [Google Scholar]
- 12.Aktories K, Barbieri JT. Bacterial cytotoxins: targeting eukaryotic switches. Nat Rev Microbiol. 2005;3:397–410. doi: 10.1038/nrmicro1150. [DOI] [PubMed] [Google Scholar]
- 13.Aktories K. Rho proteins: targets for bacterial toxins. Trends Microbiol. 1997;5:282–8. doi: 10.1016/S0966-842X(97)01067-6. [DOI] [PubMed] [Google Scholar]
- 14.Aktories K, Lang AE, Schwan C, Mannherz HG. Actin as target for modification by bacterial protein toxins. FEBS J. 2011;278:4526–43. doi: 10.1111/j.1742-4658.2011.08113.x. [DOI] [PubMed] [Google Scholar]
- 15.Barbieri JT, Riese MJ, Aktories K. Bacterial toxins that modify the actin cytoskeleton. Annu Rev Cell Dev Biol. 2002;18:315–44. doi: 10.1146/annurev.cellbio.18.012502.134748. [DOI] [PubMed] [Google Scholar]
- 16.Fiorentini C, Falzano L, Travaglione S, Fabbri A. Hijacking Rho GTPases by protein toxins and apoptosis: molecular strategies of pathogenic bacteria. Cell Death Differ. 2003;10:147–52. doi: 10.1038/sj.cdd.4401151. [DOI] [PubMed] [Google Scholar]
- 17.Gruenheid S, Finlay BB. Microbial pathogenesis and cytoskeletal function. Nature. 2003;422:775–81. doi: 10.1038/nature01603. [DOI] [PubMed] [Google Scholar]
- 18.Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- 19.Le Clainche C, Drubin DG. Actin lessons from pathogens. Mol Cell. 2004;13:453–4. doi: 10.1016/s1097-2765(04)00088-7. [DOI] [PubMed] [Google Scholar]
- 20.Downing KH, Nogales E. Tubulin structure: insights into microtubule properties and functions. Curr Opin Struct Biol. 1998;8:785–91. doi: 10.1016/s0959-440x(98)80099-7. [DOI] [PubMed] [Google Scholar]
- 21.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 22.Mandelkow E, Mandelkow EM. Microtubular structure and tubulin polymerization. Curr Opin Cell Biol. 1989;1:5–9. doi: 10.1016/s0955-0674(89)80029-8. [DOI] [PubMed] [Google Scholar]
- 23.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–26. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 24.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 25.Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422:753–8. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- 26.Uchiya K, Tobe T, Komatsu K, Suzuki T, Watarai M, Fukuda I, Yoshikawa M, Sasakawa C. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol Microbiol. 1995;17:241–50. doi: 10.1111/j.1365-2958.1995.mmi_17020241.x. [DOI] [PubMed] [Google Scholar]
- 27.Demers B, Sansonetti PJ, Parsot C. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 1998;17:2894–903. doi: 10.1093/emboj/17.10.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida S, Katayama E, Kuwae A, Mimuro H, Suzuki T, Sasakawa C. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 2002;21:2923–35. doi: 10.1093/emboj/cdf319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida S, Handa Y, Suzuki T, Ogawa M, Suzuki M, Tamai A, Abe A, Katayama E, Sasakawa C. Microtubule-severing activity of Shigella is pivotal for intercellular spreading. Science. 2006;314:985–9. doi: 10.1126/science.1133174. [DOI] [PubMed] [Google Scholar]
- 30.Davis J, Wang J, Tropea JE, Zhang D, Dauter Z, Waugh DS, Wlodawer A. Novel fold of VirA, a type III secretion system effector protein from Shigella flexneri. Protein Sci. 2008;17:2167–73. doi: 10.1110/ps.037978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Germane KL, Ohi R, Goldberg MB, Spiller BW. Structural and functional studies indicate that Shigella VirA is not a protease and does not directly destabilize microtubules. Biochemistry. 2008;47:10241–3. doi: 10.1021/bi801533k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 33.Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–21. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 34.Celli J, Deng W, Finlay BB. Enteropathogenic Escherichia coli (EPEC) attachment to epithelial cells: exploiting the host cell cytoskeleton from the outside. Cell Microbiol. 2000;2:1–9. doi: 10.1046/j.1462-5822.2000.00033.x. [DOI] [PubMed] [Google Scholar]
- 35.Garmendia J, Frankel G, Crepin VF. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun. 2005;73:2573–85. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect Immun. 2001;69:4027–33. doi: 10.1128/IAI.69.6.4027-4033.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw RK, Smollett K, Cleary J, Garmendia J, Straatman-Iwanowska A, Frankel G, Knutton S. Enteropathogenic Escherichia coli type III effectors EspG and EspG2 disrupt the microtubule network of intestinal epithelial cells. Infect Immun. 2005;73:4385–90. doi: 10.1128/IAI.73.7.4385-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuzawa T, Kuwae A, Yoshida S, Sasakawa C, Abe A. Enteropathogenic Escherichia coli activates the RhoA signaling pathway via the stimulation of GEF-H1. EMBO J. 2004;23:3570–82. doi: 10.1038/sj.emboj.7600359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- 40.Howell B, Larsson N, Gullberg M, Cassimeris L. Dissociation of the tubulin-sequestering and microtubule catastrophe-promoting activities of oncoprotein 18/stathmin. Mol Biol Cell. 1999;10:105–18. doi: 10.1091/mbc.10.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benais-Pont G, Punn A, Flores-Maldonado C, Eckert J, Raposo G, Fleming TP, Cereijido M, Balda MS, Matter K. Identification of a tight junction-associated guanine nucleotide exchange factor that activates Rho and regulates paracellular permeability. J Cell Biol. 2003;160:729–40. doi: 10.1083/jcb.200211047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luperchio SA, Schauer DB. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 2001;3:333–40. doi: 10.1016/s1286-4579(01)01387-9. [DOI] [PubMed] [Google Scholar]
- 43.Deng W, Li Y, Vallance BA, Finlay BB. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect Immun. 2001;69:6323–35. doi: 10.1128/IAI.69.10.6323-6335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mundy R, Petrovska L, Smollett K, Simpson N, Wilson RK, Yu J, Tu X, Rosenshine I, Clare S, Dougan G, Frankel G. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun. 2004;72:2288–302. doi: 10.1128/IAI.72.4.2288-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardwidge PR, Deng W, Vallance BA, Rodriguez-Escudero I, Cid VJ, Molina M, Finlay BB. Modulation of host cytoskeleton function by the enteropathogenic Escherichia coli and Citrobacter rodentium effector protein EspG. Infect Immun. 2005;73:2586–94. doi: 10.1128/IAI.73.5.2586-2594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guttman JA, Samji FN, Li Y, Deng W, Lin A, Finlay BB. Aquaporins contribute to diarrhoea caused by attaching and effacing bacterial pathogens. Cell Microbiol. 2007;9:131–41. doi: 10.1111/j.1462-5822.2006.00773.x. [DOI] [PubMed] [Google Scholar]
- 47.Schachter J. Overview of human diseases. In: Barron AL, editor. Microbiology of Chlamydia. Boca Raton, FL: CRC Press; 1988. pp. 153–65. [Google Scholar]
- 48.Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–90. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grieshaber SS, Grieshaber NA, Hackstadt T. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J Cell Sci. 2003;116(Pt 18):3793–802. doi: 10.1242/jcs.00695. [DOI] [PubMed] [Google Scholar]
- 50.Scidmore MA. Recent advances in Chlamydia subversion of host cytoskeletal and membrane trafficking pathways. Microbes Infect. 2011;13:527–35. doi: 10.1016/j.micinf.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alzhanov DT, Weeks SK, Burnett JR, Rockey DD. Cytokinesis is blocked in mammalian cells transfected with Chlamydia trachomatis gene CT223. BMC Microbiol. 2009;9:2. doi: 10.1186/1471-2180-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Lesser CF, Lory S. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature. 2008;456:112–5. doi: 10.1038/nature07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Archuleta TL, Du Y, English CA, Lory S, Lesser C, Ohi MD, Ohi R, Spiller BW. The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J Biol Chem. 2011;286:33992–8. doi: 10.1074/jbc.M111.258426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. Edwardsiella tarda – virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012;14:26–34. doi: 10.1016/j.micinf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 55.Xie HX, Yu HB, Zheng J, Nie P, Foster LJ, Mok YK, Finlay BB, Leung KY. EseG, an effector of the type III secretion system of Edwardsiella tarda, triggers microtubule destabilization. Infect Immun. 2010;78:5011–21. doi: 10.1128/IAI.00152-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhle V, Jackel D, Hensel M. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic. 2004;5:356–70. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 57.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–43. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Dussurget O. New insights into determinants of Listeria monocytogenes virulence. Int Rev Cell Mol Biol. 2008;270:1–38. doi: 10.1016/S1937-6448(08)01401-9. [DOI] [PubMed] [Google Scholar]
- 59.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–40. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 60.Stavru F, Archambaud C, Cossart P. Cell biology and immunology of Listeria monocytogenes infections: novel insights. Immunol Rev. 2011;240:160–84. doi: 10.1111/j.1600-065X.2010.00993.x. [DOI] [PubMed] [Google Scholar]
- 61.Kuhn M, Goebel W. Molecular studies on the virulence of Listeria monocytogenes. Genet Eng (NY) 1995;17:31–51. [PubMed] [Google Scholar]
- 62.Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–75. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- 63.Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci USA. 2002;99:161–6. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–47. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 65.Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5:463–9. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 66.Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-γ S in nerve terminals. Nature. 1995;374:186–90. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 67.Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA. Dynamin 2 binds γ-tubulin and participates in centrosome cohesion. Nat Cell Biol. 2004;6:335–42. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- 68.Tanabe K, Takei K. Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot-Marie-Tooth mutant. J Cell Biol. 2009;185:939–48. doi: 10.1083/jcb.200803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henmi Y, Tanabe K, Takei K. Disruption of microtubule network rescues aberrant actin comets in dynamin2-depleted cells. PLoS One. 2011;6:e28603. doi: 10.1371/journal.pone.0028603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way? J Cell Sci. 2001;114(Pt 21):3795–803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- 71.Liu BP, Chrzanowska-Wodnicka M, Burridge K. Microtubule depolymerization induces stress fibers, focal adhesions, and DNA synthesis via the GTP-binding protein Rho. Cell Adhes Commun. 1998;5:249–55. doi: 10.3109/15419069809040295. [DOI] [PubMed] [Google Scholar]
- 72.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–85. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pfeuffer T, Goebel W, Laubinger J, Bachmann M, Kuhn M. LaXp180, a mammalian ActA-binding protein, identified with the yeast two-hybrid system, co-localizes with intracellular Listeria monocytogenes. Cell Microbiol. 2000;2:101–14. doi: 10.1046/j.1462-5822.2000.00034.x. [DOI] [PubMed] [Google Scholar]
- 74.Maucuer A, Camonis JH, Sobel A. Stathmin interaction with a putative kinase and coiled-coil-forming protein domains. Proc Natl Acad Sci USA. 1995;92:3100–4. doi: 10.1073/pnas.92.8.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–31. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 76.Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol. 2004;6:420–6. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi K. WAVE2 protein complex coupled to membrane and microtubules. J Oncol. 2012;2012:590531. doi: 10.1155/2012/590531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001;52:259–74. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 79.Brumell JH, Goosney DL, Finlay BB. SifA, a type III secreted effector of Salmonella typhimurium, directs Salmonella-induced filament (Sif) formation along microtubules. Traffic. 2002;3:407–15. doi: 10.1034/j.1600-0854.2002.30604.x. [DOI] [PubMed] [Google Scholar]
- 80.Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–49. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stein MA, Leung KY, Zwick M, Garcia-del Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20:151–64. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 82.Schroeder N, Mota LJ, Meresse S. Salmonella-induced tubular networks. Trends Microbiol. 2011;19:268–77. doi: 10.1016/j.tim.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 83.Ohlson MB, Huang Z, Alto NM, Blanc MP, Dixon JE, Chai J, Miller SI. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4:434–46. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med. 2005;352:2325–36. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 85.Radhakrishnan GK, Yu Q, Harms JS, Splitter GA. Brucella TIR domain-containing protein mimics properties of the toll-like receptor adaptor protein TIRAP. J Biol Chem. 2009;284:9892–8. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, Svanborg C, Miethke T. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 87.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sengupta D, Koblansky A, Gaines J, Brown T, West AP, Zhang D, Nishikawa T, Park SG, Roop RM, 2nd, Ghosh S. Subversion of innate immune responses by Brucella through the targeted degradation of the TLR signaling adapter, MAL. J Immunol. 2010;184:956–64. doi: 10.4049/jimmunol.0902008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Radhakrishnan GK, Harms JS, Splitter GA. Modulation of microtubule dynamics by a TIR domain protein from the intra-cellular pathogen Brucella melitensis. Biochem J. 2011;439:79–83. doi: 10.1042/BJ20110577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kelly CP, LaMont JT. Clostridium difficile – more difficult than ever. N Engl J Med. 2008;359:1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 91.Schwan C, Stecher B, Tzivelekidis T, van Ham M, Rohde M, Hardt WD, Wehland J, Aktories K. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009;5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–53. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–35. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 94.Durand ML, Calderwood SB, Weber DJ, Miller SI, Southwick FS, Caviness VS, Jr, Swartz MN. Acute bacterial meningitis in adults. A review of 493 episodes. N Engl J Med. 1993;328:21–8. doi: 10.1056/NEJM199301073280104. [DOI] [PubMed] [Google Scholar]
- 95.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190:1661–9. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 96.Hirst RA, Gosai B, Rutman A, Guerin CJ, Nicotera P, Andrew PW, O’Callaghan C. Streptococcus pneumoniae deficient in pneumolysin or autolysin has reduced virulence in meningitis. J Infect Dis. 2008;197:744–51. doi: 10.1086/527322. [DOI] [PubMed] [Google Scholar]
- 97.Iliev AI, Djannatian JR, Opazo F, Gerber J, Nau R, Mitchell TJ, Wouters FS. Rapid microtubule bundling and stabilization by the Streptococcus pneumoniae neurotoxin pneumolysin in a cholesterol-dependent, non-lytic and Src-kinase dependent manner inhibits intracellular trafficking. Mol Microbiol. 2009;71:461–77. doi: 10.1111/j.1365-2958.2008.06538.x. [DOI] [PubMed] [Google Scholar]
- 98.Laurent CE, Delfino FJ, Cheng HY, Smithgall TE. The human c-Fes tyrosine kinase binds tubulin and microtubules through separate domains and promotes microtubule assembly. Mol Cell Biol. 2004;24:9351–8. doi: 10.1128/MCB.24.21.9351-9358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Teckchandani AM, Birukova AA, Tar K, Verin AD, Tsygankov AY. The multidomain protooncogenic protein c-Cbl binds to tubulin and stabilizes microtubules. Exp Cell Res. 2005;306:114–27. doi: 10.1016/j.yexcr.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 100.Elm C, Braathen R, Bergmann S, Frank R, Vaerman JP, Kaetzel CS, Chhatwal GS, Johansen FE, Hammerschmidt S. Ectodomains 3 and 4 of human polymeric Immunoglobulin receptor (hpIgR) mediate invasion of Streptococcus pneumoniae into the epithelium. J Biol Chem. 2004;279:6296–304. doi: 10.1074/jbc.M310528200. [DOI] [PubMed] [Google Scholar]
- 101.Gordon SB, Molyneux ME, Boeree MJ, Kanyanda S, Chaponda M, Squire SB, Read RC. Opsonic phagocytosis of Streptococcus pneumoniae by alveolar macrophages is not impaired in human immunodeficiency virus-infected Malawian adults. J Infect Dis. 2001;184:1345–9. doi: 10.1086/324080. [DOI] [PubMed] [Google Scholar]