Abstract
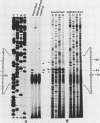
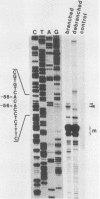
The process of trans splicing is essential to the maturation of all mRNAs in the Trypanosomatidae, a family of protozoan parasites, and to specific mRNAs in several species of nematode. In Trypanosoma brucei, a 39-nucleotide (nt) leader sequence originating from a small, 139-nt donor RNA (the spliced leader [SL] RNA) is spliced to the 5' end of mRNAs. An intermediate in this trans-splicing process is a Y structure which contains the 3' 100 nt of the SL RNA covalently linked to the pre-mRNA via a 2'-5' phosphodiester bond at the branch point residue. We mapped the branch points in T. brucei alpha- and beta-tubulin pre-mRNAs. The primary branch acceptors for the alpha- and beta-tubulins are 44 and 56 nt upstream of the 3' splice sites, respectively, and are A residues. Minor branch acceptors were detected 42 and 49 nt upstream of the alpha-tubulin splice site and 58 nt upstream of the splice site in beta-tubulin. The regions surrounding these branch points lack homology to the consensus sequences determined for mammalian cells and yeasts; there is also no conservation among the sequences themselves. Thus, the identified sequences suggest that the mechanism of branch point recognition in T. brucei differs from the mechanism of recognition by U2 RNA that has been proposed for other eucaryotes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Borst P. Discontinuous transcription and antigenic variation in trypanosomes. Annu Rev Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C. E. Structure and regulated expression of genes encoding fructose biphosphate aldolase in Trypanosoma brucei. EMBO J. 1985 Nov;4(11):2997–3003. doi: 10.1002/j.1460-2075.1985.tb04035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988 May;8(5):2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Kimmel B. E., Samson S., Wu J., Hirschberg R., Yarbrough L. R. Tubulin genes of the African trypanosome Trypanosoma brucei rhodesiense:nucleotide sequence of a 3.7-kb fragment containing genes for alpha and beta tubulins. Gene. 1985;35(3):237–248. doi: 10.1016/0378-1119(85)90002-2. [DOI] [PubMed] [Google Scholar]
- Krämer A. Analysis of RNase-A-resistant regions of adenovirus 2 major late precursor-mRNA in splicing extracts reveals an ordered interaction of nuclear components with the substrate RNA. J Mol Biol. 1987 Aug 5;196(3):559–573. doi: 10.1016/0022-2836(87)90032-5. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Layden R. E., Eisen H. Alternate trans splicing in Trypanosoma equiperdum: implications for splice site selection. Mol Cell Biol. 1988 Mar;8(3):1352–1360. doi: 10.1128/mcb.8.3.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels P. A., Poliszczak A., Osinga K. A., Misset O., Van Beeumen J., Wierenga R. K., Borst P., Opperdoes F. R. Two tandemly linked identical genes code for the glycosomal glyceraldehyde-phosphate dehydrogenase in Trypanosoma brucei. EMBO J. 1986 May;5(5):1049–1056. doi: 10.1002/j.1460-2075.1986.tb04321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram J., Perry K. L., Lizardi P. M., Lührmann R., Agabian N., Nelson R. G. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: identification of the U2, U4, and U6 RNA analogs. Mol Cell Biol. 1989 Mar;9(3):1212–1223. doi: 10.1128/mcb.9.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhich M. L., Boothroyd J. C. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol Cell Biol. 1988 Sep;8(9):3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W. J., Watkins K. P., Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell. 1986 Nov 21;47(4):517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Aebi M., Hornig H., Weissmann C., Sharp P. A. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Perry K. L., Watkins K. P., Agabian N. Trypanosome mRNAs have unusual "cap 4" structures acquired by addition of a spliced leader. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sather S., Agabian N. A 5' spliced leader is added in trans to both alpha- and beta-tubulin transcripts in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5695–5699. doi: 10.1073/pnas.82.17.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E. O., Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988 Oct 7;55(1):41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Sutton R. E., Boothroyd J. C. Evidence for trans splicing in trypanosomes. Cell. 1986 Nov 21;47(4):527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. E., Boothroyd J. C. Trypanosome trans-splicing utilizes 2'-5' branches and a corresponding debranching activity. EMBO J. 1988 May;7(5):1431–1437. doi: 10.1002/j.1460-2075.1988.tb02960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow L. S., Milhausen M., Rutter W. J., Agabian N. Tubulin genes are tandemly linked and clustered in the genome of trypanosoma brucei. Cell. 1983 Jan;32(1):35–43. doi: 10.1016/0092-8674(83)90494-4. [DOI] [PubMed] [Google Scholar]
- Tschudi C., Richards F. F., Ullu E. The U2 RNA analogue of Trypanosoma brucei gambiense: implications for a splicing mechanism in trypanosomes. Nucleic Acids Res. 1986 Nov 25;14(22):8893–8903. doi: 10.1093/nar/14.22.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C., Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988 Feb;7(2):455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C., Young A. S., Ruben L., Patton C. L., Richards F. F. Calmodulin genes in trypanosomes are tandemly repeated and produce multiple mRNAs with a common 5' leader sequence. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3998–4002. doi: 10.1073/pnas.82.12.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]