Abstract
Background
IgA nephropathy (IgAN) is a complex syndrome characterized by deposition of IgA and IgA containing immune complexes (ICs) composed of IgG and complement C3 proteins in the mesangial area of glomeruli. The low-affinity receptors for the Fc region of IgG (FcγRs) are involved in autoantibody/immune complex-induced organ injury as well as ICs clearance. The aim of the study was to associate multiple polymorphisms within FCGR gene locus with IgAN in a large Chinese cohort.
Patients and Methods
60 single nucleotide polymorphisms (SNPs) spanning a 400 kb range within FCGR gene locus were analyzed in 2100 DNA samples from patients with biopsy proven IgAN and healthy age- and sex-matched controls from the same population in Chinese.
Results
Among the 60 SNPs investigated, 15 gene polymorphisms within FCGR gene locus (25%) were associated with susceptibility to IgAN. The most significantly associated SNPs within individual genes were FCGR2B rs12118043 (p = 8.74*10−3, OR 0.76, 95% CI 0.62–0.93), and FCRLB rs4657093 (p = 2.28*10−3, OR 0.77, 95% CI 0.65–0.91). Both conditional analysis and linkage disequilibrium analysis suggested they were independent signals associated with IgAN. Associations between FCGR2B rs12118043 and proteinuria (p = 3.65×10−2) as well as gross hematuria (p = 4.53×10−2), between FCRLB rs4657093 and levels of serum creatinine (p = 2.67×10−2) as well as eGFR (p = 5.41*10−3) were also observed. Electronic cis-expression quantative trait loci analysis supported their possible functional significance, with protective genotypes correlating lower gene expressions.
Conclusion
Our data from genetic associations and expression associations revealed potentially pathogenic roles of Fc receptor gene polymorphisms in IgAN.
Introduction
IgA nephropathy (IgAN) was described histologically for the first time in 1968 by Berger and Hinglais as les dépôts intercapillaires d'IgA-IgG (intercapillary deposits of IgA-IgG)[1]. It was characterized by the deposition of IgA in the mesangial area of glomeruli, and proliferation of the glomerular mesangium with deposition of immune complexes composed of IgG and complement C3 proteins. Current data indicates that at least four hits contribute to development of IgA nephropathy: aberrant glycosylation of IgA1, synthesis of antibodies directed against galactose-deficient IgA1, binding of the galactose-deficient IgA1 by the anti-glycan/glycopeptides antibodies to form immune complexes (ICs), and accumulation of these complexes in the glomerular mesangium to initiate renal injury[2]. But the presence of circulating IgA1-containing ICs is not unique to patients with IgAN. IgA1-IgG ICs can also be detected in persons without apparent renal disease[3], [4], [5]. Anyhow, the pathogenic importance of ICs has been widely recognized, with plenty of evidence such as, serum levels of IgG antibodies specific for galactose deficient IgA1 correlated with disease severity[3]; the pathogenic circulating IgA1-IgG ICs in patients with IgAN are relatively large (>800 kD)[6]; ICs from patients with IgAN containing galactose-deficient IgA1 bind to the cells more efficiently than do uncomplexed IgA1 or ICs from healthy controls[7]; complexes with galactose- deficient IgA1 induce cultured human mesangial cells to proliferate, secrete extracellular matrix components, release cytokines and further interfere mesangial cell-podocyte crosstalk[8]. However, factors influencing the formation/composition of these ICs and the intrinsic mechanisms leading to cell activation and glomerular damage were still not clearly elucidated.
Molecular mechanisms of autoantibody/immune complex-induced organ injury as well as ICs clearance often involve two main components, namely the low-affinity receptors for the Fc region of IgG (FcγRs) and the complement system[9], [10]. Several lines of functional evidences were emerging to the important role of complement in IgAN, especially alternative pathway[11], [12], [13], [14]. Recent genome association studies (GWAS) also indicated that genetic variants of CFH/CFHR were associated with IgAN, further highlighting the inherited and pathogenic role of complement pathway in IgAN[15]. Fcγ Rs contribute to the regulation of a multitude of immune and inflammatory responses. Polymorphisms in the genes encoding FcγRs (FCGR) have been associated with susceptibility to a number of autoimmune or inflammatory diseases. However, few studies have been conducted to address the functional role of FcγRs as well as FCGR gene polymorphisms in IgAN. Up to date, only two genetic studies involving less than 200 patients with IgAN had been reported to associate reported FCGR gene variants with IgAN, but with contradictory conclusions [16], [17]. So in the current study, we aimed to associate multiple polymorphisms within FCGR gene locus with IgAN in a large Chinese cohort comprised by more than 2000 samples to further determine its genetic role in IgAN.
Materials and Methods
Patients and Controls
The study population consisted of 1,200 IgAN cases and 900 healthy controls of Chinese Han ancestry from north of China. All of the patients with IgAN were confirmed by renal biopsy. Patients with secondary IgAN or with other comorbid renal diseases were excluded. Clinical and laboratory data at the time of diagnosis were collected for each patient. Written informed consent was obtained from each patient. This study complied with the Declaration of Helsinki and was approved by the medical ethics committee of Peking University First Hospital.
DNA extraction
Genomic DNA was isolated from whole peripheral blood using a modified salt extraction technique. DNA concentration and quality [optical density (OD) 260/OD 280 and OD 260/OD 230 measurements] were determined by a Nanodrop ND1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Genotyping
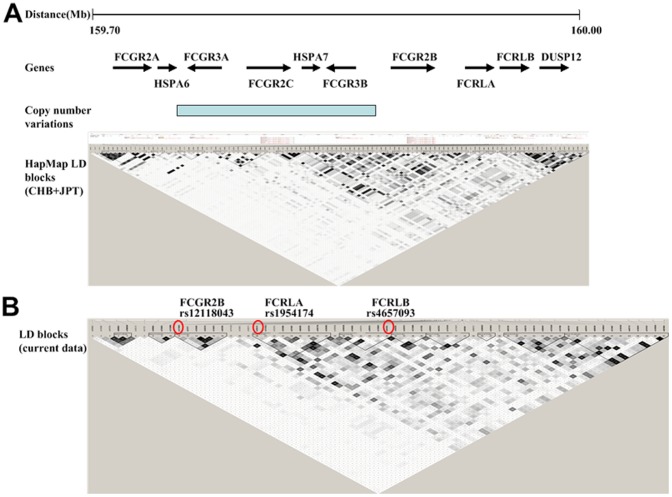
Genes encoding low-affinity FcγRs including FCGR2A, FCGR3A, FCGR2C, FCGR3B and FCGR2B locate in 1q23 spanning a 200-Kb region ( Figure 1A ). As genes within the locus were of low but complex linkage disequilibrium (LD), an extended region with 100 kb upstream and 100 kb downstream of the FCGR gene locus was analyzed. SNPs were genotyped on a customized Illumina Human 610-Quad BeadChip platform as previously reported[18]. The genotyping call rate was 99.9%. A total of 60 SNPs were analyzed in the current association study.
Figure 1. Lack of extensive haplotype blocks with FCGR gene loci in 1q23.
( Figure 1A ) The linkage disequilibrium (LD) blocks were derived from the HapMap3 Asian CHB and JPT dataset (http://www.hapmap.org). CHB: 80 Han Chinese from Beijing, China; JPT: 82 Japanese in Tokyo, Japan. The locus was complicated by multiple kinds of genetic variations including single-nucleotide polymorphisms (SNPs) as well as copy number variations with low linkage disequilibrium. ( Figure 1B ) The linkage disequilibrium (LD) blocks of SNPs in the current study. The SNPs associated with IgAN were marked by circle.
Cis-eQTL (cis-expression quantative trait loci) analysis in HapMap samples
Normalized mRNA data from Epstein–Barr virus (EBV)-transformed lymphoblastoid cell lines of HapMap3 CHB (80 Han Chinese from Beijing, China) and JPT (82 Japanese in Tokyo, Japan) population were obtained from the database of the Gene Expression Variation (GENEVAR) project at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/humgen/genevar/)[19].
Statistical analysis
The SNPs meeting the quality control criteria of less than 1% overall missing data, consistency with Hardy-Weinberg Equilibrium genotype frequency expectations (P>0.05) were included. Association analysis was performed using the trend test in PLINK controlling population stratification. Odd ratio (OR) values were presented for the minor allele of a SNP. Linkage disequilibrium (LD) was tested using Haploview (version 4.2, http://www.broad.mit.edu/mpg/haploview) as well as the EM algorithm within PLINK. The association between SNPs and CNV was determined by chi-square test and Person's correlation coefficients were calculated for LD estimation[20], [21]. Power was calculated by Power and Sample Size Calculations Software (version 3.0, http://biostat.mc.vanderbilt.edu/PowerSampleSize). Spearman's coefficient were calculated to correlate genotypes and gene expressions in cis-eQTL analysis as reported[19].
Results
Fc receptor gene polymorphisms associated with susceptibility to IgAN
As can be seen from Table 1 , among the 60 SNPs investigated, 15 SNPs within FCGR gene locus (25%) were associated with susceptibility to IgAN, suggesting likely true associations. Among SNPs within the classical FCGR genes, rs12118043A showed statistically significant disease association (p = 8.74*10−3, OR 0.76, 95% CI 0.62–0.93). A SNP of rs4657093C (p = 2.28*10−3, OR 0.77, 95% CI 0.65–0.91) within FCRLB showed the most significant association signal in the extended region (Table S1). Power calculations indicated that we had at least 99% power to detect loci with allelic frequencies >0.10 and relative risk >1.5 assuming an α-level of 0.05 (P<0.05) in the current study.
Table 1. Associations between FCGR gene polymorphisms and susceptibility to IgAN.
| SNP | Bp | Gene | Function class | Minor Allele | MAF case/control (%) | Trend test p values | Allele OR (95% CI) |
| rs4657039 | 159729352 | intergenic | G | 24.37/27.77 | 1.28*10−2 | 0.84 (0.73–0.96) | |
| rs12118043 | 159913448 | FCGR2B | intron | A | 8.42/10.81 | 8.74*10−3 | 0.76 (0.62–0.93) |
| rs1954173 | 159944408 | FCRLA | intron | G | 16.00/18.42 | 3.86*10−2 | 0.84 (0.72–0.99) |
| rs1954174 | 159944431 | FCRLA | intron | T | 42.20/38.69 | 2.19*10−2 | 1.16 (1.02–1.31) |
| rs2333749 | 159951245 | FCRLA | nearGene-3′ | T | 22.36/25.22 | 3.08*10−2 | 0.85 (0.74–0.97) |
| rs10917750 | 159952215 | intergenic | C | 15.62/18.85 | 5.89*10−3 | 0.80 (0.68–0.94) | |
| rs10494356 | 159953885 | intergenic | G | 42.67/39.30 | 2.82*10−2 | 1.15 (1.02–1.30) | |
| rs7549830 | 159955055 | intergenic | C | 50.50/46.90 | 2.07*10−2 | 1.16 (1.02–1.31) | |
| rs1891019 | 159958057 | FCRLB | nearGene-5′ | T | 47.61/51.72 | 8.46*10−3 | 0.85 (0.75–0.96) |
| rs4657093 | 159959627 | FCRLB | intron | C | 13.91/17.35 | 2.28*10−3 | 0.77 (0.65–0.91) |
| rs1891020 | 159961078 | FCRLB | intron | A | 15.95/18.81 | 1.52*10−2 | 0.82 (0.70–0.96) |
| rs12079477 | 159961481 | FCRLB | intron | G | 48.11/51.72 | 2.09*10−2 | 0.87 (0.77–0.98) |
| rs1417582 | 159962712 | FCRLB | intron | T | 16.01/18.92 | 1.38*10−2 | 0.82 (0.70–0.96) |
| rs1503813 | 159967303 | intergenic | G | 18.30/21.01 | 2.83*10−2 | 0.84 (0.72–0.98) | |
| rs905594 | 160007898 | ATF6 | intron | C | 37.43/34.48 | 4.93*10−2 | 1.14 (1.00–1.29) |
Linkage disequilibrium and conditional analysis suggested independent associations
The LD statistics based on haplotype frequencies estimated via the EM algorithm within PLINK showed the most significantly associated SNPs rs12118043, rs1954174, and rs4657093 within FCGR2B, FCRLA, FCRLB were in low LD ( Figure 1B ). rs12118043 has an r2 = 0.016 with rs1954174, rs12118043 has an r2 = 0.001with rs4657093, and rs1954174 has an r2 = 0.123 with rs4657093. rs12118043 (p = 5.00*10−3) remained significantly associated with IgAN after conditional logistic regression incorporating rs4657093, suggesting they were independently associated with IgAN. However, the association with rs1954174 became non-significant in conditional analysis (p = 0.17).
Association analysis between FCGR3B copy numbers and FCGR2B rs12118043 genotypes in HapMap samples indicated weak linkage disequilibrium
Fc receptor genes likely arose by segmental duplications during evolution and it was reported that copy number variations in 1q23 involved FCGR3A, FCGR2C and FCGR3B [20], [22], [23], [24], [25]. To determine whether the effect of FCGR2B rs12118043A in risk of IgAN originated from its independent contribution or was in LD with CNVs at this locus, we derived data of FCGR3B copy numbers and FCGR2B rs12118043 genotypes from HapMap samples. As it was widely accepted that it was difficult to genotype FCGR3B copy numbers accurately by a single method, the FCGR3B copy numbers we applied were from an integrated suite of five assays[21], which provided the bases of reliability and precision for further analysis. As can be seen from Table 2 , FCGR2B rs12118043 risk genotypes associated with lower FCGR3B copy numbers in Caucasians (p = 0.03) but not in people from Asia or Africa. When individual genotypes were coded as 1, 2, and 3, represented homozygote AA, heterozygote AC, and homozygote CC, respectively. The square of Pearson's correlation coefficient (r2) calculated for copy number of FCGR3B and FCGR2B rs12118043 genotypes were 0.01 (p = 0.35) in HapMap Asians and 0.07 (p = 0.02) in Caucasians respectively. The data indicated that FCGR2B rs12118043A was in weak LD with FCGR3B low copy numbers, especially in peoples of Asian and African ancestry, which was also consistent to previous reports including several other different SNPs[20].
Table 2. Associations between FCGR3B copy numbers and FCGR2B rs12118043 genotypes in HapMap samples.
| FCGR2B rs12118043 Genotype | ||||||
| FCGR3B CN | CHB+JPT (n = 87) | CEU (n = 80) | YRI (n = 78) | |||
| AA+AC | CC | AA+ AC | CC | AA+ AC | CC | |
| 0 | –– | –– | 1(3.1) | –– | –– | –– |
| 1 | 1(8.3) | 8(10.7) | 3(9.4) | 1(2.0) | –– | 11(14.1) |
| 2 | 7(58.3) | 44(58.7) | 26(81.2) | 34(69.4) | –– | 59(75.6) |
| 3 | 3(25.0) | 23(30.7) | 2(6.2) | 12(24.5) | –– | 6(7.7) |
| 4 | 1(8.3) | –– | –– | 2(4.1) | –– | 2(2.6) |
| P values | p = 0.14 | p = 0.03 | ||||
As samples of rs12118043 genotype AA were few, dominant model with AA+AC versus CC was applied. Likelihood ratio test was applied as more than 2 cells had expected count less than 5 in every Chi-square test.
Fc receptor gene polymorphisms associated with severity of IgAN
The above data indicated IgAN associated SNPs FCGR2B rs12118043 and FCRLB rs4657093 may impact IgAN susceptibility independently. We further checked their correlations with severity of IgAN accordingly. The parameters included blood pressure, serum creatinine, urine protein, uric acid, estimated glomerular filtration rate (eGFR) calculated based on MDRD formula modified for Chinese population[26], and Hass pathology grade classification[27]. It was indicated that FCGR2B rs12118043 associated with proteinuria, with protective genotypes correlating lower levels of proteinuria (AA+AC vs. CC 1.54±1.13 g/day vs. 2.00±1.99g/day, p = 3.65×10−2) and lower frequency of gross hematuria (26.6% vs. 34.0%, p = 4.53×10−2). FCRLB rs4657093 protective genotypes associated with smaller age of onset (31.31±10.91 vs. 32.96±11.28, p = 2.58×10−2), and better renal function including higher levels of eGFR (CC+CT vs. TT 91.62±26.74 ml/min*1.73 m2 vs. 80.07±27.41 ml/min*1.73 m2, p = 5.41*10−3) and lower levels of serum creatinine (92.56±36.51 µmol/L vs. 107.67±67.79 µmol/L, p = 2.67×10−2) ( Table 3 ).
Table 3. Associations between Fc receptor gene polymorphisms and severity of IgAN.
| Clinical Parameters | FCGR2B rs12118043 | FCRLB rs4657093 | ||||
| AA+AC | CC | p | CC+CT | TT | p | |
| Age (year) | 33.27±11.29 | 32.41±11.20 | 0.33 | 31.31±10.91 | 32.96±11.28 | 2.58×10−2 |
| Gender (Male %) | 53.9 | 54.8 | 0.82 | 46.6 | 45.0 | 0.63 |
| SBP (mmHg) | 125±17 | 124±18 | 0.61 | 123±18 | 124±17 | 0.14 |
| DBP (mmHg) | 78±12 | 79±13 | 0.84 | 78±13 | 79±13 | 0.25 |
| eGFR(ml/min/1.73 m2) | 84.35±22.96 | 82.05±28.44 | 0.62 | 91.62±26.74 | 80.07±27.41 | 5.41*10−3 |
| Scr (umol/L) | 93.94±106.62 | 106.62±67.31 | 0.23 | 92.56±36.51 | 107.67±67.79 | 2.67×10−2 |
| 24 hour UTP (g/day) | 1.54±1.13 | 2.00±1.99 | 3.65×10−2 | 1.73±1.38 | 1.98±1.99 | 0.38 |
| Gross Hematuria (%) | 26.6 | 34.0 | 4.53×10−2 | 36.9 | 31.4 | 0.08 |
| Uric Acid (umol/L) | 351.55±104.34 | 347.84±112.17 | 0.68 | 343.53±106.79 | 350.08±112.38 | 0.39 |
| LDL-c (mmol/L) | 2.87±1.22 | 3.14±5.21 | 0.48 | 3.39±7.86 | 3.00±3.12 | 0.24 |
| Serum IgA (g/L) | 2.86±1.01 | 2.83±1.08 | 0.89 | 2.81±0.95 | 2.85±1.10 | 0.82 |
| HASS Grade (%) | 0.77 | 0.55 | ||||
| I | 39.7 | 38.0 | 37.3 | 38.7 | ||
| II | 9.6 | 6.1 | 9.7 | 5.4 | ||
| III | 37.0 | 38.5 | 37.3 | 38.7 | ||
| IV | 5.5 | 8.4 | 8.2 | 7.7 | ||
| V | 8.2 | 8.9 | 7.5 | 9.4 | ||
SBP: systolic blood pressure; DBP: diastolic blood pressure; eGFR: estimated glomerular filtration rate (eGFR) calculated based on MDRD formula modified for Chinese population; Scr: serum creatinine; 24 hour UTP: 24 hours total urine protein; LDL-c: low density lipoprotein cholesterol
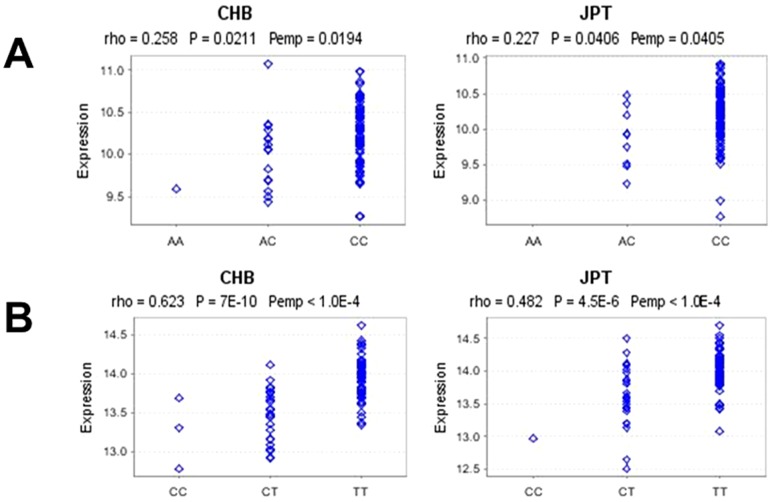
Cis-eQTL (cis-expression quantative trait loci) analysis supported possible functional significance of Fc receptor gene polymorphisms
A common hypothesis held that common SNPs impact disease by altering abundance of gene transcripts. We thus checked correlations between genotypes and gene expressions in cis (cis eQTL or local eQTL maps to the location whose expression levels are associated with genetic variation located physically close to the gene)[19]. By analyzing correlations between genotypes and nearby gene expression within a 1 Mb distance, we observed significant associations between genotypes of FCGR2B rs12118043 and FCGR2B expression ( Figure 2A ), and between genotypes of FCRLB rs4657093 and FCRLA expression ( Figure 2B ) in HapMap3 Asians (p<0.05 both in CHB and JPT populations), in which the protective alleles always associated with lower gene expressions. The data further supported FCGR2B rs12118043 and FCRLB rs4657093 were likely true disease associated SNPs.
Figure 2. Cis-eQTL (cis-expression quantative trait loci) analysis of Fc receptor gene polymorphisms.
( Figure 2A ) associations between genotypes of FCGR2B rs12118043 and FCGR2B expression. ( Figure 2B ) associations between genotypes of FCRLB rs4657093 and FCRLA expression. eQTLs have been studied in lymphoblastoid cell lines (LCLs) from the HapMap3 Asian populations. The distance from the genomic location of the transcription start site (TSS) to SNP genomic location was less than 1 Mb. Spearman's rank correlation coefficients (rho) and p values were presented. The data was derived from Genevar project (http://www.sanger.ac.uk/resources/software/genevar). CHB: 80 Han Chinese from Beijing, China; JPT: 82 Japanese in Tokyo, Japan.
Discussion
FcγRs are now recognized as the dominant molecules responsible for coupling the recognition of antigens by IgG antibodies to the cellular effector pathways of macrophages, neutrophils, natural killer cells and mast cells[28]. They are a critically involved in the maintenance of peripheral tolerance, regulating dendritic cell maturation and plasma cell survival[29]. Gene polymorphisms of FCGR gene family have been associated with multiple immune-related diseases including human autoimmune, infectious or malignant diseases. Novel therapeutic strategies targeting FcγRs especially on FCGR gene polymorphisms are emerging [28], [30], [31], [32]. Fc receptor-like (FcRL) proteins are a family of cellular receptors homologous to FcγRs that are preferentially expressed on B lineage cells. The extracellular ligands as well as functions of these receptors are still unknown or controversial [33], [34], [35], [36]. However, several genetic studies also revealed FCRL gene variants were associated with multiple immune-related diseases, no matter in genome-wide association studies or in candidate gene based studies [37], [38], [39], [40], [41]. All the above highlighted the central bridging roles of FcγRs and FcRLs in immunity with their genetic variants to make up a sort of susceptibility background.
IgAN is a complex syndrome characterized by deposition of IgA and IgA containing immune complexes composed of IgG and complement C3 proteins in the mesangial area of glomeruli. As FcγRs and FcRLs were expressed in B cells and mesangial cells and they were the effector molecules in mediating antibody/immune complex effects, the roles of FcγRs and FcRLs in IgAN will be of particular interest. In the present study, we investigated 60 SNPs in 1q23 spanning an extended region with 100 kb upstream and 100 kb downstream of the FCGR gene locus. We observed that 15 gene polymorphisms within FCGR gene locus (25%) were associated with susceptibility to IgAN. The most significantly associated SNPs were rs12118043, rs1954174, and rs4657093 within FCGR2B, FCRLA, and FCRLB respectively. Both conditional analysis and linkage disequilibrium analysis suggested they were independent signals associated with IgAN. Interestingly, LD between FCGR2B rs12118043 and FCGR3B copy numbers was higher in Caucasians than that in Asians and Africans, supporting complex genetic architectures in the gene locus. Integration of different genetic variants in different populations will reveal more information about disease pathogenesis. Future studies involving copy number variations in the gene locus will be deserved, especially in Asians and Africans. In addition, we observed associations between FCGR2B rs12118043 and proteinuria as well as gross hematuria, between FCRLB rs4657093 and levels of serum creatinine as well as eGFR, which indicated that the variants not only impacted disease susceptibility but also disease severity. At last, cis-eQTL analysis supported their possible functional significance, with protective genotypes correlating lower gene expressions. As that was observed in infectious diseases [31], [42], FCGR2B protective genotypes correlated lower gene expression and lower frequency of gross hematuria, latter of which was significantly associated mucosal infection and was an important indicator of episodes of IgAN. Receptor deficiency may down-regulate responses mediated by the receptor. For example, as FcγR2b was the only one inhibitory receptor to suppress downstream events such as cellular proliferation, phagocytosis, and inflammatory cytokine release, FCGR2B rs12118043 protective genotypes may have better humoral immunity, i.e. produce less autoantibodies and mediate balanced inflammation[31]. The data above from two different sorts of associations – genetic association and expression association, suggested the associations were likely true. Anyhow, the association in IgAN was relatively weak and more detailed pathogenesis was still not clear. Further assays in gene-target animal models or cell lines will be welcomed, which will explain more about the specific role of FcγRs in IgAN.
In conclusion, we investigated multiple SNPs spanning a 400 kb range within FCGR gene locus in 2100 Chinese, and observed significant associations between FCGR2B, FCRLA, and FCRLB gene variants and IgAN. Both conditional analysis and linkage disequilibrium analysis suggested they were independent signals associated with IgAN. Genetic associations and expression associations suggested the Fc receptor gene polymorphisms play potentially pathogenic roles in IgAN. Future studies involving more genetic variants in diverse populations and investigations of specific pathogenesis will be deserved.
Supporting Information
Genotype counts of investigated SNPs in IgAN patients and controls.
(DOC)
Acknowledgments
We thank our collaborators of Ali G Gharavi from Department of Medicine, Columbia University College of Physicians and Surgeons, New York, USA for genotyping sorting and assistance. The authors thank all the members of the laboratory for technical assistance and the patients and their families for their cooperation and for giving consent to participate in this study.
Funding Statement
This work was supported by grants from the Major State Basic Research Development Program of China (973 program, No. 2012CB517700), National Natural Science Foundation of China (No. 81200524), Research Fund of Beijing Municipal Science and Technology for the Outstanding PhD Program (20121000110), The Foundation of Ministry of Education of China (20120001120008), and Natural Science Fund of China to the Innovation Research Group (81021004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berger J, Hinglais N (1968) [Intercapillary deposits of IgA-IgG]. J Urol Nephrol (Paris) 74: 694–695. [PubMed] [Google Scholar]
- 2. Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, et al. (2011) The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22: 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Fan R, Zhang Z, Brown R, Hall S, et al. (2009) Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tissandie E, Morelle W, Berthelot L, Vrtovsnik F, Daugas E, et al. (2011) Both IgA nephropathy and alcoholic cirrhosis feature abnormally glycosylated IgA1 and soluble CD89-IgA and IgG-IgA complexes: common mechanisms for distinct diseases. Kidney Int 80: 1352–1363. [DOI] [PubMed] [Google Scholar]
- 5. Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, et al. (1999) Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest 104: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Novak J, Tomana M, Matousovic K, Brown R, Hall S, et al. (2005) IgA1-containing immune complexes in IgA nephropathy differentially affect proliferation of mesangial cells. Kidney Int 67: 504–513. [DOI] [PubMed] [Google Scholar]
- 7. Novak J, Vu HL, Novak L, Julian BA, Mestecky J, et al. (2002) Interactions of human mesangial cells with IgA and IgA-containing immune complexes. Kidney Int 62: 465–475. [DOI] [PubMed] [Google Scholar]
- 8. Novak J, Raskova KL, Suzuki H, Tomana M, Matousovic K, et al. (2011) IgA1 immune complexes from pediatric patients with IgA nephropathy activate cultured human mesangial cells. Nephrol Dial Transplant 26: 3451–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mayadas TN, Tsokos GC, Tsuboi N (2009) Mechanisms of immune complex-mediated neutrophil recruitment and tissue injury. Circulation 120: 2012–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jancar S, Sanchez CM (2005) Immune complex-mediated tissue injury: a multistep paradigm. Trends Immunol 26: 48–55. [DOI] [PubMed] [Google Scholar]
- 11. Hashimoto A, Suzuki Y, Suzuki H, Ohsawa I, Brown R, et al. (2012) Determination of severity of murine IgA nephropathy by glomerular complement activation by aberrantly glycosylated IgA and immune complexes. Am J Pathol 181: 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oortwijn BD, Eijgenraam JW, Rastaldi MP, Roos A, Daha MR, et al. (2008) The role of secretory IgA and complement in IgA nephropathy. Semin Nephrol 28: 58–65. [DOI] [PubMed] [Google Scholar]
- 13. Roos A, Rastaldi MP, Calvaresi N, Oortwijn BD, Schlagwein N, et al. (2006) Glomerular activation of the lectin pathway of complement in IgA nephropathy is associated with more severe renal disease. J Am Soc Nephrol 17: 1724–1734. [DOI] [PubMed] [Google Scholar]
- 14. Kim SJ, Koo HM, Lim BJ, Oh HJ, Yoo DE, et al. (2012) Decreased circulating C3 levels and mesangial C3 deposition predict renal outcome in patients with IgA nephropathy. PLoS One 7: e40495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiryluk K, Novak J, Gharavi AG (2012) Pathogenesis of Immunoglobulin A Nephropathy: Recent Insight from Genetic Studies. Annu Rev Med. [DOI] [PMC free article] [PubMed]
- 16. Xu G, He Q, Shou Z, Wang H, Zhang X, et al. (2007) NA1/NA2 heterozygote of Fcgr3b is a risk factor for progression of IgA nephropathy in Chinese. J Clin Lab Anal 21: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka Y, Suzuki Y, Tsuge T, Kanamaru Y, Horikoshi S, et al. (2005) FcgammaRIIa-131R allele and FcgammaRIIIa-176V/V genotype are risk factors for progression of IgA nephropathy. Nephrol Dial Transplant 20: 2439–2445. [DOI] [PubMed] [Google Scholar]
- 18. Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, et al. (2011) Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, et al. (2012) Patterns of cis regulatory variation in diverse human populations. PLoS Genet 8: e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niederer HA, Willcocks LC, Rayner TF, Yang W, Lau YL, et al. (2010) Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet 19: 3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hollox EJ, Detering JC, Dehnugara T (2009) An integrated approach for measuring copy number variation at the FCGR3 (CD16) locus. Hum Mutat 30: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, et al. (2009) Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat 30: E640–E650. [DOI] [PubMed] [Google Scholar]
- 23. Zhou XJ, Lv JC, Bu DF, Yu L, Yang YR, et al. (2010) Copy number variation of FCGR3A rather than FCGR3B and FCGR2B is associated with susceptibility to anti-GBM disease. Int Immunol 22: 45–51. [DOI] [PubMed] [Google Scholar]
- 24. Zhou XJ, Lv JC, Qin LX, Yang HZ, Yu F, et al. (2011) Is FCGR2A a susceptibility gene to systemic lupus erythematosus in Chinese? Lupus 20: 1198–1202. [DOI] [PubMed] [Google Scholar]
- 25. McKinney C, Merriman TR (2012) Meta-analysis confirms a role for deletion in FCGR3B in autoimmune phenotypes. Hum Mol Genet 21: 2370–2376. [DOI] [PubMed] [Google Scholar]
- 26. Ma YC, Zuo L, Chen JH, Luo Q, Yu XQ, et al. (2006) Modified glomerular filtration rate estimating equation for Chinese patients with chronic kidney disease. J Am Soc Nephrol 17: 2937–2944. [DOI] [PubMed] [Google Scholar]
- 27. Lv J, Zhang H, Zhou Y, Li G, Zou W, et al. (2008) Natural history of immunoglobulin A nephropathy and predictive factors of prognosis: a long-term follow up of 204 cases in China. Nephrology (Carlton) 13: 242–246. [DOI] [PubMed] [Google Scholar]
- 28. Nimmerjahn F, Ravetch JV (2008) Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 8: 34–47. [DOI] [PubMed] [Google Scholar]
- 29. Espeli M, Clatworthy MR, Bokers S, Lawlor KE, Cutler AJ, et al. (2012) Analysis of a wild mouse promoter variant reveals a novel role for FcgammaRIIb in the control of the germinal center and autoimmunity. J Exp Med 209: 2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nimmerjahn F, Ravetch JV (2011) FcgammaRs in health and disease. Curr Top Microbiol Immunol 350: 105–125. [DOI] [PubMed] [Google Scholar]
- 31. Willcocks LC, Carr EJ, Niederer HA, Rayner TF, Williams TN, et al. (2010) A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc Natl Acad Sci U S A 107: 7881–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith KG, Clatworthy MR (2010) FcgammaRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol 10: 328–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ehrhardt GR, Cooper MD (2011) Immunoregulatory roles for fc receptor-like molecules. Curr Top Microbiol Immunol 350: 89–104. [DOI] [PubMed] [Google Scholar]
- 34. Davis RS (2007) Fc receptor-like molecules. Annu Rev Immunol 25: 525–560. [DOI] [PubMed] [Google Scholar]
- 35. Santiago T, Kulemzin SV, Reshetnikova ES, Chikaev NA, Volkova OY, et al. (2011) FCRLA is a resident endoplasmic reticulum protein that associates with intracellular Igs, IgM, IgG and IgA. Int Immunol 23: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Masuda K, Mori H, Ohara O, Nakayama M, Wang JY, et al. (2010) Defining the immunological phenotype of Fc receptor-like B (FCRLB) deficient mice: Confounding role of the inhibitory FcgammaRIIb. Cell Immunol 266: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thabet MM, Wesoly J, Slagboom PE, Toes RE, Huizinga TW (2007) FCRL3 promoter 169 CC homozygosity is associated with susceptibility to rheumatoid arthritis in Dutch Caucasians. Ann Rheum Dis 66: 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeng Z, Duan Z, Zhang T, Wang S, Li G, et al. (2012) Association of FCRL4 polymorphisms on disease susceptibility and severity of ankylosing spondylitis in Chinese Han population. Clin Rheumatol 31: 1449–1454. [DOI] [PubMed] [Google Scholar]
- 39. Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, et al. (2011) Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 7: e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu X, Pan CM, Zhao SX, Liang J, Gao GQ, et al. (2011) A genome-wide association study identifies two new risk loci for Graves' disease. Nat Genet 43: 897–901. [DOI] [PubMed] [Google Scholar]
- 41. Bajpai UD, Swainson LA, Mold JE, Graf JD, Imboden JB, et al. (2012) A functional variant in FCRL3 is associated with higher Fc receptor-like 3 expression on T cell subsets and rheumatoid arthritis disease activity. Arthritis Rheum 64: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu XP, Wu JQ, Zhu LP, Wang X, Xu B, et al. (2012) Association of Fcgamma receptor IIB polymorphism with cryptococcal meningitis in HIV-uninfected Chinese patients. PLoS One 7: e42439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype counts of investigated SNPs in IgAN patients and controls.
(DOC)