Significance
Mitochondrial fission is critical for mammalian cell division, mitophagy, and development. Fission initiates via recruitment of dynamin-related GTPases to the mitochondrial surface. In yeast and human, the recruitment utilizes adaptors that differ in sequence and predicted structure. Key unresolved issues are whether these adaptors function independently in membrane recruitment and whether a single adaptor and GTPase are sufficient to catalyze scission. We show that three human adaptors work interchangeably with a single mitochondrial dynamin to accomplish fission. We also show that an adaptor alters the architecture of the dynamin polymer in a manner that could facilitate membrane constriction and severing.
Keywords: Drp1/Dnm1, Fis1/Mdv1, Mff/MiD49/MiD51, mitochondrial fission dynamin
Abstract
Mitochondrial fission is mediated by the dynamin-related GTPases Dnm1/Drp1 (yeast/mammals), which form spirals around constricted sites on mitochondria. Additional membrane-associated adaptor proteins (Fis1, Mdv1, Mff, and MiDs) are required to recruit these GTPases from the cytoplasm to the mitochondrial surface. Whether these adaptors participate in both GTPase recruitment and membrane scission is not known. Here we use a yeast strain lacking all fission proteins to identify the minimal combinations of GTPases and adaptors sufficient for mitochondrial fission. Although Fis1 is dispensable for fission, membrane-anchored Mdv1, Mff, or MiDs paired individually with their respective GTPases are sufficient to divide mitochondria. In addition to their role in Drp1 membrane recruitment, MiDs coassemble with Drp1 in vitro. The resulting heteropolymer adopts a dramatically different structure with a narrower diameter than Drp1 homopolymers assembled in isolation. This result demonstrates that an adaptor protein alters the architecture of a mitochondrial dynamin GTPase polymer in a manner that could facilitate membrane constriction and severing activity.
Dynamin-related proteins (DRPs) are self-assembling GTPases that regulate lipid-remodeling events at different cellular membranes (1). Two of these DRPs, Dnm1 (yeast) and Drp1 (human), play conserved roles in mitochondrial fission, which is important for biological processes including mitochondrial inheritance during cell division (2, 3), clearance of defective mitochondria via mitophagy (4–7), and mammalian development (8, 9).
In vivo, both the Dnm1 and Drp1 GTPases assemble from the cytoplasm into structures that encircle mitochondria at sites of future fission (10–13). In vitro, addition of GTP to Dnm1-lipid tubules is sufficient to constrict synthetic liposomes (14, 15). However, a recent study revealed that mitochondrial constriction in yeast and mammals occurs at sites where endoplasmic reticulum (ER) tubules circumscribe mitochondria (16). This ER-mediated mitochondrial constriction occurs before Dnm1 or Drp1 recruitment, suggesting that DRPs act after the initial constriction event to complete membrane fission. Neither Dnm1 nor Drp1 has been shown to catalyze membrane scission independently in vivo or in vitro.
A variety of adaptor proteins localized to the outer mitochondrial membrane (OMM) play important but poorly understood roles in Dnm1/Drp1 recruitment and function. The membrane recruitment step is understood best in yeast, where Dnm1 binds to the fungal-specific adaptor mitochondrial division protein 1 (Mdv1) (17, 18), which in turn binds to the tail-anchored fission protein 1 (Fis1) protein (19). Fluorescence microscopy studies show that Mdv1 colocalizes with Dnm1 at sites of mitochondrial fission (20). In vitro, Mdv1 interacts with the GTP-bound form of Dnm1 and stimulates Dnm1 self-assembly (21).
Fis1 is conserved in humans (hFis1) but does not appear to recruit Drp1 to mitochondria. Instead, Drp1 recruitment is mediated by mitochondrial fission factor (Mff), another tail-anchored protein (22, 23). Two additional human proteins, the orthologs mitochondrial dynamics proteins 49 and 51 (MiD49 and MiD51), are N-terminally anchored in the OMM and also play a role in Drp1 recruitment (24, 25). Neither Mff nor the MiD proteins is related by sequence or predicted secondary structure to Mdv1. The Mff and MiD49/51 proteins form rings surrounding mitochondria, suggesting that they coassemble with Drp1 (24), but their specific roles in Drp1 assembly and membrane scission are not well understood. Thus, major questions—whether adaptor proteins participate in lipid remodeling and membrane scission and whether they act independently or in concert in vivo—remain unanswered.
Here we use a yeast strain devoid of fission proteins to identify the minimal combination of DRPs and adaptors sufficient for mitochondrial fission. We provide evidence that Fis1 is dispensable for mitochondrial membrane scission. We also demonstrate that Mdv1, Mff, and MiDs paired individually with their respective DRPs are interchangeable, in that each is sufficient to catalyze fission. Importantly, coassembly of an MiD protein with Drp1 dramatically decreases the diameter of the Drp1 structures formed. This result provides a direct demonstration that an adaptor protein can alter the architecture of a DRP assembly in a manner that could facilitate their membrane constriction and severing ability.
Results
Requirements of Individual Yeast Proteins for Mitochondrial Fission.
In WT yeast, opposing fission and fusion events maintain branched mitochondrial tubules positioned at the cell cortex (Fig. S1A, WT). When fission is disrupted, fusion continues unopposed, and cells contain mitochondrial nets or a single, interconnected mitochondrion, which often collapses to one side of the cell (Fig. S1A, mutant) (10, 12). To determine the minimal protein requirements for fission, we generated a yeast “tester” strain lacking Dnm1, the Mdv1 adaptor, and Fis1. This strain also lacked a paralog of the Mdv1 adaptor, carbon catabolite repression associated factor 4 (Caf4), which was shown previously to be dispensable for fission in vivo (26). This tester strain was viable but exhibited severe mitochondrial fission defects (Table S1 and Fig. S1A). Expression of WT Dnm1 or of Dnm1 tethered to the outer mitochondrial membrane by its N or C terminus was unable to rescue mitochondrial fission defects in the tester strain (Table S1). Previous studies showed that pairwise combinations of cytoplasmic Dnm1 expressed with either WT Fis1 or Mdv1 also failed to rescue mitochondrial morphology (18, 19).
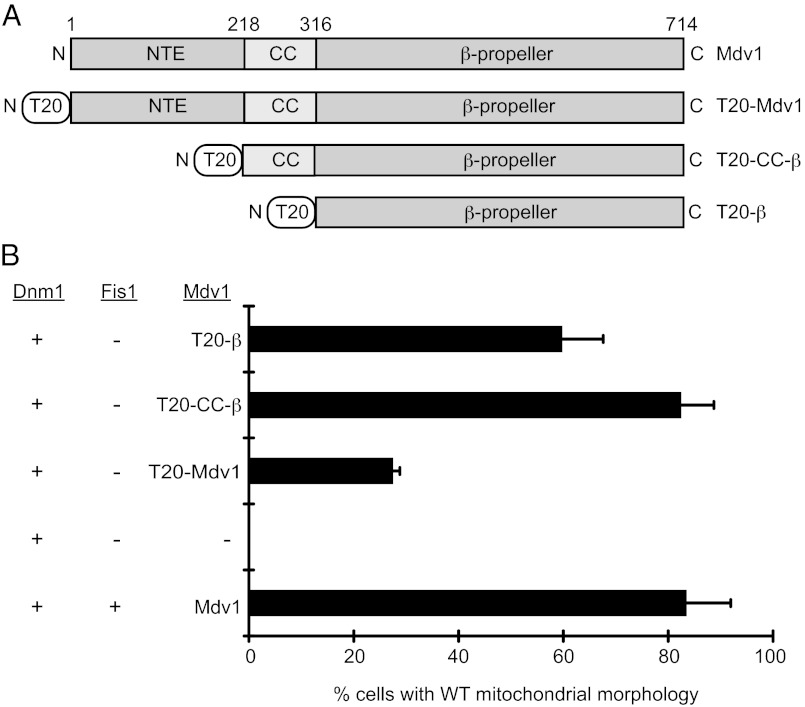
To determine whether Fis1 was necessary for post–Dnm1-recruitment steps in fission, we expressed combinations of WT Dnm1 together with WT or mitochondrial membrane-tethered Mdv1 in the tester strain. Mdv1 contains three domains, an N-terminal extension (NTE) that binds Fis1 (27), a middle domain that dimerizes Mdv1 via an antiparallel coiled-coil (CC) (28), and a predicted β-propeller domain (β) that interacts with Dnm1 (27, 29) (Fig. 1A). Full-length and truncated forms of Mdv1 were tethered to the outer mitochondrial membrane by the translocase of outer membrane (Tom20) anchor (labeled T20, Fig. 1A). Immunoblotting of whole-cell extracts confirmed that all proteins were expressed stably in vivo (Fig. S1 B and C). Mitochondrial morphology then was quantified to assess the ability of different protein combinations to restore WT mitochondrial fission and morphology. Normal mitochondrial morphology was restored in 80% of the cells by expressing cytoplasmic Dnm1 together with WT Mdv1 and Fis1 (Fig. 1B). Surprisingly, although WT mitochondrial morphology was restored in strains expressing Dnm1 plus all three tethered forms of Mdv1, the full-length construct was not the most efficient. The lack of an Fis1-binding partner for the NTE domain in the full-length Mdv1 construct may affect the conformation of the protein and be responsible for this effect. Consistent with this idea, the most efficient rescue occurred upon expression of the tethered Mdv1 CC plus β-propeller domain (lacking the NTE domain) (Fig. 1B). The mitochondrial morphology rescue observed in these studies suggests that soluble Dnm1 and tethered forms of Mdv1 are sufficient to catalyze fission in the absence of Fis1.
Fig. 1.
Fis1 is dispensable for mitochondrial fission. (A) Domain structure of WT Mdv1 and Mdv1 constructs fused to the N-terminal transmembrane anchor of yeast T20. NTE, CC, and predicted β domains are shown. (B) Quantification of mitochondrial morphology in cells expressing the indicated fission proteins. All values are mean ± SEM; n ≥ 300. Representative images of WT and fission mutant mitochondria scored are shown in Fig. S1. JSY strains 8614, 9234, 9801, 9802, and 9803 were used.
Fis1 Is Not Essential for Dnm1 Assembly into Fission Complexes or Membrane Scission.
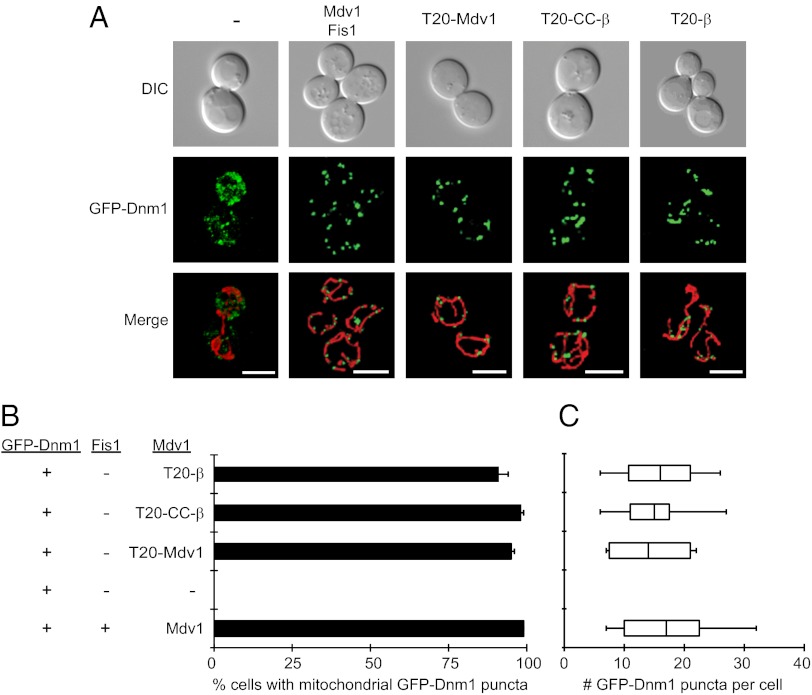
When cells lack Mdv1 and Fis1, GFP-Dnm1 cannot be recruited to mitochondria and instead remains in the cytoplasm (Fig. 2 A and B). When WT Mdv1 and Fis1 are present, GFP-Dnm1 assembles into punctate fission complexes distributed evenly along mitochondrial tubules (Fig. 2 A and B). Consistent with their ability to rescue fission defects, all three forms of tethered Mdv1 were able to recruit GFP-Dnm1 to mitochondria in the absence of Fis1 (Fig. 2 A and B). The percentage of cells in the population containing GFP-Dnm1 complexes (Fig. 2B) and the number of complexes per cell (Fig. 2C) were similar to that observed in WT although fission complexes formed by two of the tethered Mdv1 proteins (T20-Mdv1 and T20-β) were less functional than Mdv1 T20-CC-β (Fig. 1B).
Fig. 2.
Dnm1 fission complexes assemble on mitochondria in the absence of Fis1. (A) Representative images of GFP-Dnm1 puncta on mitochondria. Differential interference contrast (DIC), GFP-Dnm1, and merged mito-RFP (mitochondrial matrix-targeted dsRed) images are shown. (Scale bar, 5 µm.) (B) Quantification of the number of cells in a population containing punctate GFP-Dnm1 fission complexes on mitochondria. All values are mean ± SEM; n ≥ 300. (C) Box-and-whisker plots showing the number of mitochondrial GFP-Dnm1 puncta per cell in the indicated strains. n ≥ 10 cells. The y axes for B and C are the same. JSY strains 9493, 9548, 9804, 9805, and 9806 were used.
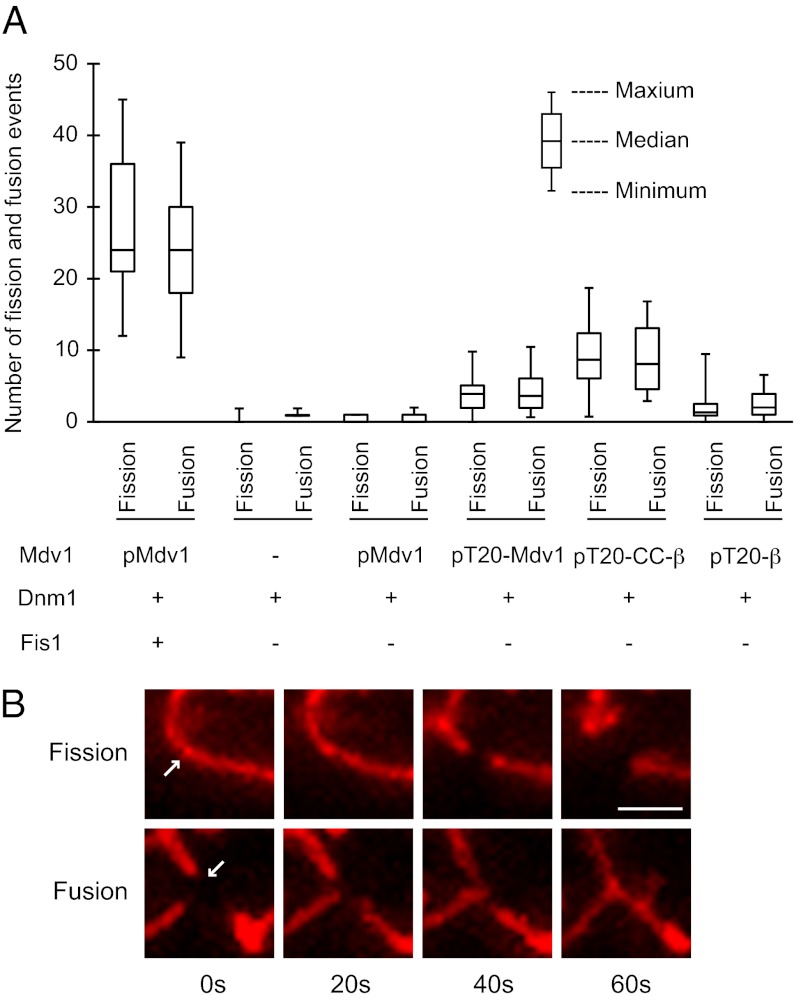
In vivo, yeast mitochondrial fission and fusion are coordinated, each process occurring approximately once every 2 min (30). Although the molecular basis of this coordination is unknown, the balance is critical for robust mitochondrial function (31). Quantification of fission and fusion events in time-lapse imaging studies confirmed that these processes were balanced in our WT yeast strain (Fig. 3A). We next determined whether the balance of fission and fusion was altered when fission occurred without Fis1 in the tester strain. When cells expressed only cytoplasmic Dnm1, unopposed fusion formed interconnected mitochondria with few or no free tips. As a consequence, once the system achieved steady state, neither fission nor fusion was observed (Fig. 3A). Similar results were obtained for the strain expressing WT Dnm1 and Mdv1 in the absence of Fis1. In contrast, when cells expressed cytoplasmic Dnm1 and tethered forms of Mdv1, fission and fusion events again were balanced, although the number of fission and fusion events was reduced (Fig. 3A), most likely because the tethered proteins were less functional than WT (Fig. 1B). Representative images of fission and fusion events in these cells are shown in Fig. 3B. Together, our results demonstrate that after Mdv1 and Dnm1 are recruited to the mitochondrial surface, Fis1 is not essential for the assembly of functional fission complexes and the subsequent membrane scission event. Moreover, a balance between fission and fusion is achieved in these strains.
Fig. 3.
Mitochondrial fission and fusion events in cells lacking Fis1. (A) Box-and-whisker plot showing the distribution of fission (n ≥ 20 cells) and fusion (n ≥ 20 cells) events in WT and strains expressing the indicated fission proteins. Total fission or fusion events per cell for a 15-min interval are indicated. (B) Representative mito-RFP–labeled mitochondria are shown undergoing fission (Upper, arrow) and fusion (Lower, arrow) in cells expressing Dnm1 and tethered Mdv1 (T20-β). (Scale bar, 1 µm.)
Mff or MiDs Are Sufficient to Recruit Human Drp1 to Mitochondria and Catalyze Fission.
The hFis1, Mff, and MiD49/51 adaptors are all expressed in mammalian cells. As a consequence, it has been difficult to determine definitively whether these adaptors work individually or in concert to influence fission complex assembly or mitochondrial division after Drp1 membrane recruitment. To address this issue, we individually expressed OMM-tethered forms of each adaptor protein with soluble Drp1 (variant 3, NCBI reference sequence number NP_005681.2) in the yeast tester strain. To maintain their appropriate membrane topologies, the cytoplasmic domains of hFis1 and Mff were targeted using the yeast C-terminal Fis1 anchor (yTM), and the MiD proteins were targeted using the yeast N-terminal T20 anchor (Fig. 4A). These mammalian proteins were expressed stably in yeast (Fig. S1 D–H).
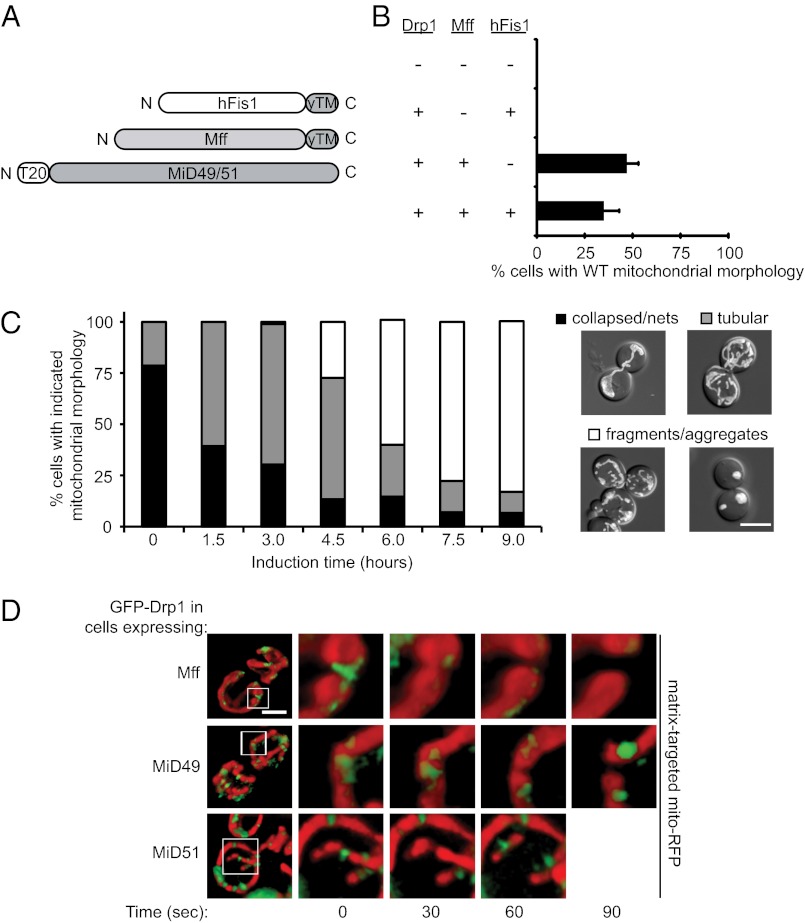
Fig. 4.
Mammalian Drp1 and mitochondrial-tethered adaptor proteins rescue mitochondrial fission defects in yeast. (A) Domain structures of hFis1 and Mff fused to the C-terminal outer membrane anchor of yeast Fis1 (yTM). Human MiD49 or MiD51 were targeted to the mitochondrial outer membrane via fusion to the N-terminal transmembrane anchor of yeast T20. (B) Quantification of mitochondrial morphology in cells expressing Mff plus the indicated fission proteins and mito-OMGFP. All values are mean ± SEM; n ≥ 300. (C) Quantification of mitochondrial morphologies observed in cells during induction of Drp1 and MiD51 from the MET25 promoter. The merged DIC and mito-OMGFP images to the right of the graph are representative of the mitochondrial morphology categories scored. (Scale bar, 5 µm.) (D) Time-lapse images of GFP-Drp1 at fission sites in cells expressing Mff (Top), MiD49 (Middle), or MiD51 (Bottom). Boxed regions mark fission sites imaged at 30-s intervals in each row. (Scale bar, 2 µm.)
In the absence of Drp1, expression of any of the three tethered forms shown in Fig. 4A did not rescue fission defects (Table S1). In addition, mitochondrial fission was not rescued by expression of Drp1 alone (Table S1) or by Drp1 with tethered hFis1 (Fig. 4B). The latter result is consistent with a previous report that hFis1 is not essential for Drp1 recruitment in mammalian cells (23). In contrast, expression of soluble Drp1 with tethered Mff was sufficient for partial rescue of mitochondrial fission and WT mitochondrial morphology in vivo (Fig. 4B). Expressing tethered hFis1 in addition to Mff had little effect on this rescue. Thus, hFis1 does not appear to impact fission mediated by Drp1 and tethered Mff in this system.
We also examined mitochondrial fission rescue in a tester strain expressing Drp1 and MiD51 from the repressible methionine requiring (MET25) promoter. Before induction (Fig. 4C, 0 h), ∼79% of the cells in the population contained collapsed mitochondria and interconnected nets characteristic of a fission defect (Fig. 4C, black). The remaining 21% contained WT tubular mitochondria (Fig. 4C, gray) because the MET25 promoter is repressed inefficiently under these conditions and produces low levels of both Drp1 and MiD51 proteins. WT mitochondrial morphology increased to 61 and 69% after 1.5 or 3.0 h of MiD51 induction, respectively, suggesting that cytoplasmic Drp1 and tethered MiD51 are sufficient to catalyze fission in the yeast tester strain. Upon further induction (4.5–9 h), the percentage of cells containing tubular mitochondrial morphology was reduced, and the percentage of the population containing fragmented and aggregated mitochondrial membranes increased steadily from 27 to 83% (Fig. 4C, white). This aggregated mitochondrial phenotype also was observed when MiD49 and MiD51 were overexpressed in mammalian cells (24). Similar results were obtained when the experiment was performed with MiD49 in place of MiD51 (Fig. S1I).
Time-lapse imaging studies confirmed that the changes in mitochondrial morphology observed upon expression of Drp1 with Mff, MiD49, or MiD51 were caused by mitochondrial fission. In our tester strain expressing functional GFP-Drp1 and Mff, MiD49 or MiD51, green Drp1 puncta were observed on RFP-labeled mitochondrial tubules at sites where fission occurred (Fig. 4D). In control studies, hFis1 was not able to recruit GFP-Drp1 to mitochondria (Fig. S1J), as is consistent with our observation that these two proteins do not support fission (Fig. 4B). Together, these results establish that Drp1 is able to function with multiple adaptors to catalyze mitochondrial membrane fission.
Effect of Mff and MiD Adaptors on GTP Hydrolysis by Drp1.
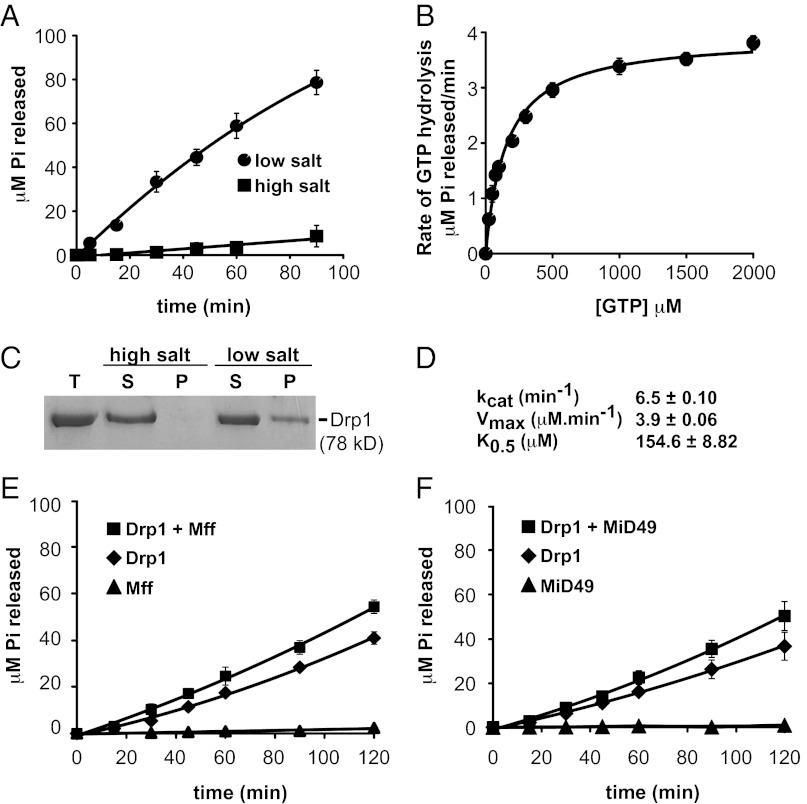
To determine whether mammalian adaptor proteins altered the kinetic properties of Drp1, we purified untagged versions of all three proteins (full-length Drp1 and the Mff and MiD49 cytoplasmic domains). Analytical ultracentrifugation studies are consistent with Drp1 and Mff forming homodimers, whereas MiD49 behaves as a monomer in solution (Fig. S2). As is characteristic of self-assembling GTPases in the dynamin family, Drp1 pelleted in low, but not high, ionic strength buffer in a standard sedimentation assay (Fig. 5C). GTP-hydrolysis by Drp1 also increased up to 15-fold in low ionic strength buffer, indicating that self-assembly stimulated GTP hydrolysis (Fig. 5A). Under assembly-stimulated conditions (low ionic strength, Fig. 5 B and D), the catalytic activity of Drp1 (kcat = 6.5/min) was similar to that reported for the yeast mitochondrial dynamin Dnm1 (21). However, the Drp1 catalytic activity shown here is 7.6 times greater than that reported previously for a calmodulin-binding peptide (CBP)-Drp1 fusion protein (32). It is possible that the N-terminal CBP tag on Drp1 or the bacterial expression system used to purify the CBP-Drp1 fusion protein contributed to the lower activity observed in the study by Chang et al. (32). Importantly, the addition of Mff or MiD49 only modestly increased the assembly-driven GTP hydrolysis activity of Drp1 (Fig. 5 E and F). Thus, in this minimal in vitro system, these adaptors do not act as classical GTPase effectors to enhance nucleotide hydrolysis by Drp1.
Fig. 5.
Effects of Mff and MiD49 on Drp1 GTPase activity. (A) Time course of GTP hydrolysis by Drp1 (0.6 μM) measured in 100 μM GTP, 37 °C at high (500 mM KCl) and low (50 mM KCl) ionic strength. (B) Steady-state kinetics of Drp1 (0.6 μM) GTP hydrolysis measured at low ionic strength (50 mM KCl), 37 °C. (C) A Coomasie blue-stained gel showing velocity sedimentation of Drp1 at high and low ionic strength. P, pellet; S, supernatant; T, total. (D) Drp1 kinetic parameters determined as described in B and Materials and Methods. kcat, turnover number; Vmax, maximal rate of hydrolysis; K0.5, substrate concentration at which velocity is one-half maximal. (E and F) GTP hydrolysis by Drp1 (0.1 μM) measured in 200 μM GTP and 50 mM KCl at 37 °C in the presence and absence of the indicated adaptor proteins. Cytoplasmic domains of Mff (E) or MiD49 (F) purified from yeast were included at 0.5 μM.
MiD49 Coassembles with Drp1 and Reduces Polymer Diameter.
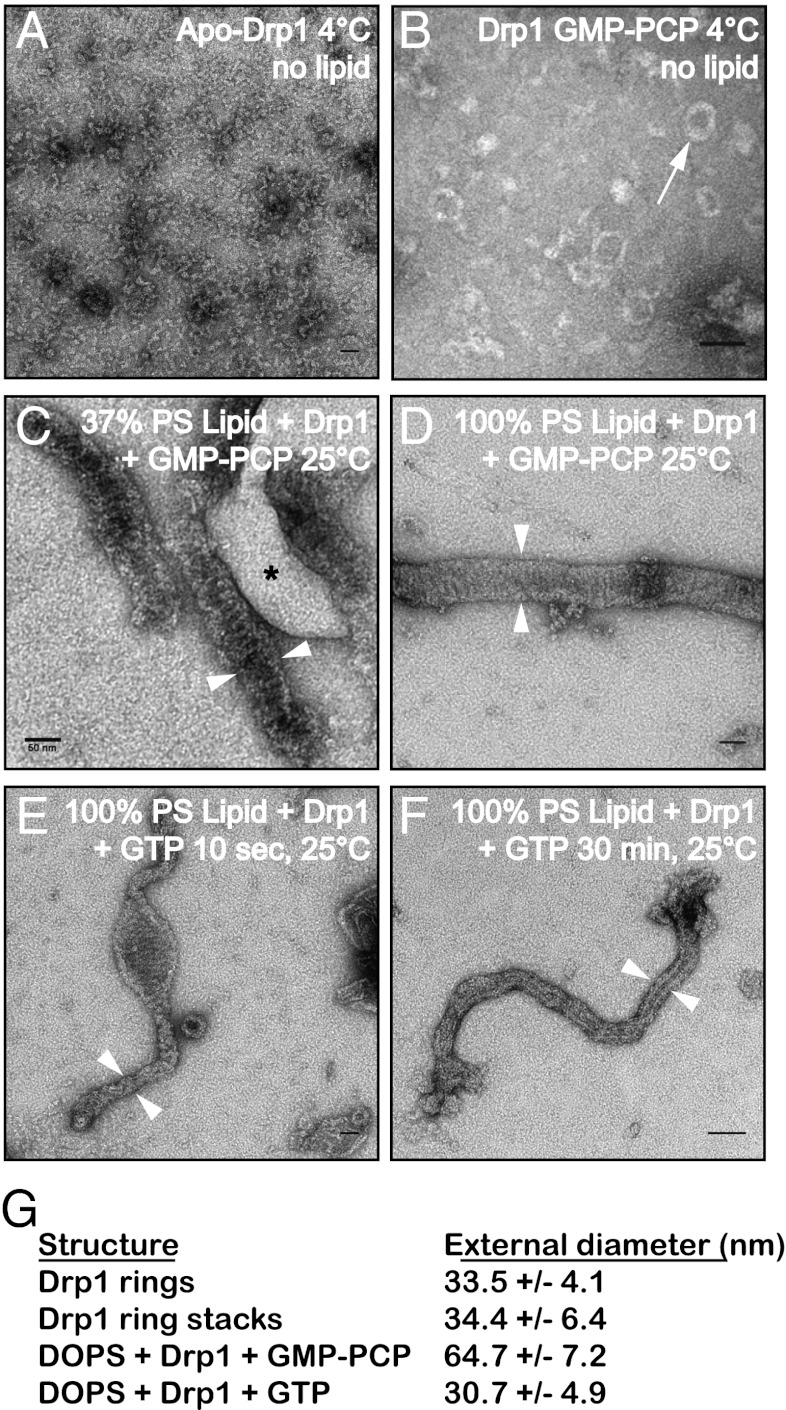
We used negative staining transmission electron microscopy (TEM) to analyze the structures formed by Drp1 in vitro. At low temperature, apo-Drp1 (without nucleotide) did not assemble into well-ordered structures (Fig. 6A). When the nonhydrolyzable analog β,γ-methyleneguanosine 5′-triphosphate (GMP-PCP) was added, Drp1 assembled into rings with an average external diameter of 33.5 ± 4.1 nm (Fig. 6 B and G). Raising the temperature to 25 °C in the presence of GMP-PCP produced Drp1 spirals (34.4 ± 6.4 nm). Intrinsically, Drp1 did not bind to liposomes containing moderate concentrations of anionic lipids (e.g., 37% phosphatidyl-serine; asterisk in Fig. 6C). Drp1 was able to deform nonphysiological liposomes made from purely anionic lipids (e.g., 100% 1,2-dioleoyl-sn-glycero-3-phospho-L-serine, DOPS) in the presence of GMP-PCP at 25 °C, forming tubes with ordered striations along their length (Fig. 6D). Interestingly, the diameter of these Drp1 tubes was significantly smaller (64.7 ± 7.2 nm) than that reported previously for Dnm1 assembled on lipids (109–121 nm) (14, 15). Constriction of these structures occurred upon exposure to GTP (Fig. 6 E and F), generating polymers with an average diameter of 30.7 ± 4.9 nm (Fig. 6G).
Fig. 6.
Drp1 self-assembly induces lipid tubulation and constriction in vitro. (A–F) Transmission electron micrographs of negatively stained Drp1 assemblies. (A) Drp1 protomers do not assemble in the absence of nucleotide at 4 °C. (B) Drp1 assembles into limited rings in the presence of GMP-PCP at low temperature (white arrow). (C) At 25 °C, Drp1 forms spirals or stacks of rings in the presence of GMP-PCP that exclude liposomes containing molar 37% PS (asterisk). (D) Drp1 assembles around liposomes containing molar 100% PS in the presence of GMP-PCP at 25 °C. (E and F) Drp1-decorated lipid tubes assembled in the presence of GMP-PCP were imaged after treatment with 1 mM GTP for 10 s (E) or 30 min (F). (G) Average external diameters of Drp1 structures as indicated by white arrowheads in C–F. For all measurements, n = 50. (Scale bars, 50 nm.)
We also analyzed the interaction of Drp1 with MiD49. In the presence of GMP-PCP, Drp1 self-assembles and pellets in a sedimentation assay (Fig. 7A). Although the cytoplasmic domain of MiD49 alone (MiD49∆TM) remains in the supernatant fraction, the adaptor sediments in the presence of Drp1, consistent with the idea that the two proteins bind to each other and may coassemble (Fig. 7A). These findings were confirmed using a flotation assay (in the presence of GMP-PCP). His-tagged MiD49∆TM alone (His-MiD49∆TM) was able to bind and float with liposomes containing nickel-modified lipids after centrifugation in a sucrose step gradient (Fig. 7B, Upper, lane 1). This fractionation pattern was dependent upon the presence of nickel (nickel-nitrilotriacetic acid, Ni-NTA) lipids and did not occur when membranes lacking the Ni-NTA moiety were substituted in the experiment (Fig. 7B, Lower, lane 1). Although Drp1 has a weak affinity for the Ni-NTA liposomes on its own (Fig. 7B, Upper, lane 4), the fraction of Drp1 bound to these liposomes increased visibly in the presence of the MiD49 adaptor (Fig. 7B, Upper, lane 7). In control experiments, MiD49 and Drp1 (alone or in combination) did not float with electrostatically neutral lipids (Fig. 7B, Lower, lanes 6 and 9).
Fig. 7.
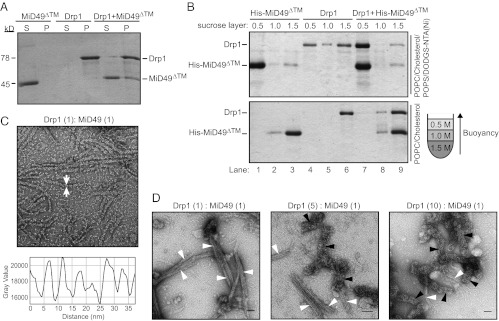
MiD49 copolymerizes with Drp1 and decreases polymer diameter. (A) MiD49∆TM (lacking the transmembrane domain) cosediments with Drp1. (B) liposomes containing DO DGS-NTA(Ni) (Upper) decorated with His-tagged MiD49∆TM promote flotation of Drp1 in a sucrose step gradient. Charge-neutral POPC/cholesterol liposomes (Lower) bind His-tagged MiD49∆TM poorly and do not promote Drp1 flotation. As depicted in the cartoon gradient at the right, protein-bound liposomes float to the top of the 0.5 M sucrose layer. (C) (Upper) In the presence of MiD49∆TM, Drp1 forms ordered polymers (arrows) with a diameter of 14.9 ± 1.5 nm. (Lower) Periodicity (∼5 nm) measured along the length of the Drp1:MiD49 polymers. (D) Effect of Drp1:MiD49 (molar:molar) ratios on polymer assembly. Decreasing MiD49 concentration reduces the formation of narrow (14.9-nm) polymers (white arrowheads) and increases the diameter of larger (34.4-nm) polymers (black arrowheads).
Negative transmission electron microscopy staining revealed a dramatic effect of MiD49 on Drp1 polymer formation. At 25 °C in the presence of GMP-PCP, Drp1 plus MiD49 formed extended, uniform polymers with distinct striations (Fig. 7C, Upper). These polymers had an average external diameter of 14.9 ± 1.5 nm, which is less than half the diameter of ring stacks formed by Drp1 alone (34.4 ± 6.4 nm) (Fig. 6G). Measurement of pixel intensity along the length of these structures revealed a highly regular periodicity of ∼5 nm (Fig. 7C, Lower). In control studies, MiD49 did not assemble reproducibly into similar polymers in the presence or absence of GTP or GTP analogs.
To investigate further the nature of these narrower polymers, we examined assembly in the presence of different Drp1:MiD49 (molar:molar) ratios. Incubation of Drp1 and MiD49 at a 1:1 ratio in the presence of GMP-PCP produced mainly polymers with the smaller average diameter (Fig. 7D, white arrowheads,14.9 ± 1.5 nm). These polymers often associated laterally into bundles. When a Drp1:MiD49 ratio of 5:1 or 10:1 was examined, fewer narrow polymers were observed (Fig. 7D, white arrowheads), with a concomitant increase in polymers of larger diameter (black arrowheads). Both the appearance and the diameter of the latter spirals were similar to those formed by Drp1 alone (Fig. 6C). These data are consistent with the idea that coassembly of MiD49 with Drp1 is stoichiometric and suggest that MiD49 copolymerizes with Drp1 rather than simply nucleating assembly of a Drp1 homopolymer.
Discussion
The adaptor proteins studied here were shown originally to mediate the recruitment of the Dnm1 or Drp1 GTPases to mitochondria, but their postrecruitment roles in mitochondrial fission were not clear. In this study, we demonstrate that individual adaptor-GTPase pairs act after recruitment to catalyze membrane division in vivo. In the case of Drp1, coassembly with one of these adaptors increases the order and dramatically decreases the diameter of the polymers formed.
The identification of Fis1 and Dnm1/Drp1 in yeast and mammals initially suggested that the basic molecular machinery for mitochondrial fission was conserved during evolution. Although the role of yeast Fis1 in Mdv1-Dnm1 recruitment to mitochondria has never been questioned, data supporting a function for mammalian Fis1 in Drp1 recruitment was contradictory. This issue was resolved recently by the demonstration that it is human Mff, rather than Fis1, that acts as the mitochondrial receptor for Drp1 (23). Soon after, MiD49 and MiD51 also were reported to mediate Drp1 mitochondrial recruitment (24, 25). We show here that yeast Fis1 is dispensable for fission when the Mdv1 adaptor is membrane-tethered, allowing Dnm1 recruitment to mitochondria. Moreover, expression of human Fis1 and Drp1 in yeast was not sufficient to rescue defects in mitochondrial fission. Thus, Fis1 has not been conserved throughout evolution because of an essential role in Dnm1/Drp1-mediated membrane scission. What, then, is the conserved function of Fis1? Although mitochondrial fission proteins also have been implicated in peroxisome division and mitophagy in yeast and mammals (33–35), Fis1 appears to be dispensable for peroxisome fission in human cells (23) and for mitophagy in yeast (36, 37). In addition, it was suggested recently that mammalian Fis1 interacts directly with MiD51 (also called mitochondrial elongation factor 1, or MIEF1) to regulate fission negatively (25). Further studies clearly are necessary to determine whether Fis1 has a conserved function(s) in organelle division.
Our findings show unambiguously that a single type of adaptor protein is sufficient for mitochondrial membrane scission by human Drp1. Why, then, do mammalian cells simultaneously express Mff, MiD49, and MiD51? Studies to date have not identified significant differences in the mitochondrial fission events mediated by these different adaptors (23–25). However, the assays used in these studies (morphological quantification and fixed time-point analysis) would fail to detect significant temporal, spatial, or mechanistic differences in Drp1 recruitment, assembly, and/or membrane scission that are specific to each adaptor. In addition, the physiological circumstances (i.e., apoptosis, mitophagy) in which each adaptor is activated might differ. Documented posttranslational modifications of Drp1 including phosphorylation (3, 38–42), sumoylation (43–46), nitrosylation (47), and ubiquitination (48–52) also could influence the identity of the adaptor used for fission, as could posttranslational modifications of the adaptors themselves. Finally, it is possible that multiple adaptors work together with Drp1 at a single division site. Such cooperation has been documented for the paralogous adaptors Mdv1 and Caf4 in yeast (53), and it seems likely that the MiD49 and MiD51 paralogs also will prove to have the capacity to function with Drp1 at the same fission site in mammals.
Distances of ≤1 nm between opposing lipid bilayers are thought to be necessary for initiation of inner leaflet hemifusion and subsequent membrane scission (54, 55). Taking into account the diameter of a lipid bilayer (∼5 nm) (56, 57) and mitochondria’s double membrane, the average external diameter we measured for Drp1-lipid tubules (30.7 nm) (Fig. 6G) would not produce a luminal distance small enough to initiate fission of both the inner and outer mitochondrial membranes. This problem could be overcome by coassembling MiD49 with Drp1, because the ∼15-nm average external diameter of the Drp1:MiD49 copolymer is sufficiently narrow to drive fission. Like the copolymers formed by coincubation of dynamin-1 with endophilin (58) or amphiphysin (59), the MiD49:Drp1 copolymers shown here also change the structural properties of a dynamin GTPase polymer. In the case of N-BAR (Bin–Amphiphysin–Rvs domain with an additional predicted N-terminal amphipathic helix) proteins and dynamin-1, the hybrid coat has a different diameter and different pitch. A detailed understanding of how MiD49 alters structural features of the Drp1 polymer and the functional consequences of the hybrid assembly for the fission process requires further study.
Our findings clarify the individual functions of mitochondrial adaptors and challenge the notion that these proteins act solely to recruit and stimulate assembly of the DRPs Dnm1 and Drp1 on the correct cellular membrane. Instead, coassembly of adaptors with DRPs may work generally to change the physical properties of the resulting polymers in a manner that promotes or regulates membrane scission. For example, coassembly could prevent promiscuous fission by altering contacts between adjacent turns of the DRP helix, thereby inhibiting mechano-chemical conformational changes that lead to constriction and fission. Such an inhibited state may be regulatory, delaying constriction until a signaling event or another factor is recruited. Alternatively, as shown here, the copolymer may have different geometric properties and be able to form a more compact state that promotes membrane constriction and fission. Regardless of the mechanism, we suggest that the ability to modulate polymer geometry will prove to be a common function of mitochondrial dynamin adaptors.
Materials and Methods
Strains and Plasmids.
Yeast strains and plasmids used in this study are listed in Tables S2 and S3. Standard yeast and bacterial techniques were used for construction and growth of strains (60, 61). Details of plasmid construction can be found in SI Materials and Methods.
Fluorescence Microscopy.
Mitochondrial morphologies were quantified as described previously (28, 62, 63) in the indicated strains expressing mitochondrial-matrix–targeted fast-folding RFP (mito-ffRFP also referred to as “mito-RFP”) or OMM-targeted GFP (mito-OMGFP; Fig. 4). Unless noted in the text, Mdv1, Mff, and MiD49/51 proteins were expressed from the MET25 promoter and integrated at the MDV1 locus. Dnm1 and GFP-Dnm1 were expressed from the native promoter and locus. Drp1 (variant 3) and GFP-Drp1 were expressed from the MET25 and copper homeostasis 1 (CUP1) promoters, respectively, on pRS416.
Overnight cultures were grown at 30 °C in the appropriate selective synthetic dextrose medium containing 100 µg/mL methionine, were diluted to 0.2 OD600 in medium containing 10 µg/mL methionine, and were grown for 3–5 h. For Fig. 4C, cells were grown as described above but were diluted continually into synthetic dextrose medium lacking methionine (to maintain an OD600 between 0.2 and 1.0) and were scored at the indicated times. Mitochondrial phenotypes and formation of GFP-Dnm1 puncta were scored in 100 cells, and the data shown represent the average ± SEM of at least three independent experiments. Images were acquired and processed as described (28).
Time-Lapse Imaging.
For single-color time-lapse imaging (Fig. 3), cells expressing mito-RFP were grown in selective synthetic dextrose medium were applied to Lab-Tek II Chamber wells (Thermo Scientific) treated with Concavalin A (2 mg/mL; Sigma) and were maintained at 30 °C. Z stacks (0.2-µm optical sections) of fields of cells were acquired every 7 s over a 20-min time course using a 3i Marianas Live Cell Imaging microscope workstation (Intelligent Imaging Innovations) equipped with dual ultra-sensitive Cascade II 512B EMCCD cameras (Roper Scientific) configured with a Roper Dual-cam and Sutter DG-4 Illuminator (Sutter Instruments) with a 100×, 1.45 NA Plan-Apochromat objective (Zeiss). Data were deconvolved and analyzed using SlideBook 4.2 software (Intelligent Imaging Innovations). Substacks containing fission events were isolated from the entire stack to minimize signal background and were assembled in Photoshop CS3 (Adobe). Brightness and contrast were adjusted using only linear operations applied to the entire image. For quantification, only cells that underwent one or more fission or fusion events during the time course were selected for analysis. The results were expressed as the number of fission or fusion events per cell during a 15-min interval.
For two-color time-lapse imaging (Fig. 4D), cells expressing GFP-Drp1, mito-RFP, and the indicated mammalian adaptor (MiD49, MiD51, or Mff) were grown in selective synthetic dextrose medium and applied to a Y04c microfluidic chamber (CellASIC Corp.). Injection of cells and medium was controlled by an ONIX Microfluidic Perfusion System and ONIX FG version 2.6 software (CellASIC Corp.). Z stacks of cells (0.3-µm optical sections) were imaged every 30 s over a 30-min time course using an Observer Z1 microscope (Zeiss) equipped with HE GFP (set 38) and mRFP (set 63) shift-free filter sets, an Axiocam MRm Rev.3 camera, and a 100×, 1.4 NA Plan-Apochromat objective (Zeiss). GFP and DsRed channels were acquired sequentially using AxioVision 4.8 software (Zeiss), and data were deconvolved and analyzed using AxioVision 4.6 software (Zeiss). Substacks containing fission events were isolated and assembled as described above.
Protein Production.
Human Drp1 (isoform 3; NP_005681.2), MiD49 (accession number Q96C03), and Mff (accession number Q9GZY8) constructs, each containing an N-terminal PreScission protease cleavage site and a FLAG-One-STrEP tag (IBA), were expressed in JSY9612. Overnight cultures were diluted into selective synthetic dextrose medium containing 1 mM CuSO4 to induce expression (final OD600 of 0.2) and were grown in a Belco fermentor at 30 °C for 24 h. After harvesting, cell pellets were snap frozen in liquid nitrogen as small droplets and were pulverized in a freezer mill (3 min × 15 cycles). All subsequent purification steps were performed at 4 °C. Cell powders were dissolved in either high ionic strength lysis buffer for hDrp1 [100 mM Tris⋅Cl (pH 8.0), 500 mM NaCl, 1 mM DTT, 1 mM EDTA] or low ionic strength buffer for MiD49 or Mff [100 mM Tris⋅Cl (pH 8.0), 150 mM NaCl, 1 mM DTT, 1 mM EDTA] containing Protease Inhibitor Mixture III (Calbiochem). The lysates were clarified by centrifugation at 30,000 × g for 1 h, filtered (0.45 µm), loaded onto a 5-mL StrepTrap HP column (GE Healthcare), washed with 1 L lysis buffer, and cleaved in the column with PreScission protease (GE Healthcare) for 16 h. Drp1 was dialyzed against 20 mM Hepes (pH 7.4), 150 mM KCl, 2 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA, snap frozen in liquid nitrogen, and stored at −80 °C. Mff and MiD49 were purified further by size-exclusion chromatography (Sepharose 200; GE Healthcare), dialyzed against 20 mM Hepes (pH 7.4), 150 mM KCl, 2 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA, and stored at 4 °C. Equilibrium sedimentation analysis performed on purified protein indicated that Drp1 variant 3 is a dimer in high ionic strength buffer [observed molecular weight (MWobs)/calculated molecular weight (MWcal) = 2.17], the cytoplasmic domain of Mff is a dimer (MWobs/MWcal = 2.25), and the cytoplasmic domain of MiD49 is a monomer (MWobs/MWcal = 0.99) (Fig. S2).
GTPase Assay.
Inorganic phosphate release was measured using the malachite green phosphate assay (POMG-25H; BioAssay Systems) as described by the manufacturer and by Leonard et al. (64). For the time-course analysis, Drp1 (0.6 µM) was assayed at 37 °C in high (500 mM KCl) or low (50 mM KCl) ionic strength buffer containing 20 mM Hepes (pH 7.4), 2 mM MgCl2, 1 mM DTT, and 100 µM GTP. Reactions were halted at the indicated times by diluting 20 µL in 25 mM EDTA (final concentration) in a microtiter plate. Although 25 mM EDTA was sufficient to halt the reaction, we found that higher EDTA concentrations lowered the signal generated by the malachite reagent during the development step. Samples were incubated at room temperature with 20 µL Malachite Reagent (BioAssay Systems) and 60 µL water for 30 min, and the absorbance at 600 nm was measured using a Modulus Microplate Reader (Turner BioSystems). For the steady-state kinetic analysis, GTP assays were performed at 37 °C in reactions containing 0.6 µM Drp1, 20 mM Hepes (pH 7.4), 50 mM KCl, 2 mM MgCl2, and 1 mM DTT containing variable GTP concentrations (0, 25, 50, 75, 100, 200, 300, 500, 1,000, 1,500, and 2,000 µM). Fixed volumes were removed every 5 min for 70 min, quenched by EDTA, and developed as described above. The kcat and the substrate concentration at which velocity is one-half maximal (k0.5) were calculated in GraphPad Prism using nonlinear regression curve fitting. For time-course analyses with adaptor proteins, GTPase assays were performed in reactions containing Drp1 (0.1 µM) plus or minus Mff (0.5 µM) or MiD49 (0.5 µM) at 37 °C in 50 µM KCl, 20 mM Hepes (pH 7.4), 2 mM MgCl2, 1 mM DTT, and 200 µM GTP. In all experiments, data shown are the average ± SEM values obtained from triplicate samples analyzed at the same time. Each experiment was repeated three times.
Velocity Sedimentation and Flotation Assays.
For the velocity sedimentation assay (Fig. 5C), 1.25 µM Drp1 was incubated in either low ionic strength buffer [20 mM Hepes ( pH 7.4), 50 mM KCl, 2 mM MgCl2, 1 mM DTT] or high ionic strength buffer [20 mM Hepes (pH 7.4), 500 mM KCl, 2 mM MgCl2, 1 mM DTT] for 1 h at 37 °C. The reactions were spun down at 40,000 rpm (TLA 100 rotor) in a Beckman Optima MAX Ultracentrifuge for 1 h at 25 °C. Supernatants were removed, and pellet fractions were resuspended in an equal volume of buffer. Then 25 µL of total, supernatant, and pellet fractions were separated on 10% SDS/PAGE gels and stained with Coomassie Brilliant Blue dye. For the velocity sedimentation in Fig. 7A, 1.25 µM MiD49∆TM, 1.25 µM Drp1, or 1.25 µM of both proteins was dialyzed at 25 °C into 20 mM Hepes (pH 7.4), 25 mM KCl, 2 mM MgCl2, 1 mM DTT, and 200 µM GMP-PCP for 6 h. Samples were pelleted and processed for SDS/PAGE as described above.
Liposome Preparation.
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), cholesterol, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhodamine-PE; ammonium salt), 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt; Ni2+-NTA-DOGS), and DOPS were purchased from Avanti Polar Lipids in chloroform. Four types of mixtures were prepared: (i) POPC, POPS, cholesterol, Rhodamine-PE were mixed in a molar ratio 44.4:37:18.5:0.007 [37% phosphatidylserine (PS) liposomes; Fig. 6C]; (ii) POPC, POPS, cholesterol, and Ni2+-NTA-DOGS were mixed in a molar ratio 44.4:31:18.5:6 (nickel liposomes); (iii) 100% DOPS (100% PS liposomes; Fig. 6 D–F); and (iv) POPC:cholesterol were mixed in a molar ratio 80:20 (neutral liposomes). For formulation 3 (100% PS), we used the dioleoyl form of phosphatidyl-serine to preserve fluidity and solubility of the liposomes.
Chloroform was evaporated by gentle vortexing under a steady stream of nitrogen gas to make a thin lipid film around the walls of glass vials. These films were dried under vacuum for 1 h at room temperature. Dried lipid films then were resolubilized in absolute hexane. The hexane also was evaporated under streaming nitrogen while vortexing, followed by a second round of desiccation for 3–4 h at room temperature. Lipid films were suspended in aqueous buffer [20 mM Hepes (pH 7.5), 100 mM KCl] by vortexing at room temperature. Aliquots from the liposome preparation were stored at −80 °C.
Flotation Assays.
Liposomes and proteins were mixed in a molar ratio of 1,000:1 (lipid:protein) in Beckman polycarbonate centrifuge tubes. After 1-h incubation at 4 °C, the mixtures were homogenized with 300 µL 2 M sucrose in 20 mM Hepes (pH7.5), 100 mM KCl, 1 mM GMP-PCP (∼1.5-M final sucrose concentration). Two additional layers of 1 M (150 µL) and 0.5 M (300 µL) sucrose in the same buffer were overlaid carefully (in that order) on top of the homogenized mixture. The mixtures were spun using a TLS-55 rotor in a Beckman centrifuge for 1 h at 54,000 rpm (4 °C). After the spin, the liposomes migrated to interfaces between individual sucrose layers and could be seen as turbid bands. The interfaces between individual sucrose layers were collected by pipetting and analyzed by 10% SDS/PAGE and Coomassie Brilliant Blue staining.
In Vitro Membrane Binding and Tubulation Reactions.
For Drp1 tubulation reactions, DOPS liposomes were mixed with protein (1:1, mass:mass). After 1 h, GMP-PCP was added, and the sample was incubated for 4 h at room temperature. The effects of GTP hydrolysis were analyzed in two ways. First, after adsorption of the lipid and protein mixtures to EM grids, the sample was washed in 1 mM GTP followed immediately by blotting and staining. Second, a stock of 10 mM GTP was added to lipid-protein mixtures to a final concentration of 1 mM GTP for 30 min before the mixtures were applied to EM grids for staining.
For Drp1-MiD49 copolymerization, proteins were mixed 1:1 (mass:mass) with or without liposomes and were dialyzed overnight against 20 mM Hepes, 25 mM KCl, 200 mM GMP-PCP, 2 mM MgCl2, and 1 mM DTT.
EM.
For negative-stain EM, carbon-coated copper grids were glow discharged for 15 s. Then 5 µL of the sample was added to the surface, blotted, and stained with 1% uranyl acetate. Images were acquired using an FEI Tecnai T12 electron microscope equipped with a LaB6 filament and operated at 120 kV. Magnifications of 21,000–42,000× were recorded on a Gatan CCD.
Supplementary Material
Acknowledgments
We thank J. Macfarlane and S. Safran for cloning expertise and assistance with GTPase assays; M. Ryan and A. van der Bliek for providing MiD and Mff plasmids; J. M. McCaffery for use of the John Hopkins University Integrated Imaging Facility; and M. Kay, T. Formosa, and members of the J.M.S. and A.F. laboratories for stimulating discussions. This work was supported by National Institutes of Health (NIH) Grants GM53466 and GM84970 (to J.M.S.). D.M.E. and the University of Utah Protein Interaction Core Facility are supported by NIH Grant GM82545.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300855110/-/DCSupplemental.
References
- 1.Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5(2):133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Gorsich SW, Shaw JM. Importance of mitochondrial dynamics during meiosis and sporulation. Mol Biol Cell. 2004;15(10):4369–4381. doi: 10.1091/mbc.E03-12-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282(15):11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 4.Parone PA, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE. 2008;3(9):e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13(5):589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108(25):10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11(8):958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 9.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186(6):805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleazard W, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1(5):298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4(5):815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 12.Otsuga D, et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143(2):333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147(4):699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170(7):1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18(1):20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerveny KL, McCaffery JM, Jensen RE. Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol Biol Cell. 2001;12(2):309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151(2):353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151(2):367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naylor K, et al. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem. 2006;281(4):2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 21.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325(5942):874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19(6):2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer CS, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12(6):565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, et al. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30(14):2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170(2):237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158(3):445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koirala S, et al. Molecular architecture of a dynamin adaptor: Implications for assembly of mitochondrial fission complexes. J Cell Biol. 2010;191(6):1127–1139. doi: 10.1083/jcb.201005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerveny KL, Jensen RE. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell. 2003;14(10):4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunnari J, et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8(7):1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14(10):1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang CR, et al. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285(42):32494–32503. doi: 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch A, et al. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278(10):8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 34.Kuravi K, et al. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 19):3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;278(19):17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 36.Mendl N, et al. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci. 2011;124(Pt 8):1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17(1):87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Cereghetti GM, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105(41):15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282(30):21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 40.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8(10):939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han XJ, et al. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182(3):573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, et al. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44(4):532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10(7):748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Figueroa-Romero C, et al. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009;23(11):3917–3927. doi: 10.1096/fj.09-136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177(3):439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zunino R, Braschi E, Xu L, McBride HM. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J Biol Chem. 2009;284(26):17783–17795. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324(5923):102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horn SR, et al. Regulation of mitochondrial morphology by APC/CCdh1-mediated control of Drp1 stability. Mol Biol Cell. 2011;22(8):1207–1216. doi: 10.1091/mbc.E10-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178(1):71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7(10):1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: Implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286(13):11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonashiro R, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25(15):3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Q, Koirala S, Perkins EM, McCaffery JM, Shaw JM. The mitochondrial fission adaptors Caf4 and Mdv1 are not functionally equivalent. PLoS ONE. 2012;7(12):e53523. doi: 10.1371/journal.pone.0053523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bashkirov PV, et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135(7):1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336(6088):1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Bose PS, Sigworth FJ. Using cryo-EM to measure the dipole potential of a lipid membrane. Proc Natl Acad Sci USA. 2006;103(49):18528–18533. doi: 10.1073/pnas.0608714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leforestier A, Lemercier N, Livolant F. Contribution of cryoelectron microscopy of vitreous sections to the understanding of biological membrane structure. Proc Natl Acad Sci USA. 2012;109(23):8959–8964. doi: 10.1073/pnas.1200881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundborger A, et al. An endophilin-dynamin complex promotes budding of clathrin-coated vesicles during synaptic vesicle recycling. J Cell Sci. 2011;124(Pt 1):133–143. doi: 10.1242/jcs.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1(1):33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 60.Green M, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2012. [Google Scholar]
- 61.Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology. Vol 350. San Diego: Academic, Inc.; 2002. [Google Scholar]
- 62.Amiott EA, Cohen MM, Saint-Georges Y, Weissman AM, Shaw JM. A mutation associated with CMT2A neuropathy causes defects in Fzo1 GTP hydrolysis, ubiquitylation, and protein turnover. Mol Biol Cell. 2009;20(23):5026–5035. doi: 10.1091/mbc.E09-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bui HT, Karren MA, Bhar D, Shaw JM. A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment. J Cell Biol. 2012;199(4):613–622. doi: 10.1083/jcb.201207079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leonard M, Song BD, Ramachandran R, Schmid SL. Robust colorimetric assays for dynamin’s basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.