Abstract
The accurate determination of the risk of cancer recurrence is an important unmet need in the management of prostate cancer. Patients and physicians must weigh the benefits of currently available therapies against the potential morbidity of these treatments. Herein we describe the development of a gene expression-based continuous risk index and a validation of this test in an independent, blinded cohort of post-radical prostatectomy (RP) patients. A gene expression signature, prognostic for prostate-specific antigen (PSA) recurrence, was identified through a bioinformatic analysis of the expression of 1,536 genes in malignant prostate tissue from a training cohort of consecutive patients treated with RP. The assay was transferred to a real-time RT-PCR platform, and a continuous risk index model was constructed based on the expression of 32 genes. This 32-gene risk index model was validated in an independent, blinded cohort of 270 RP patients. In multivariate analyses, the risk index was prognostic for risk of PSA recurrence and had added value over standard prognostic markers such as Gleason score, pathologic tumor stage, surgical margin status, and presurgery PSA (hazard ratio, 4.05; 95% confidence interval, 1.50–10.94; P = 0.0057). Furthermore, RP patients could be stratified based on the risk of PSA recurrence and the development of metastatic disease. The 32-gene signature identified here is a robust prognostic marker for disease recurrence. This assay may aid in postoperative treatment selection and has the potential to impact decision making at the biopsy stage.
Prostate cancer remains a significant health problem, with more than 217,000 men diagnosed in the United States in 2010, and is the second leading cause of male cancer mortality (1). Management of prostate cancer is challenging, given the protracted and variable nature of the disease. A key clinical need is to identify patients with indolent vs. aggressive disease more accurately, as treatment choices may affect quality of life and survival significantly. For clinically localized and locally advanced cancer, radical prostatectomy (RP) is an established and accepted treatment; ∼90,000 RPs are performed each year (2). The standard of care following RP surgery has been debated, as the overall 10-y prostate-specific antigen (PSA) recurrence rate is ∼33% (3–5). For patients with adverse pathologic features, three randomized clinical studies have demonstrated that adjuvant radiotherapy reduces the risk of PSA recurrence (6–8). However, even in these high-risk populations, 36–56% of patients in the control arms did not experience PSA recurrence. In the Southwest Oncology Group (SWOG) 8794 trial, the number of men treated with adjuvant radiotherapy to prevent one case of metastatic disease at a median follow-up of 12.6 y was 12.2 (9). In contrast, RP is considered primary curative therapy in pathologically organ-confined disease, and adjuvant therapy generally is not recommended for these patients. However, in retrospective cohort studies of patients with pathologic tumor (pT) stage 2 disease, 10-y rates of PSA recurrence ranged from ∼7% to 20% (10, 11). Thus, identification of patients with organ-confined disease likely to experience disease recurrence represents a significant clinical need.
In clinical practice, variables such as Gleason score, pT stage, surgical margin status, and serum PSA are used to predict the risk of progression both alone and in combination (12–15) and have improved the ability to predict outcomes. They remain insufficient, however, in quantifying the risk of recurrence in many patients. The utility of objective, unique gene expression-based signatures in the prognosis and management of various cancers recently gained acceptance within the oncologic community (16–18). Herein, we describe the development and independent validation of a gene expression-based prognostic index that stratifies RP patients based on the risk of biochemical failure (BCF) and metastasis. In addition, we assessed the feasibility and applicability of the assay on prostate needle biopsy tissue obtained before RP. This prostate cancer recurrence risk index may improve upon the current standard of care for assessing risk of recurrence and metastasis in patients with prostate cancer.
Results
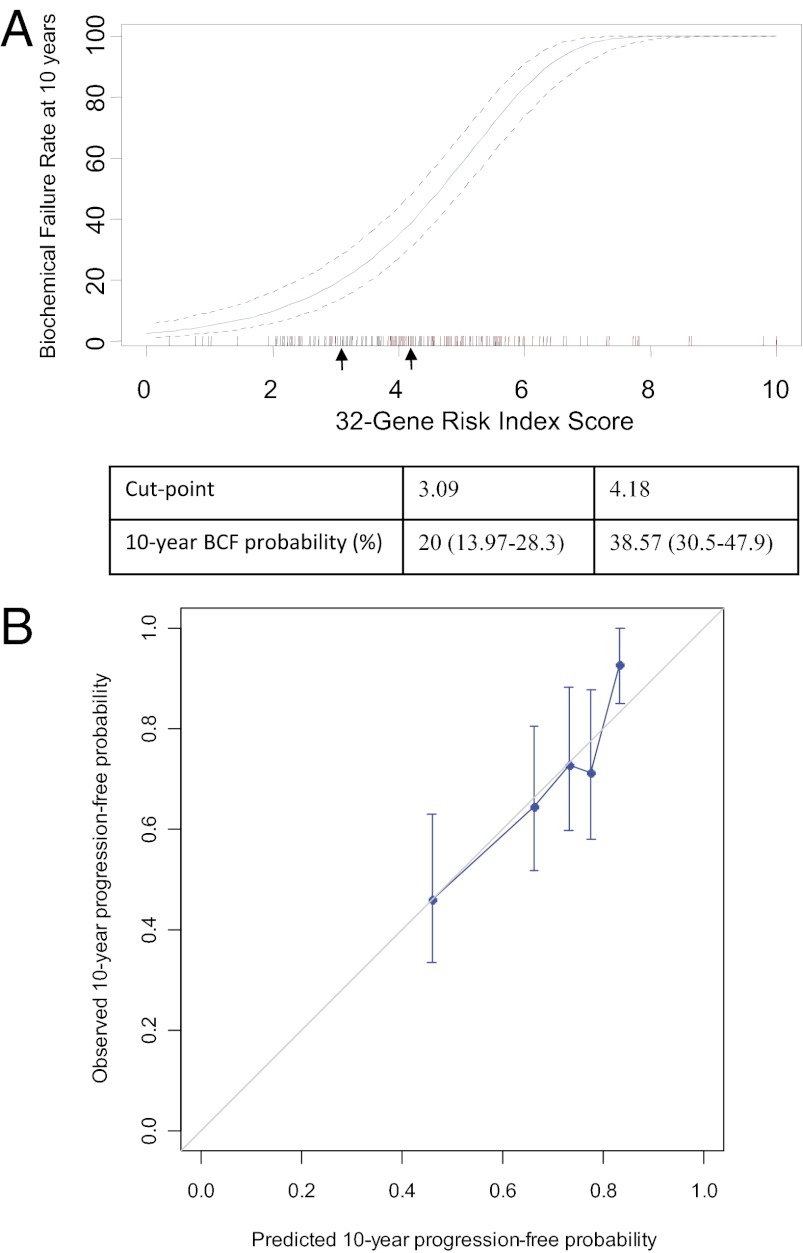
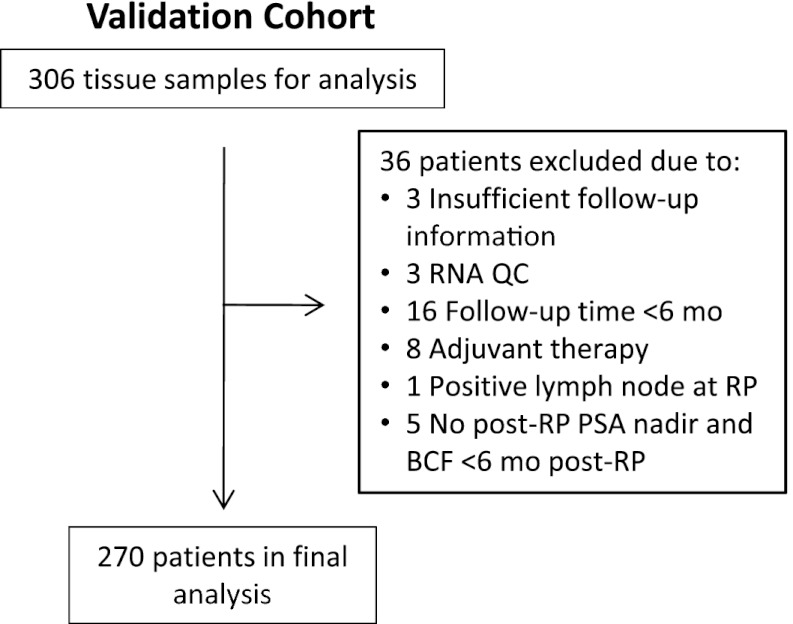
The identification of the 32-gene signature is described in SI Results. To allow for individualized recurrence risk assessment, a continuous risk index was constructed using discovery cohort data (Fig. 1A and Table S1). The 32-gene risk index was validated in an independent, blinded set of RP tissue samples. Of 306 samples evaluated, 270 were eligible for final analysis (Fig. 2). Demographics and clinical characteristics for this validation cohort are reported in Table 1.
Fig. 1.
Risk index model and calibration curve. Ten-year predicted risk of BCF based on the discovery cohort. Arrows indicate proof-of-concept cut points. The cut points were set at 3.09 and 4.18, corresponding to 10-y BCF rates of ∼20% and 40% in the discovery cohort. (A) Red hash marks indicate patients with a BCF event; black hash marks indicate patients without a BCF event. (B) Calibration curve assessing agreement between the predicted and actual rates of biochemical progression-free probability. The diagonal 45° line represents an ideal model with perfect prediction.
Fig. 2.
Validation cohort flow chart. Tissue samples for the validation cohort were obtained from consecutive patients who underwent RP surgery as part of their clinical care at MGH from January 1996 to December 1997.
Table 1.
Demographics and patient characteristics for the validation cohort
| Characteristic | Validation cohort (n = 270) |
| Age, y | |
| Mean (SD) | 61.96 (7.06) |
| Range | 37.00–79.00 |
| Presurgery PSA, ng/mL | |
| Mean (SD) | 7.06 (5.68) |
| Range | 0.80–52.4 |
| PSA unknown, no. (%) | 36 (13.3) |
| Gleason score at RP, no. (%) | |
| ≤6 | 110 (41) |
| 7 | 120 (44) |
| ≥8 | 40 (15) |
| pT stage, no. (%) | |
| 2 | 221 (82) |
| 3 | 49 (18) |
| Surgical margin, no. (%) | |
| Negative | 204 (76) |
| Positive | 66 (24) |
| Salvage therapy after BCF, no. (%) | |
| No | 225 (83) |
| Yes | 45 (17) |
| BCF event, no. | |
| No | 195 |
| Yes | 75 |
| Time to BCF event, y, median (range) | 4.28 (0.70–11.54) |
| Follow-up time, no BCF event, y, median (range) | 9.43 (0.53–13.49) |
| Metastasis event, no. | |
| No | 253 |
| Yes | 17 |
| Time to metastasis event, y, median (range) | 7.54 (2.09–12.96) |
| Follow-up time, no metastasis event, y, median (range) | 10.35 (0.47–14.67) |
| Death (all-cause) event, no. | |
| No | 229 |
| Yes | 41 |
| Time to death (all-cause), y, median (range) | 9.84 (1.41–13.13) |
| Follow-up time, no death event, y, median (range) | 12.74 (10.74–14.18) |
We first evaluated the 32-gene risk index (Table S2) in the context of commonly used prognostic factors for post-RP prostate cancer patients, including Gleason score, pT stage, surgical margin status, and presurgery PSA. In a univariate analysis, the 32-gene risk index, Gleason score, surgical margin, and presurgery PSA all were prognostic for BCF (Table 2). Fitting a restricted cubic spline of the 32-gene risk index in the Cox model demonstrated that a nonlinear term was not significant, indicating that a simple linear model was appropriate. A multivariate model demonstrated that the 32-gene risk index was prognostic for risk of BCF and had added value in the context of the standard prognostic tests [hazard ratio (HR), 4.05; 95% confidence interval (CI), 1.50–10.94; P = 0.0057; Tables 3 and 4]. Surgical margin status was the only other factor with significant prognostic value in the multivariate analysis [Note: Similar results were observed in a separate univariate and multivariate analysis excluding the 36 patients with missing presurgery PSA values, rather than imputing values (Tables S3–S5)].
Table 2.
Univariate Cox regression analysis of BCF in the validation cohort
| Variable | HR (95% CI) | P value |
| 32-Gene risk index | 8.16 (4.07–16.37) | <0.0001 |
| Gleason score | <0.0001 | |
| 7 vs. ≤6 | 1.91 (1.08–3.38) | 0.03 |
| ≥8 vs. ≤6 | 5.07 (2.76–9.31) | <0.0001 |
| pT stage (3 vs. 2) | 1.62 (0.95–2.75) | 0.08 |
| Margin (pos. vs. neg.) | 2.40 (1.50–3.83) | 0.0002 |
| Log(1 + baseline PSA) | 1.66 (1.23–2.24) | 0.0009 |
The HR calculated for the 32-gene index is based on a five-unit change.
Table 3.
Multivariate Cox regression analysis of BCF in the validation cohort (with 32-gene risk index)
| Variable | HR (95% CI) | P value |
| 32-Gene risk index | 4.05 (1.50–10.94) | 0.0057 |
| Gleason score | 0.16 | |
| 7 vs. ≤6 | 1.31 (0.72–2.40) | 0.3778 |
| ≥8 vs. ≤6 | 2.15 (0.97–4.77) | 0.0583 |
| pT stage (3 vs. 2) | 0.85 (0.48–1.50) | 0.5647 |
| Margin (pos. vs. neg.) | 2.33 (1.40–3.86) | 0.0011 |
| Log(1+baseline PSA) | 1.19 ( 0.87–1.63) | 0.2841 |
The HR calculated for the 32-gene index is based on a five-unit change.
Table 4.
Multivariate Cox regression analysis of BCF in the validation cohort (without 32-gene risk index)
| Variable | HR (95% CI) | P value |
| Gleason score | <0.0001 | |
| 7 vs. ≤6 | 1.51 (0.83–2.73) | 0.1773 |
| ≥8 vs. ≤6 | 3.95 (2.08–7.50) | <0.0001 |
| pT stage (3 vs. 2) | 0.96 (0.55–1.68) | 0.8836 |
| Margin (pos. vs. neg.) | 2.10 (1.28–3.43) | 0.0032 |
| Log(1+baseline PSA) | 1.31 (0.96–1.79) | 0.0871 |
The HR calculated for the 32-gene index is based on a five-unit change.
The discrimination ability of the risk index was quantified by calculating the concordance index, which provides the probability that in a randomly selected pair of patients in which one had a BCF event and one did not, the patient who had a BCF event would be assigned the worse predicted risk. The risk index had a concordance index of 0.6905. To assess calibration—the agreement between predicted outcome and actual outcome—we generated a calibration curve (Fig. 1B). The risk index was similar to the ideal curve (a 45° line indicating perfect agreement) across each quintile of risk index-predicted progression-free probabilities.
The incremental value of the 32-gene index was assessed by modeling the risk index with three postoperative nomograms (details in SI Materials and Methods, Table S6) (15, 19, 20). The 32-gene risk index enhanced the predictive ability of all three nomograms. The concordance index scores were 0.6869, 0.6973, and 0.6972 using the nomograms alone, and 0.7154, 0.7132, and 0.7219 for the models including the 32-gene risk score.
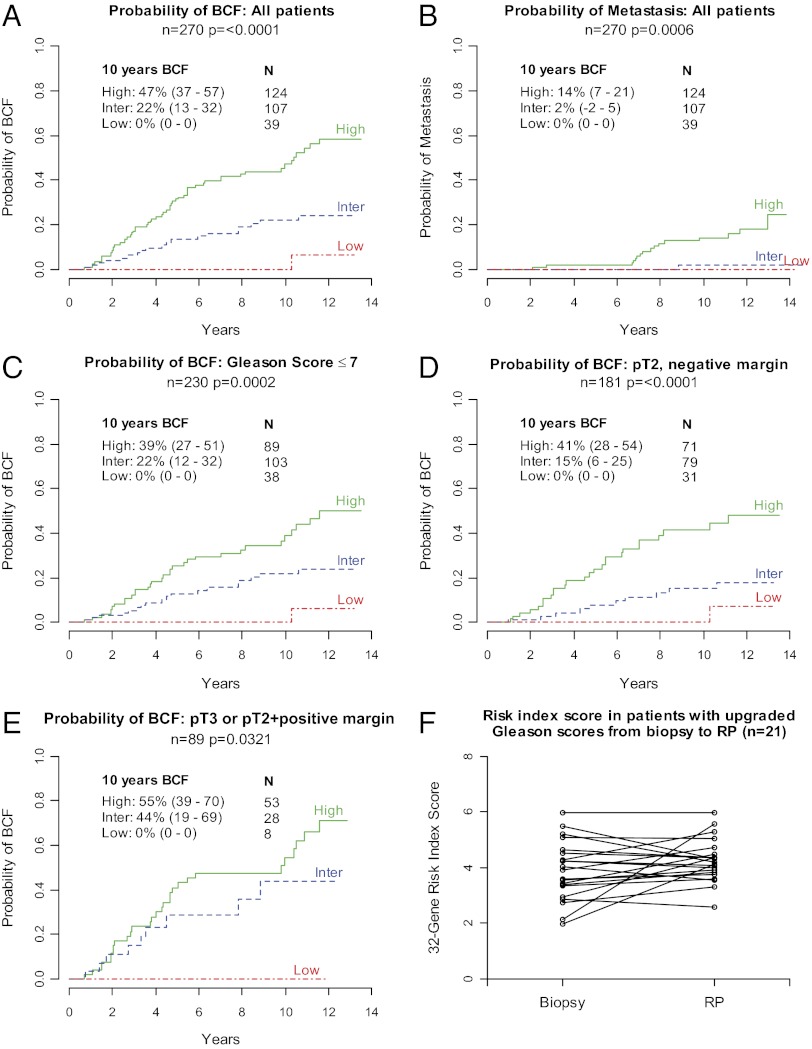
Using proof-of-concept cut points establishing low-, intermediate-, and high-risk groups (Fig. 1A), the risk index classified 39 patients (14%) as low risk, 107 patients (40%) as intermediate risk, and 124 patients (46%) as high risk in the validation cohort. Ten-year BCF probabilities were significantly different among the low-, intermediate-, and high-risk groups (0%, 22%, and 47%, respectively, P < 0.0001; Fig. 3A). Next, we examined several clinically relevant subgroups of patients. In patients with Gleason scores ≤7 (n = 230), the risk index classified 38 patients (16%) as low risk, 103 patients (45%) as intermediate risk, and 89 patients (39%) as high risk. Ten-year BCF probabilities were 0%, 22%, and 39%, respectively, and were significantly different among groups (P = 0.0002; Fig. 3C). Furthermore, in patients with a Gleason score of 7, those with a 4 + 3 pattern (n = 20) had significantly higher 32-gene risk index scores than patients with a 3 + 4 pattern (n = 100; 5.5 vs. 4.2, P = 0.00015). In patients with pT3 disease or positive surgical margins (n = 89), those classified as high risk (n = 53, 60%) had a 10-y BCF probability of 55%, whereas no patient classified as low risk (n = 8, 9%) experienced BCF (Fig. 3E). In patients with pT2 disease and a negative surgical margin (n = 181), the risk index classified 71 patients (39%) as high risk; these patients had a 10-y BCF probability of 41%. Notably, this group included 20 patients (28%) with a Gleason score of 6, a cohort that would be considered very low risk by traditional factors (pT2, negative surgical margin, Gleason score = 6); however, of the 20 patients identified by the 32-gene risk index score as high risk, 5 patients (25%) experienced BCF. In comparison, 31 patients (17%) with pT2 disease and a negative surgical margin were classified as low risk, and none of these patients experienced BCF in the first 10 y post-RP (Fig. 3D). In a multivariate analysis of patients with pT2 disease and a negative surgical margin, a high- vs. low-risk risk index score was associated with a significant increased risk of BCF (HR, 8.0; 95% CI, 1.04–61.55; P < 0.05). Additional subgroup analyses are presented in the Figs. S1–S3.
Fig. 3.
Cumulative incidence of BCF and metastasis, and feasibility of the 32-gene risk index in needle biopsy samples. (A and C–E) Cumulative incidence of BCF in all patients, patients with Gleason scores ≤7, patients with pT2 disease and a negative surgical margin, and patients with pT3 disease or a positive surgical margin. (B) Cumulative incidence of metastasis in all patients. (F) Thirty-two–gene risk index scores in patients with upgraded Gleason scores between biopsy and RP. Analyses are from the validation cohort.
Next, we assessed the ability of the 32-gene test to predict the risk of metastasis. Seventeen patients in the validation cohort had post-RP metastasis events. The risk categories defined based on risk of BCF significantly stratified patients based on risk of metastasis. The 10-y probability of metastasis was 14% in the high-risk group, 2% in the intermediate-risk group, and 0% in the low-risk group (P = 0.0006; Fig. 3B).
Net reclassification improvement (NRI) analyses revealed that for reclassification based on metastatic events, the 32-gene index performed numerically better than all nomograms, and significantly better than the 1999 nomogram (P = 0.045; Table S7). For reclassification based on BCF, the 32-gene risk index performed numerically better than the nomograms; however, the NRI was not statistically significant (P > 0.05; Table S7).
The feasibility and applicability of the 32-gene risk index assay on core needle biopsy tissue were assessed in patients with available needle biopsy tissue (n = 79). There was a strong concordance correlation between 32-gene risk index scores on biopsy and RP samples from the same patients (P < 0.0001, r = 0.66). Next, we evaluated the prognostic ability of the 32-gene assay on biopsy tissue in the context of other clinicopathologic variables available at the biopsy stage (biopsy Gleason score, clinical tumor (cT) stage, proportion of cores positive, highest percent tumor content, and PSA). The 32-gene risk index score (from biopsy tissue) was prognostic for BCF following RP surgery in both univariate (HR, 4.92; 95% CI, 1.45–16.78; P = 0.0107) and multivariate (HR, 7.57; 95% CI, 1.15–49.49; P = 0.0345; Tables 5 and 6) Cox regression analyses. PSA was the only other factor with significant prognostic value in the multivariate analysis. Finally, to assess the ability of the 32-gene risk index to provide consistent prognostic information in the context of the multifocal nature of prostate cancer, we analyzed a subset of patients (n = 21) who had Gleason scores of 6 at biopsy and an upgraded score of 7, 8, or 9 at RP. The 32-gene risk index scores were similar between biopsy and RP (P = 0.11; Fig. 3F).
Table 5.
Univariate Cox regression analysis of post-RP BCF using 32-gene risk index scores from needle biopsy tissue samples
| Variable | HR (95% CI) | P value |
| 32-Gene risk index | 4.92 (1.45–16.78) | 0.0107 |
| Gleason score at biopsy | 0.0162 | |
| 7 vs. ≤6 | 3.21 (2.96–3.48) | 0.0052 |
| ≥8 vs. ≤6 | 2.80 (0.78–10.06) | 0.1155 |
| cT stage (2 vs. 1) | 0.83 (0.25–2.76) | 0.7638 |
| Proportion of cores positive (>⅓ vs. ≤⅓) | 1.03 (0.48–2.23) | 0.9306 |
| Highest percent positive core | 1.00 (0.88–1.13) | 0.9955 |
| Log(1+baseline PSA) | 2.09 (1.19–3.65) | 0.0098 |
Validation cohort, n = 79.
The HR calculated for the 32-gene index is based on a five-unit change. All variables are pre-RP values.
Table 6.
Multivariate Cox regression analysis of post-RP BCF using 32-gene risk index scores from needle biopsy tissue samples
| Variable | HR (95% CI) | P value |
| 32-Gene risk index | 7.57 (1.15–49.49) | 0.0345 |
| Gleason score at biopsy | 0.0474 | |
| 7 vs. ≤6 | 2.30 (0.83–6.36) | 0.1090 |
| ≥8 vs. ≤6 | 0.46 (0.07–3.21) | 0.4345 |
| cT stage (2 vs. 1) | 1.60 (0.41–6.30) | 0.5011 |
| Proportion of cores positive (>⅓ vs. ≤⅓) | 1.08 (0.43–2.73) | 0.8669 |
| Highest percent positive core | 0.90 (0.76–1.06) | 0.2109 |
| Log(1+baseline PSA) | 2.15 (1.10–4.20) | 0.0253 |
Validation cohort, n = 79.
The HR calculated for the 32-gene index is based on a five-unit change. All variables are pre-RP values.
Discussion
We identified a 32-gene prognostic signature for prostate cancer progression that includes genes from multiple functional families—including transcription factors, cell cycle genes, metabolic process genes, and genes with unknown functions—and developed a continuous risk index that allows for individualized risk assessment based on the gene expression profile of the tumor. The assay and risk index were validated in an independent, blinded cohort of 270 RP patients. The 32-gene risk index was an independent predictor of BCF after RP and added significant prognostic value to standard clinical and pathologic variables. The only markers in the multivariate analysis that were predictive of BCF were the 32-gene risk index score and surgical margin (Tables 3 and 4).
The 32-gene risk index described herein may provide additional prognostic information in several clinically relevant subgroups of patients. In RP patients, stratification of risk is used to determine the need for adjuvant therapy as well as the intensity of follow-up required. The current standard of care for adjuvant radiotherapy is based on the presence or absence of adverse pathologic features (21). As expected, the risk index identified most patients with pT3 disease or positive surgical margins as high risk; however, the risk index also identified a low-risk subset of patients—none of whom experienced a BCF event during the follow-up period (Fig. 3E). Conversely, in patients with pT2 disease and a negative surgical margin, who are considered to have a low risk of disease recurrence following RP, the 32-gene risk index provided a statistically significant stratification of patients and identified a high-risk group with a 41% probability of BCF (Fig. 3D). Notably, 28% of the patients characterized as high risk by the 32-gene risk index had a Gleason score of 6, indicating they likely would not have been assessed as high risk using conventional clinical parameters. Thus, clinicians may be able to integrate information from the 32-gene risk index to help make decisions about whether individual patients should be considered candidates for adjuvant therapy and more or less intensive follow-up. Although this study did not investigate the molecular mechanism underlying the 32-gene index predictions, we speculate that the gene expression profile can provide information regarding the aggressiveness of tumors not captured by traditional prognostic variables.
Another significant issue in prostate cancer management is the assessment of risk and treatment selection after a positive biopsy (active surveillance vs. radical treatment). An estimated 23–42% of all patients diagnosed with prostate cancer do not require treatment (22), and recent studies have demonstrated that 48 men must be treated to prevent one prostate cancer-related death (23). Although the 32-gene risk index was developed and validated using RP samples, a molecular-based prognostic test has potential clinical utility in malignant biopsy samples. As proof of concept, we examined matched biopsy and RP specimens and found a significant correlation between the risk index scores obtained in these two specimens. Several key obstacles in creating a biopsy tissue-based prognostic test are the multifocal nature of prostate cancer, the potential for misclassification due to sampling, and Gleason score heterogeneity. To assess the ability of the risk index to predict PSA recurrence consistently in this context, we identified a clinically relevant subgroup from our validation cohort—patients with a Gleason score of 6 on their biopsy tissue sample and a score of 7–9 on the RP tissue sample. The risk index scores were similar between biopsy and RP, indicating that the molecular signature identified by the 32-gene risk index may provide consistent data in the context of the multifocal nature of prostate cancer and the heterogeneity of Gleason scores. Finally, the Cox regression analyses of the 32-gene index scores derived from core biopsy tissue provide preliminary evidence that the risk index has prognostic ability above other clinicopathologic variables available at biopsy, including Gleason score, cT stage, number of positive cores, and highest percent positive core. It is important to note that the prognostic strength of some factors in these Cox analyses may be underestimated as a result of the smaller sample size in the biopsy cohort (n = 79): for example, the cohort included just seven patients with Gleason scores ≥8. If confirmed in larger follow-up studies including untreated patients, such a prognostic biomarker will be useful in stratifying patients who are at minimal risk for progression and therefore candidates for active observation.
In recent years, gene expression-based prognostic signatures have become standard of care in breast cancer; however, to date, genomic markers have not been incorporated into standard clinical practice in prostate cancer. Several prognostic gene expression signatures have been described (24–26), whereas several groups have described unsuccessful efforts to identify prognostic gene expression-based signatures (27, 28). Recently, a prognostic marker based on the expression of 31 cell cycle proliferation genes was described and examined in two cohorts: patients who had undergone RP and patients diagnosed by use of transurethral resection of the prostate (TURP) in the United Kingdom and were managed conservatively (26). In the post-RP cohort, the signature significantly stratified patients based on risk and added value to standard clinicopathologic variables in a multivariate model. The clinical utility of the signature is unclear as the lowest-risk group presented had a 10-y risk of biochemical recurrence of 23.7%, a threshold likely too high to impact treatment decisions. Whether the signature can define a clinically relevant lower-risk group alone or in combination with other clinicopathologic variables has not been reported. The TURP cohort had more aggressive and advanced disease at diagnosis (i.e., patients were all symptomatic); thus, prostate cancer mortality could be modeled. However, the disease progression at diagnosis and the conservative treatment approach for this cohort are unusual in contemporary clinical practice. The authors note the need for validation of the signature using needle biopsy tissue in patients treated according to current practice standards.
Limitations of our study are as follows: First, it is not representative of all patients who receive therapy for prostate cancer, as it includes only patients treated with RP; second, the samples were obtained from a single institution. The strengths of the study include the low number of patients lost to follow-up, a long follow-up time (∼14 y), and uniformity of the surgery due to select surgeons performing the procedure in a high-volume center. The 32-gene risk index requires further validation in a multi-institutional cohort or clinical trial setting. In addition, the utility of the risk index in providing prognostic information at the biopsy stage will be assessed.
Materials and Methods
Patients and Tissue Specimens.
Tissue samples for the discovery (n = 209) and validation (n = 306) cohorts were obtained from consecutive patients who underwent RP surgery as part of their clinical care at Massachusetts General Hospital (MGH; Boston, MA) from September 1993 to September 1995 and January 1996 to December 1997, respectively (Fig. 2, Table 1, and Tables S1, S8 and S9). In addition, malignant tissue samples from presurgery prostate core needle biopsies were obtained from patients in the validation cohort whose initial biopsies were performed at MGH (n = 79). Medical records were reviewed, and clinical data captured included demographic information, presurgery PSA, pT stage, surgical margin status, Gleason score, time to BCF, time to metastasis, and overall survival (Table 1). For the biopsy cohort, additional data captured included cT stage, number of positive and negative cores, and highest percent tumor content. The Gleason score was rereviewed by a single urological pathologist (C.-L.W.) based on the modified Gleason classification (29). BCF was defined as a post-RP detectable serum PSA. The detection threshold was the generally accepted, contemporary threshold value at the time, above which a serum PSA could be detected by a laboratory-available standard PSA assay. From 1993 to 2001, this detection limit decreased from 0.5 ng/mL to 0.1 ng/mL. Most patients (87%) had a second, confirmatory PSA test on record. Metastasis events were captured based on imaging reports and clinical notes in the medical record indicating metastatic disease. Metastasis-free data points were defined as follow-up visits with no indication of metastasis. Survival information was captured from MGH medical records and social security database inquiries. In the absence of a confirmed death event, the last confirmed patient follow-up visit in which a PSA test was obtained was used as the censored time in overall survival analyses. Exclusion criteria included neoadjuvant or adjuvant therapy before BCF and lymph node metastasis present at RP. Additional patients not included were those with no follow-up clinical information, no tumor tissue available, and insufficient RNA quantity or quality. For the validation cohort, patients also were required to have a documented post-RP PSA nadir and at least one PSA measurement >6 mo post-RP. The study was approved by the institutional review board at MGH. Informed and signed consent was obtained from all patients for the use of their tissue at the time of surgery.
All tissues were formalin fixed and paraffin embedded. Tumor samples were sectioned into ∼7-µm–thick tissue sections, and the area of the tissue with the highest histologic tumor grade was marked by a pathologist (C.-L.W.) for isolation and analysis. Tissue was isolated by manual macrodissection. The lower limit of tumor tissue accepted was 70%. Tissue samples were processed and RNA isolated as described in SI Materials and Methods.
Discovery of a Prognostic Gene Set.
Gene expression profiling was performed on tissue samples from the discovery cohort via the cDNA-mediated annealing, selection, extension, and ligation assay (Illumina) (30, 31). In total, 1,536 genes were selected for profiling based on a review of the scientific literature for candidate prostate cancer-related genes and pilot studies using gene expression profiling of prostate cancer tissue of different Gleason grades compared with nonmalignant prostate tissue. See SI Materials and Methods and SI Results for additional details on the methods and statistical analyses of the gene expression data.
Construction of a Continuous Risk Index.
A continuous risk index was constructed using gene expression data from the real-time RT-PCR assays and clinical information from the discovery cohort. The discovery cohort was divided into a training set (n = 134) and a test set (n = 61; Fig. S1). Principal components were calculated in the training set and fitted into a Cox regression model. Significant principal components were retained for risk index calculation. The final coefficients for individual genes were a combination of coefficients from the principal components and coefficients for principal components from the Cox model. A continuous risk model was constructed and scaled into a risk index with a range of 0–10. Individual risk scores were calculated as the level of expression of each gene multiplied by the corresponding coefficient and normalized to the 10-point scale. The performance of the risk model was evaluated using the test set (Fig. S1). Finally, the entire discovery cohort (n = 195) was used to construct the final continuous risk index. Cut points establishing low-, intermediate-, and high-risk groups were designated a priori corresponding to 10-y BCF rates of ∼20% and 40% in the discovery cohort (cut points: 3.09 and 4.18). These cut points were chosen as proof of concept based on clinical judgment of potential thresholds for clinical utility (e.g., conservative approach vs. adjuvant therapy).
Statistical Analyses.
The primary clinical endpoint was time to BCF. Demographics and patient characteristics were analyzed by descriptive statistics: t tests were used for continuous variables; Fisher’s exact test was used for categorical variables. Cumulative incidence was used to estimate probability of BCF and metastasis. Differences among risk groups were tested by Gray’s K-sample test (32). Univariate and multivariate Cox proportional hazards regression analyses were used to evaluate independent prognostic factors associated with BCF. The HR for the 32-gene index was based on a five-unit change. Missing presurgery PSA values (n = 36) were imputed by the predictive mean matching method (33, 34); predictor variables included Gleason score, pT stage, lymph node metastasis, and surgical margin. A P value less than 0.05 was considered indicative of statistical significance, and all tests were two-sided. Calibration was assessed visually by dividing patients into quintiles of the risk index-predicted freedom from BCF probabilities, then plotting the mean risk index-predicted freedom from recurrence probability against the Kaplan–Meier estimated freedom from BCF for each quintile. Discrimination was quantified by the concordance index, which is identical to the nonparametric area under the receiver operating characteristic (ROC) curve in the binary setting. Comparison of 32-gene risk score with the three nomogram-based predictions was done by NRI using the same risk probability thresholds. A value x% of NRI indicates that, compared with individuals without an event (BCF or metastatic event), individuals with an event are x% more likely to be classified into a higher-risk group by the 32-gene risk score than by nomogram. A concordance correlation was used to examine the agreement between core biopsy tissue-derived prostate cancer risk scores and RP-derived risk scores. All analyses were performed using the R statistical software package, version 2.11.1 (www.r-project.org). Additional details are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Hongying Yi, PhD, for statistical support; Jingmei Su, Yen Tran, and J. Todd Tuggle for study assistance; Sandra Kirley, BA, for collection of data and preparation of tissue samples; and Dennis Sgroi, MD, and Donald S. Kaufman, MD, for advice. C.-L.W. received research funding from bioTheranostics, Inc. John and Claire Bertucci Prostate Cancer Foundation provided funding support.
Footnotes
Conflict of interest statement: C.-L.W. received research support from bioTheranostics, Inc. for this study. W.S.M., C.-L.W., and M.W.K. have served on an advisory board for bioTheranostics, Inc. C.J.C. has served as a consultant for bioMerieux and bioTheranostics. B.E.S, R.S., Y.Z., C.A.S., and M.G.E. are employees and stockholders of bioTheranostics, Inc.
Data from the discovery phase of this study were presented, in part, at the 2007 American Society of Clinical Oncology Prostate Cancer Symposium.
*This Direct Submission article had a prearranged editor.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/projects/geo (accession no. GSE44353).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215870110/-/DCSupplemental.
References
- 1.American Cancer Society . Atlanta: American Cancer Society; 2010. Cancer Facts and Figures 2010. [Google Scholar]
- 2. Agency for Healthcare Research and Quality (2011) HCUPnet, Healthcare Cost and Utilization Project. Available at http://hcupnet.ahrq.gov. Accessed May 13, 2011.
- 3.Amling CL, et al. Long-term hazard of progression after radical prostatectomy for clinically localized prostate cancer: Continued risk of biochemical failure after 5 years. J Urol. 2000;164(1):101–105. [PubMed] [Google Scholar]
- 4.Han M, Partin AW, Pound CR, Epstein JI, Walsh PC. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy. The 15-year Johns Hopkins experience. Urol Clin North Am. 2001;28(3):555–565. doi: 10.1016/s0094-0143(05)70163-4. [DOI] [PubMed] [Google Scholar]
- 5.Hull GW, et al. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. J Urol. 2002;167(2 Pt 1):528–534. doi: 10.1016/S0022-5347(01)69079-7. [DOI] [PubMed] [Google Scholar]
- 6.Bolla M, et al. European Organization for Research and Treatment of Cancer Postoperative radiotherapy after radical prostatectomy: A randomised controlled trial (EORTC trial 22911) Lancet. 2005;366(9485):572–578. doi: 10.1016/S0140-6736(05)67101-2. [DOI] [PubMed] [Google Scholar]
- 7.Thompson IM, Jr, et al. Adjuvant radiotherapy for pathologically advanced prostate cancer: A randomized clinical trial. JAMA. 2006;296(19):2329–2335. doi: 10.1001/jama.296.19.2329. [DOI] [PubMed] [Google Scholar]
- 8.Wiegel T, et al. Phase III postoperative adjuvant radiotherapy after radical prostatectomy compared with radical prostatectomy alone in pT3 prostate cancer with postoperative undetectable prostate-specific antigen: ARO 96-02/AUO AP 09/95. J Clin Oncol. 2009;27(18):2924–2930. doi: 10.1200/JCO.2008.18.9563. [DOI] [PubMed] [Google Scholar]
- 9.Thompson IM, et al. Adjuvant radiotherapy for pathological T3N0M0 prostate cancer significantly reduces risk of metastases and improves survival: Long-term followup of a randomized clinical trial. J Urol. 2009;181(3):956–962. doi: 10.1016/j.juro.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Psutka SP, et al. Men with organ-confined prostate cancer and positive surgical margins develop biochemical failure at a similar rate to men with extracapsular extension. Urology. 2011;78(1):121–125. doi: 10.1016/j.urology.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Freedland SJ, Partin AW, Epstein JI, Walsh PC. Biochemical failure after radical prostatectomy in men with pathologic organ-confined disease: pT2a versus pT2b. Cancer. 2004;100(8):1646–1649. doi: 10.1002/cncr.20145. [DOI] [PubMed] [Google Scholar]
- 12.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90(10):766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 13.Greene KL, et al. Validation of the Kattan preoperative nomogram for prostate cancer recurrence using a community based cohort: Results from cancer of the prostate strategic urological research endeavor (capsure) J Urol. 2004;171(6 Pt 1):2255–2259. doi: 10.1097/01.ju.0000127733.01845.57. [DOI] [PubMed] [Google Scholar]
- 14.Makarov DV, et al. Updated nomogram to predict pathologic stage of prostate cancer given prostate-specific antigen level, clinical stage, and biopsy Gleason score (Partin tables) based on cases from 2000 to 2005. Urology. 2007;69(6):1095–1101. doi: 10.1016/j.urology.2007.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson AJ, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2005;23(28):7005–7012. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 17.Dowsett M, et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J Clin Oncol. 2010;28(11):1829–1834. doi: 10.1200/JCO.2009.24.4798. [DOI] [PubMed] [Google Scholar]
- 18.Albain KS, et al. Breast Cancer Intergroup of North America Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17(5):1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 20.Kattan MW, et al. Preoperative and postoperative nomograms incorporating surgeon experience for clinically localized prostate cancer. Cancer. 2009;115(5):1005–1010. doi: 10.1002/cncr.24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohler J, et al. NCCN clinical practice guidelines in oncology: Prostate cancer. J Natl Compr Canc Netw. 2010;8(2):162–200. doi: 10.6004/jnccn.2010.0012. [DOI] [PubMed] [Google Scholar]
- 22.Draisma G, et al. Lead time and overdiagnosis in prostate-specific antigen screening: Importance of methods and context. J Natl Cancer Inst. 2009;101(6):374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schröder FH, et al. ERSPC Investigators Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360(13):1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 24.Stephenson AJ, et al. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104(2):290–298. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talantov D, et al. Gene based prediction of clinically localized prostate cancer progression after radical prostatectomy. J Urol. 2010;184(4):1521–1528. doi: 10.1016/j.juro.2010.05.084. [DOI] [PubMed] [Google Scholar]
- 26.Cuzick J, et al. Transatlantic Prostate Group Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011;12(3):245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sboner A, et al. Molecular sampling of prostate cancer: A dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. ISUP Grading Committee The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29(9):1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 30.Fan JB, et al. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 2004;14(5):878–885. doi: 10.1101/gr.2167504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibikova M, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol. 2004;165(5):1799–1807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16(3):1141–1154. [Google Scholar]
- 33.Little RJA. Missing data adjustments in large surveys. J Bus Econ Stat. 1988;6:287–296. [Google Scholar]
- 34.Van Buuren S, Groothuis-Oudshoorn K. MICE: Multivariate imputation by chained equations in R. J Stat Softw. 2011;45(3):1–67. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.