Abstract
Statins are cholesterol-lowering drugs that inhibit 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) reductase, the rate-limiting enzyme in the synthesis of cholesterol via the mevalonate pathway. This pathway also produces coenzyme Q (a component of the respiratory chain), dolichols (important for protein glycosylation), and isoprenoids (lipid moieties responsible for the membrane association of small GTPases). We previously showed that the nematode Caenorhabditis elegans is useful to study the noncholesterol effects of statins because its mevalonate pathway lacks the sterol synthesis branch but retains all other branches. Here, from a screen of 150,000 mutagenized genomes, we isolated four C. elegans mutants resistant to statins by virtue of gain-of-function mutations within the first six amino acids of the protein ATFS-1, the key regulator of the mitochondrial unfolded protein response that includes activation of the chaperones HSP-6 and HSP-60. The atfs-1 gain-of-function mutants are also resistant to ibandronate, an inhibitor of an enzyme downstream of HMG-CoA reductase, and to gliotoxin, an inhibitor acting on a subbranch of the pathway important for protein prenylation, and showed improved mitochondrial function and protein prenylation in the presence of statins. Additionally, preinduction of the mitochondrial unfolded protein response in wild-type worms using ethidium bromide or paraquat triggered statin resistance, and similar observations were made in Schizosaccharomyces pombe and in a mammalian cell line. We conclude that statin resistance through maintenance of mitochondrial homeostasis is conserved across species, and that the cell-lethal effects of statins are caused primarily through impaired protein prenylation that results in mitochondria dysfunction.
The mevalonate pathway of cholesterol biosynthesis is the primary pathway by which cholesterol is synthesized de novo in mammals and is also essential for the synthesis of other important molecules, such as the short lipids containing prenyl groups that are attached to small GTPases and target them to membranes; dolichol-P, an intermediate during protein glycosylation; and coenzyme Q (CoQ), a soluble antioxidant that is also part of the respiratory chain in mitochondria (1, 2). The rate-limiting step of the mevalonate pathway is catalyzed by 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) reductase and the inhibitors of this enzyme (statins) are widely prescribed to lower blood cholesterol levels. Whereas the great majority of patients tolerate statins very well, some patients experience adverse side effects including muscle pains or even rhabdomyolysis (3). Statins also have anti-inflammatory effects (4) and are promising anticancer agents (5). The molecular basis for many of the noncholesterol-mediated effects of statins are poorly understood, which curtails their usefulness and makes it impossible at present to predict which patients are at risk for developing adverse effects.
Caenorhabditis elegans is eminently suited to study the cholesterol-independent effects of statins because it lacks the sterol synthesis branch of the mevalonate pathway but retains the other branches (Fig. 1A) (6–9). In C. elegans, statins cause decreased protein prenylation, induction of the endoplasmic reticulum unfolded protein response (UPRer), growth arrest, and lethality; these are all on-target effects of statins because they can be abrogated by the inclusion of mevalonate in the culture medium (9). To identify cellular processes that can compensate for a reduced output from the mevalonate pathway, we isolated C. elegans mutants that are resistant to statins and found that these are all gain-of-function (gof) mutations in the protein ATFS-1, the key regulator of the mitochondrial unfolded protein response (UPRmt) (10–13).
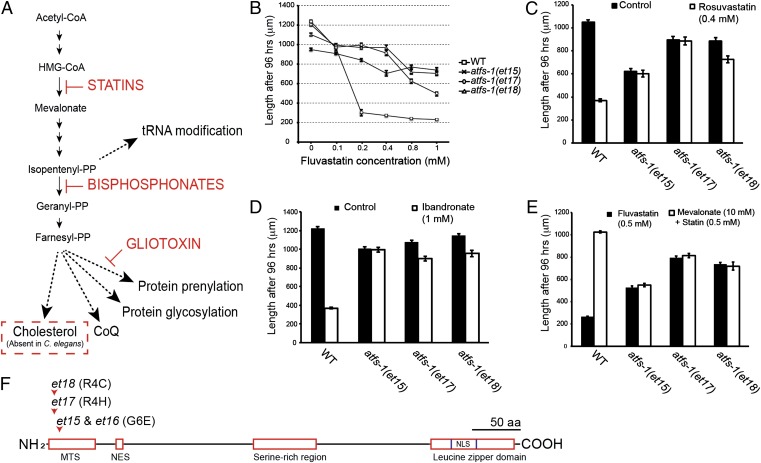
Fig. 1.
Mutations in atfs-1 confer resistance to inhibitors of the mevalonate pathway. (A) Simplified version of the mevalonate pathway, its subbranches, and sites of action of various inhibitors. The cholesterol branch of the pathway is absent in C. elegans. (B) Fluvastatin dose–response curve of different atfs-1 alleles. The unique atfs-1 alleles confer resistance to rosuvastatin (C) and to ibandronate (D). (E) Mevalonate abrogates the effects of fluvastatin on wild-type worms, but has no effect on three gain-of-function atfs-1 mutants. (F) Schematics of the ATFS-1 protein indicating the position and nature of the four atfs-1 alleles isolated in this study (et15–et18). MTS, mitochondrial targeting sequence; NES, nuclear export signal; NLS, nuclear localization signal.
Results
Isolation of Four Statin-Resistant C. elegans Mutants.
We screened ∼150,000 randomly mutagenized haploid genomes and isolated four mutant alleles (et15–et18) that confer statin resistance in C. elegans (et15 and et16 are molecularly identical, and only data from et15 will be shown in most experiments because all four alleles behaved similarly). The isolated mutants are resistant to two statins (fluvastatin and rosuvastatin) and to ibandronate (which inhibits farnesyl diphosphate synthase, i.e., several steps downstream of HMG-CoA reductase) (Fig. 1 B–D). These results indicate that all four mutants can compensate for inhibition of the mevalonate pathway. The statin-resistant mutants also improved the growth of a lethal HMG-CoA reductase null mutant but only when small doses of mevalonate were also provided, suggesting that some residual HMG-CoA reductase activity persists in statin-treated worms (Fig. S1A). Conversely, providing the mutants with mevalonate did not improve their growth or their resistance to fluvastatin, which is consistent with the fact that they grow as well on 0.5 mM statins as they do on control plates (Fig. 1E). Finally, there is no evidence that the mutant alleles et15-et18 confer general resistance against xenobiotics because they exhibited no resistance to several tested growth inhibitors (Fig. S1 B–D).
Gain-of-Function Alleles of ATFS-1 Confer Statin Resistance.
Using a gene identification strategy based on outcrossing and whole-genome sequencing (14, 15) (Fig. S2), we found that all four statin-resistant mutants carried substitution mutations in the mitochondrial targeting signal (MTS) of the protein ATFS-1, specifically at amino acid positions 4 or 6 (Fig. 1F). ATFS-1 is a leucine zipper transcription factor that contains an MTS at its N terminus and a nuclear localization signal (NLS) at its C terminus (10–13). The primary function of ATFS-1 is to activate the UPRmt. In the absence of mitochondrial stress, ATFS-1 is effectively recruited to the mitochondria where it is degraded; during mitochondrial stress, ATFS-1 is not efficiently targeted to the mitochondria and therefore allowed to accumulate in the nucleus and activate target genes, including the mitochondrial chaperones HSP-6 and HSP-60.
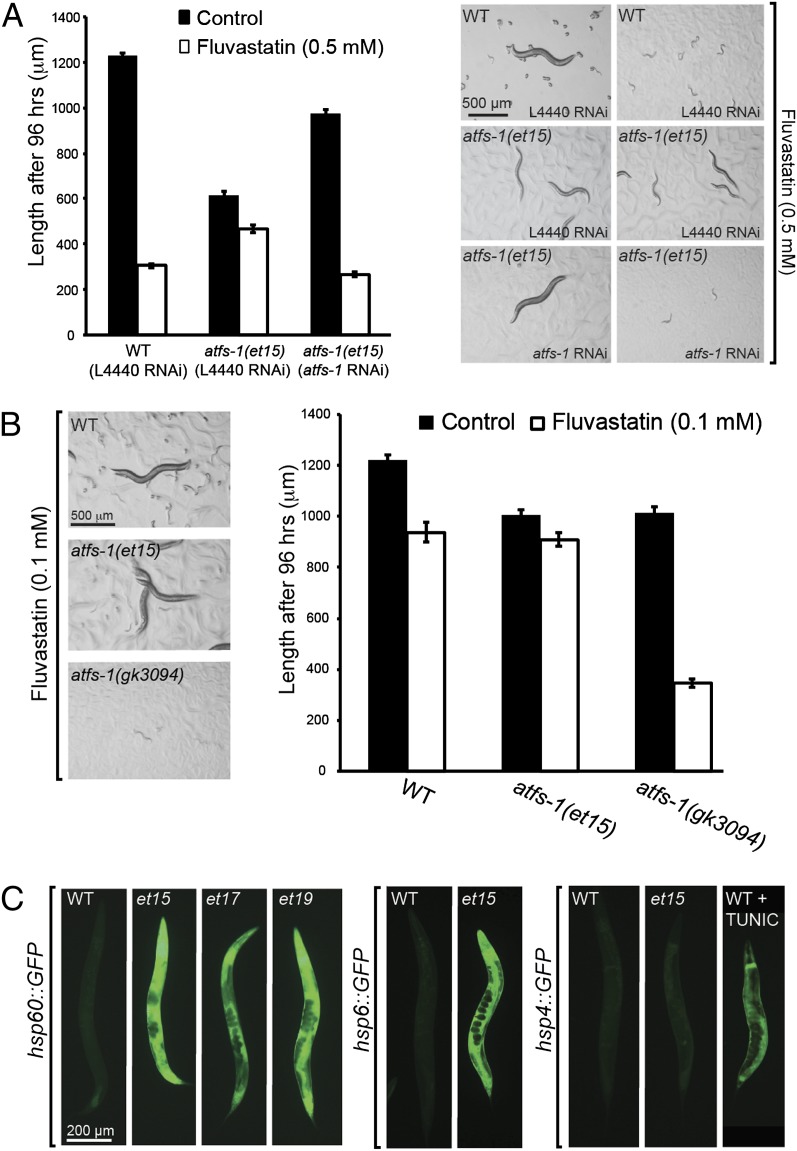
The statin-resistant atfs-1 alleles are gof mutations because: (i) RNAi against atfs-1 eliminates statin resistance in the mutants (Fig. 2A); (ii) the null atfs-1(gk3094) allele is hypersensitive to statin (Fig. 2B); (iii) the UPRmt reporters hsp-60::GFP and hsp-6::GFP (but not the UPRer reporter hsp-4::GFP) are constitutively expressed in the mutants (Fig. 2C); (iv) the statin-resistant atfs-1 mutants all grow rather poorly on normal plates (Fig. S3) and this phenotype is abrogated by treating the mutants with atfs-1 RNAi (Fig. 2A); and (v) atfs-1(et15) heterozygous worms are resistant to statins (79 of 80 cross-progeny from a wild-type male and an atfs-1(et15) hermaphrodite reached the L4 or adult stages within 72 h on 0.5 mM fluvastatin). Constitutive activation of the UPRmt is, therefore, the most likely mechanism by which the atfs-1(gof) alleles confer resistance to statins while also reducing the growth rate on control plates. This is consistent with the improved protection of the atfs-1(et15) gof mutant against ethidium bromide (EtBr) (Fig. S4 A–C), which impairs replication and transcription of the mitochondrial genome in cultured cells thus reducing the synthesis of mitochondrially encoded polypeptides (16–19), and the improved respiration of this mutant when challenged with statin (see below).
Fig. 2.
The unique atfs-1 mutants carry gain-of-function alleles that activate the UPRmt. (A) RNAi against atfs-1 suppresses the fluvastatin resistance of the atfs-1(et15) allele. The control RNAi vector (L4440) has no effect. (B) The atfs-1(gk3094) loss-of-function mutant is hypersensitive to fluvastatin: it fails to grow on 0.1 mM, a concentration that has only a minor effect on wild-type and no effect on the atfs-1(et15) gain-of-function allele. (C) The atfs-1 alleles et15, et17, and et18 display constitutive expression of two UPRmt reporters (hsp-60::GFP and hsp-6::GFP) but not of a UPRer reporter (hsp4::GFP), which can be activated by 2 μg/mL tunicamycin (Right).
Mechanistically, we propose that the atfs-1 mutants are gof alleles because the amino acid substitutions disrupt the MTS, hence allowing for an increased nuclear localization of this protein, as is the case for mutant transgenes that are entirely lacking the MTS (11). Consistent with this model, we found that providing an atfs-1 transgene lacking the MTS to atfs-1(gk3094) null mutants confers statin resistance, whereas a similar transgene lacking both the MTS and the NLS confers no resistance at all (Fig. S5).
UPRmt Activation Protects Against Statins in Worms, Yeast, and Mammalian Cells.
Activation of the UPRmt by atfs-1 gof alleles protects against statins in C. elegans, but statin treatment by itself does not detectably activate the UPRmt, as judged using the hsp-60::gfp reporter (Fig. S4D). This is presumably because of the specific nature of the statin effect on mitochondria. We however reasoned that inducing UPRmt through other means ought to confer statin resistance in wild-type worms. UPRmt can be induced in C. elegans by exposure to EtBr, which impairs mitochondrial DNA replication, or paraquat, which causes oxidative stress (19, 20). Both treatments conferred statin resistance to wild-type C. elegans (Fig. 3 A and B and Fig. S6 A and B).
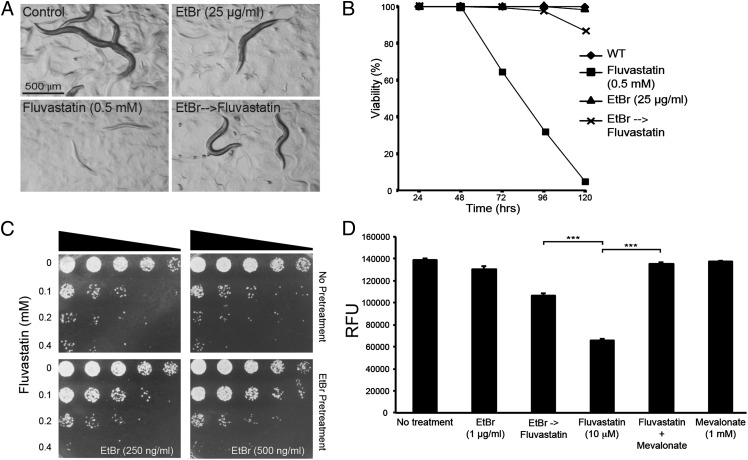
Fig. 3.
Preinduction of UPRmt using ethidium bromide protects against the adverse effects of statins in nematodes, yeast, and mammalian cells. (A and B) Worms pretreated with ethidium bromide are viable and grow into fertile adults when subsequently cultivated on 0.5 mM fluvastatin. (C) The fission yeast S. pombe tolerates higher doses of fluvastatin when pretreated with 250 ng/mL or 500 ng/mL ethidium bromide. (D) The mammalian fibroblast cell line NIH 3T3 shows better viability in the presence of 10 μM fluvastatin when it has been pretreated with 1 μg/mL ethidium bromide; bars show the average readout from the PrestoBlue cell viability assay (Invitrogen) ±SEM (n > 5 wells; ***P < 0.001).
The UPRmt may be an evolutionarily conserved mechanism to cope with the consequences of an impaired mevalonate pathway because statin resistance was also induced in the yeast Schizosaccharomyces pombe (Fig. 3C) and the mammalian fibroblast line NIH 3T3 when the mitochondrial stress response was activated using EtBr (Fig. 3D and Fig. S6C). Conservation of this phenomenon in C. elegans, S. pombe, and mammals is especially important in view of the fact that the latter two types of organisms do have the branch of the mevalonate pathway that leads to sterol synthesis (1, 21, 22), which is lacking in C. elegans. In other words, the cytotoxic effects of statins are primarily related to mitochondria homeostasis even in organisms where the main output of the pathway is considered to be sterols.
Effects of Statins on Mitochondria Are Not Related to Inhibition of CoQ Synthesis.
The fact that the UPRmt protects against the deleterious effects of statins suggests that these inhibitors interfere with mitochondria homeostasis. One mechanism for this could be that inhibiting the mevalonate pathway results in reduced synthesis of CoQ and failure of the respiratory chain in mitochondria (23, 24). Indeed, fluvastatin treatment does cause reduced respiration in C. elegans, and the atfs-1(et15) mutant respires normally even in the presence of fluvastatin (Fig. S7A). However, several lines of evidence argue against CoQ depletion accounting for the effects of statins on mitochondria: (i) supplying CoQ to C. elegans conferred no protection from the toxic effects of statins (CoQ10 was tested at 50 μg/mL and concentrations of CoQ9 ranging from 10 to 80 μg/mL were tested, all without effect); (ii) the atfs-1(et15) mutant is not resistant to rotenone, antimycin A, or sodium azide, which are inhibitors of the mitochondrial respiratory chain (25, 26) (Fig. S7 B–D); and (iii) it is well known that CoQ is dietarily available to C. elegans that are fed Escherichia coli as in our experiments (27, 28) and that very little endogenous CoQ is sufficient for a wild-type phenotype even in the absence of dietary CoQ (29).
ATFS-1 Gain-of-Function Mutants Are Resistant to Prenylation Inhibition.
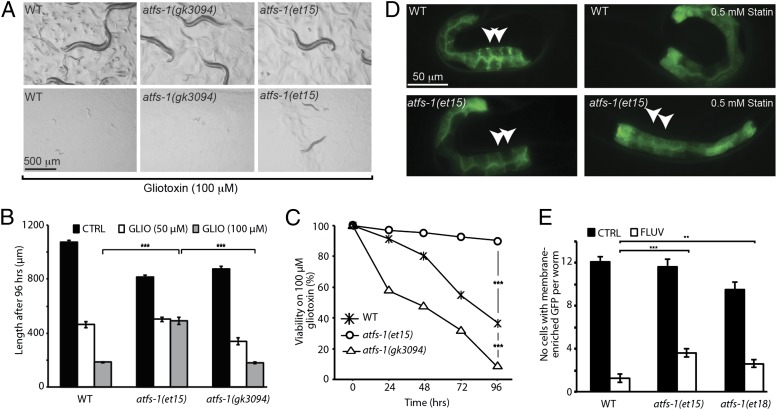
Small GTPases, especially those of the RAB family, are essential for intracellular trafficking and organelle homeostasis (5). Their activity depends on the addition of prenyl groups, i.e., farnesyl pyrophosphate or geranylgeranyl pyrophosphate, that are synthesized through the mevalonate pathway. We previously showed that statins inhibit protein prenylation in C. elegans (9) and it is therefore possible that statins impair mitochondria by preventing the prenylation of small GTPases. This hypothesis predicts that the atfs-1(gof) mutants that are resistant to statins should also be resistant to more specific inhibitors of prenylation. Gliotoxin is an inhibitor of farnesyl transferase, the enzyme that ligates farnesyl groups to the C-terminal end of small GTPases, resulting in growth defects and lethality in worms (30). Gliotoxin also suppresses the effect of an activated form of the RAS GTPase (31). We found that the atfs-1(et15) gof mutant is resistant to gliotoxin, whereas the atfs-1(gk3094) null mutant is hypersensitive (Fig. 4 A–C). These results are consistent with the hypothesis that statins exert their negative effects on mitochondria via inhibition of small GTPases. The atfs-1(et15) and atfs-1(et18) mutants were also partially resistant to the prenylation inhibitory effects of statins (Fig. 4 D and E).
Fig. 4.
The atfs-1(gof) alleles protect C. elegans against prenylation inhibition. (A) Images of wild-type (WT), atfs-1(gk3094), and atfs-1(et15) worms cultivated for 96 h on control or gliotoxin (100 μM) plates. The gain-of-function atfs-1(et15) allele confers partial resistance to gliotoxin, whereas the loss-of-function atfs-1(gk3094) allele confers hypersensitivity as assessed by measuring growth (B) or viability (C) after 96 h on 100 μM gliotoxin. (D) The atfs-1(et15) allele can partially rescue the protein prenylation defects caused by fluvastatin. The protein prenylation reporter (pGLO-1P::GFP-CAAX) shows clear membrane enrichment in untreated L1 larvae for both wild-type and atfs-1(et15). In L1 larvae from parents cultivated in the presence of 0.5 mM fluvastatin for 48 h, only the atfs-1(et15) mutant continues to show membrane enrichment (white arrowheads). (E) Quantification of the prenylation assay using pGLO-1P::GFP-CAAX. The results are presented as the number of intestinal cells showing distinct membrane enrichment. Note that both atfs-1(et15) and atfs-1(et18) show a slight but significant increase in prenylation. (n > 35 worms; **P < 0.01, ***P < 0.001).
Discussion
We found that statins disturb mitochondrial homeostasis, and that activation of the UPRmt machinery is required in C. elegans to achieve statin resistance. Furthermore, this mechanism of resistance is conserved in yeast and mammalian cells. The atfs-1(gof) mutants that we isolated by virtue of the statin resistance that they confer to C. elegans all introduce point mutations in the recently described N-terminal mitochondrial targeting signal of ATFS-1 (11). Under normal conditions, ATFS-1 is actively transported to the mitochondria, where it is rapidly degraded. During mitochondrial stress, its mitochondrial targeting efficiency decreases and more protein is available to be targeted to the nucleus, activating mitochondrial chaperone genes. We speculate that the atfs-1(gof) mutations described here suppress the mitochondrial targeting of the protein, thus improving its nuclear targeting efficiency. This is consistent with the finding that all three mutants constitutively up-regulated mitochondrial-specific chaperones (HSP-6 and HSP-60) and improved the mitochondrial function upon statin treatment.
As in other organisms, the mevalonate pathway in C. elegans is important for the synthesis of isoprenoids (farnesyl pyrophosphate and geranylgeranyl pyrophosphate), short lipid moieties that can be attached to small GTPases such as RHO, RAS, and RAB to target them to the membrane, which is essential for their proper functioning (32, 33). Inhibition of the mevalonate pathway by statins leads to depletion of these end-products in mammalian cells, which results in defective prenylation of the GTPases (33). Similarly, statins cause protein prenylation defects in C. elegans (9), which could be partially rescued in statin-resistant mutants (present study). These same mutants were also resistant to the farnesyl-transferase inhibitor gliotoxin, which specifically blocks prenylation. Altogether, these findings argue in favor of the prenylation branch being the critical branch of the mevalonate pathway that is affected by statin treatment and show that activation of the UPRmt machinery can help preserve mitochondria homeostasis in the presence of statins, enabling the cells to better use the residual output from the mevalonate pathway, hence sustaining essential GTPase prenylation (Fig. S8). How the UPRmt helps compensate for a reduced output of the mevalonate pathway is not known at present but one possibility is that it lessens the turnover rate of proteins that rely on prenylated GTPases for their transport to mitochondria.
Polymorphisms in the mitochondrial ribosome recycling factor EF-G2mt/MEF2 increase atorvastatin toxicity in yeast and mammalian cells (34), suggesting that mitochondria prone to homeostatic failure are extra sensitive to statins. This is consistent with a number of studies showing the mitochondria to be the main site of action of the noncholesterol effects of statins (3). Multiple studies have shown that statin treatment impacts on the prenylation of small GTPases (35–37), and the present study establishes a clear link between prenylation inhibition and impaired mitochondria homeostasis. It seems likely that inhibition of protein prenylation should impact many cellular processes and organelles besides the mitochondria. Indeed, statins strongly induce the UPRer in C. elegans, suggesting not only that the endoplasmic reticulum is impacted by inhibition of the mevalonate pathway, but also that the cell is able to activate an appropriate stress response (9). The mitochondrial stress caused by statins is therefore unusual in that it does not result in the activation of UPRmt even though it is evidently the required protective response.
In conclusion, our results suggest that UPRmt can help preserve mitochondria homeostasis in the presence of statins, which allows the cells to better use the residual output from the mevalonate pathway, hence sustaining essential GTPase prenylation. It would be interesting to determine whether the regulation of UPRmt varies among individuals, and whether such variation contributes to the varied susceptibility to developing side effects seen among patients receiving statin treatment.
Experimental Procedures
Nematode Strains and Maintenance.
All genotypes were maintained as described previously (38) and grown at 20 °C unless otherwise stated. The Bristol strain N2 was used as wild type (WT) in all experiments. The following alleles and transgenic lines were obtained from the Caenorhabditis Genetics Center: zcIs4[phsp4::GFP], zcIs9[hsp-60::GFP], zcIs13[hsp-6::GFP], and atfs-1(gk3094).
Mutagenesis and Isolation of Statin-Resistant Mutants.
N2 worms were mutagenized for 4 h by incubation in the presence of 0.5% ethyl methane sulfonate (EMS) according to the standard protocol (38). L1 larvae from the F2 generation were transferred to new plates containing 0.5 mM or 1.0 mM fluvastatin and then screened from days 5 to 10 to identify statin-resistant mutants, which were picked to new plates for further analysis. The isolated mutant alleles, named et15 to et18 were outcrossed 4 to 6 times before whole genome sequencing (see below), and 10 times before their phenotypic characterization and studies in experiments. Outcrossing was done by mating wild-type N2 males to a suppressor and then crossing the male progeny to wild-type hermaphrodites; individual progeny from this cross were picked to individual plates and then screened for resistance to statin. Five such cycles were carried out, amounting to 10 outcrosses.
Whole Genome Sequencing.
The genomes of suppressor mutant that had been outcrossed 4 or 6 times were sequenced to a depth of 25–40 times as previously described (15). The sequencing results were analyzed using the MAPQGene software to produce tables listing all differences between the reference N2 genome and that of the mutants and sort these differences by criteria such as noncoding substitutions, termination mutations, splice-site mutations, etc. (39). For each suppressor mutant, one or two hot spots, i.e., small genomic areas containing several mutations, were identified, which is in accordance with previous reports (14). Mutations in the hot spot that were still retained after 10 outcrosses were considered candidate statin-resistance mutations. In the case of et15–et18, it was mutations in the gene atfs-1 that conferred resistance to statins, as described above.
RNAi Feeding Experiment.
RNAi knockdown against atfs-1 was performed by feeding worms the corresponding RNAi bacterial clone from a previously published RNAi library (40). The clones were revived by culturing them overnight in LB media with ampicillin. These cultures were seeded on RNAi plates, and bacteria carrying an empty L4440 vector were used as controls. The plates were incubated for 24 h before placing five young adult worms on them. The worms were allowed to grow and reproduce until many gravid worms were present. The worms were collected and bleached to obtain a synchronized L1 larvae population. The L1 larvae were placed on appropriate plates with RNAi to determine the effects of the RNAi.
Generation of Transgenic Animals.
The plasmids pATFS-1(Δ1–32) and pATFS-1(Δ1–32 ΔNLS) bearing heat-shock–inducible mutated versions of atfs-1 have been described previously (11). Germ-line transformation was performed as described by Mello et al. (41) and the dominant rol-6(su1006) was used as a marker for transgenic worms. Plasmids were prepared with a Qiagen miniprep kit and used with the following concentrations: pRF4(rol-6) of 25 ng/µL, test plasmids of 5 ng/µL, and pBSKS (Stratagene) of 70 ng/µL.
Brood Size Assay.
At least 15 L4 transgenic or control worms were heat shocked for 1 h at 33 °C and then singled out on control or statin plates. The worms were transferred daily during the fertile period and progeny were counted 2 d after removal of the hermaphrodite.
Drug Treatment and Length Measurement.
Plates with different concentrations of fluvastatin (brand Lescol; Novartis) were made as described in a previous study (9). Briefly, 40 mg of fluvastatin was dissolved in water (2.31 mL) and spun at 5,000 × g to separate insoluble components. The solution was filter sterilized and the OD305nm was measured to determine the final concentration using a standard curve plotted from a known concentration of fluvastatin. The following compounds were also used: rosuvastatin (brand Crestor, AstraZeneca), mevalonolactone (Sigma), gliotoxin (Sigma), ibandronate (Sigma), ethidium bromide (Sigma), paraquat (Sigma), rotenone (Sigma), antimycin (Sigma), and sodium azide (Sigma). Drugs were dissolved in water except for gliotoxin, rosurvastatin, paraquat, and rotenone, which were dissolved in DMSO, and antimycin, which was dissolved in ethanol. Synchronized L1 larvae were placed on bacteria-seeded drug-containing and control plates. After 96 h, worms were mounted on glass slides, images were acquired in bright field using a Zeiss Axioplan microscope, and worm lengths were measured with ImageJ software (National Institutes of Health).
Oxygen Consumption Rate Assay.
Oxygen consumption rates were measured as in previously published protocols using a Oxytherm (Hansatech) oxygen electrode (42–44). Briefly, two crowded but not starved 60-mm diameter plates containing L4 larvae were washed three times with M9 buffer and resuspended in 1 mL of M9 buffer and then transferred into the chamber, and respiration was measured at 20 °C for at least 10 min. Samples were recovered from the chamber, centrifuged, and homogenized using a sonicator, and protein concentration was measured using a Pierce BCA Protein Assay Kit (Thermo Scientific).
Protein Prenylation Assay.
The prenylation assay was performed as described by Morck et al. (9). Briefly, young adults were placed on different drug plates and their progeny (L1 larvae) were scored for the number of GFP-enriched intestinal cells after 48 h.
Mitochondrial UPR Preinduction Experiment.
For preinduction of the UPRmt using EtBr or paraquat, five young adults were placed on EtBr plates (25 μg/mL) and allowed to reproduce. Once their progeny reached adulthood, the gravid worms were bleached to obtain synchronized L1 larvae. These larvae were again placed on EtBr plates (25 μg/mL) for 24 h. After pretreatment, the worms were transferred to either control or fluvastatin (0.5 mM) plates, and their viability was measured every 24 h until 96 h post-EtBr treatment. In the case of paraquat pretreatment assay, the synchronized L1 larvae were placed on paraquat (500 μM) plates for 24 h and then transferred to either control or fluvastatin (0.5 mM) plates. The viability of the worms was measured every 24 h until 96 h postparaquat treatment.
Mammalian Cell Culture.
The NIH 3T3 mouse embryonic fibroblast cells were maintained in DMEM with high glucose (Gibco) and 10% (vol/vol) FBS. For preinducing the mitochondrial stress response machinery, ∼2,000 3T3 cells were seeded per well on 96-well plates (Techno Plastic Products) and allowed to grow for 24 h. These cells were then treated with media containing EtBr (1 µg/mL) for 48 h. Medium was then replaced by fresh DMEM containing fluvastatin (10 μM), mevalonolactone (1 mM), or fluvastatin (10 μM) with mevalonolactone (1 mM) for 48 h. Cell viability was measured using the PrestoBlue cell viability reagent (Invitrogen) as recommended by the manufacturer.
Yeast Survival Experiment.
The wild-type S. pombe strain L972h− was cultured on YES agar or in YES broth [0.5% yeast extract, 3% (wt/vol) glucose, and 225 mg/L each of adenine, histidine, leucine, lysine, and uracil] at 30 °C. Cells were grown overnight in liquid culture with EtBr (250 or 500 ng/mL) until early or midlog phase was reached. The cells were then pelleted so as to have an OD600nm of 0.5 in 5 mL of medium. The appropriate amount of fluvastatin was added to the resuspended culture to achieve a final concentration of 0.1 mM, 0.2 mM, or 0.4 mM. The culture was grown for 24 h and then equilibrated to an OD600nm of 0.5. The equilibrated culture was serially diluted in twofold increments, spotted on YES agar plates, and incubated for 48 h at 30 °C.
Statistics.
Unless stated otherwise, columns in histograms show the average (n > 20), error bars show the SEM, and significant differences were determined using Student's t-test. In experiments monitoring survival over time, significance of the difference among genotypes was tested by survival analysis (life tables) through the Wilcoxon (Gehan) statistic (for overall and pairwise comparisons), with a 99.9% confidence level (P value ≤0.001).
Supplementary Material
Acknowledgments
We thank Peter Carlsson for comments on the manuscript, Cole Haynes for plasmids, and William Björklund for help with drug resistance assays. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was funded by the Swedish Research Council, Cancerfonden, Magnus Bergvalls Stiftelse, and Carl Tryggers Stiftelse.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218778110/-/DCSupplemental.
References
- 1.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys. 2011;505(2):131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golomb BA, Evans MA. Statin adverse effects: A review of the literature and evidence for a mitochondrial mechanism. Am J Cardiovasc Drugs. 2008;8(6):373–418. doi: 10.2165/0129784-200808060-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel TN, Shishehbor MH, Bhatt DL. A review of high-dose statin therapy: Targeting cholesterol and inflammation in atherosclerosis. Eur Heart J. 2007;28(6):664–672. doi: 10.1093/eurheartj/ehl445. [DOI] [PubMed] [Google Scholar]
- 5.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov. 2007;6(7):541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 6.Hieb WF, Rothstein M. Sterol requirement for reproduction of a free-living nematode. Science. 1968;160(3829):778–780. doi: 10.1126/science.160.3829.778. [DOI] [PubMed] [Google Scholar]
- 7.Merris M, Kraeft J, Tint GS, Lenard J. Long-term effects of sterol depletion in C. elegans: Sterol content of synchronized wild-type and mutant populations. J Lipid Res. 2004;45(11):2044–2051. doi: 10.1194/jlr.M400100-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Rauthan M, Pilon M. The mevalonate pathway in C. elegans. Lipids Health Dis. 2011;10:243. doi: 10.1186/1476-511X-10-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mörck C, et al. Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106(43):18285–18290. doi: 10.1073/pnas.0907117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haynes CM, Ron D. The mitochondrial UPR: Protecting organelle protein homeostasis. J Cell Sci. 2010;123(Pt 22):3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 11.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337(6094):587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37(4):529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 2012;8(6):e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuryn S, Le Gras S, Jamet K, Jarriault S. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics. 2010;186(1):427–430. doi: 10.1534/genetics.110.119230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarin S, Prabhu S, O’Meara MM, Pe’er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods. 2008;5(10):865–867. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zylber E, Vesco C, Penman S. Selective inhibition of the synthesis of mitochondria-associated RNA by ethidium bromide. J Mol Biol. 1969;44(1):195–204. doi: 10.1016/0022-2836(69)90414-8. [DOI] [PubMed] [Google Scholar]
- 17.King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246(4929):500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi J, et al. Effects of ethidium bromide treatment of mouse cells on expression and assembly of nuclear-coded subunits of complexes involved in the oxidative phosphorylation. Biochem Biophys Res Commun. 1990;167(1):216–221. doi: 10.1016/0006-291x(90)91753-f. [DOI] [PubMed] [Google Scholar]
- 19.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117(Pt 18):4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 20.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174(1):229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Argüelles N, et al. Design, synthesis, and docking of highly hypolipidemic agents: Schizosaccharomyces pombe as a new model for evaluating alpha-asarone-based HMG-CoA reductase inhibitors. Bioorg Med Chem. 2010;18(12):4238–4248. doi: 10.1016/j.bmc.2010.04.096. [DOI] [PubMed] [Google Scholar]
- 22.Kuranda K, François J, Palamarczyk G. The isoprenoid pathway and transcriptional response to its inhibitors in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2010;10(1):14–27. doi: 10.1111/j.1567-1364.2009.00560.x. [DOI] [PubMed] [Google Scholar]
- 23.Okello E, Jiang X, Mohamed S, Zhao Q, Wang T. Combined statin/coenzyme Q10 as adjunctive treatment of chronic heart failure. Med Hypotheses. 2009;73(3):306–308. doi: 10.1016/j.mehy.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Schaars CF, Stalenhoef AFH. Effects of ubiquinone (coenzyme Q10) on myopathy in statin users. Curr Opin Lipidol. 2008;19(6):553–557. doi: 10.1097/MOL.0b013e3283168ecd. [DOI] [PubMed] [Google Scholar]
- 25.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 26.Ishiguro H, et al. Enhancement of oxidative damage to cultured cells and Caenorhabditis elegans by mitochondrial electron transport inhibitors. IUBMB Life. 2001;51(4):263–268. doi: 10.1080/152165401753311816. [DOI] [PubMed] [Google Scholar]
- 27.Hihi AK, Gao Y, Hekimi S. Ubiquinone is necessary for Caenorhabditis elegans development at mitochondrial and non-mitochondrial sites. J Biol Chem. 2002;277(3):2202–2206. doi: 10.1074/jbc.M109034200. [DOI] [PubMed] [Google Scholar]
- 28.Jonassen T, Larsen PL, Clarke CF. A dietary source of coenzyme Q is essential for growth of long-lived Caenorhabditis elegans clk-1 mutants. Proc Natl Acad Sci USA. 2001;98(2):421–426. doi: 10.1073/pnas.021337498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branicky R, Nguyen PAT, Hekimi S. Uncoupling the pleiotropic phenotypes of clk-1 with tRNA missense suppressors in Caenorhabditis elegans. Mol Cell Biol. 2006;26(10):3976–3985. doi: 10.1128/MCB.26.10.3976-3985.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aspbury RA, Prescott MC, Fisher MJ, Rees HH. Isoprenylation of polypeptides in the nematode Caenorhabditis elegans. Biochim Biophys Acta. 1998;1392(2-3):265–275. doi: 10.1016/s0005-2760(98)00040-x. [DOI] [PubMed] [Google Scholar]
- 31.Hara M, Han M. Ras farnesyltransferase inhibitors suppress the phenotype resulting from an activated ras mutation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1995;92(8):3333–3337. doi: 10.1073/pnas.92.8.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowell DN, Huizinga DH. Protein isoprenylation: The fat of the matter. Trends Plant Sci. 2009;14(3):163–170. doi: 10.1016/j.tplants.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 33.McTaggart SJ. Isoprenylated proteins. Cell Mol Life Sci. 2006;63(3):255–267. doi: 10.1007/s00018-005-5298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callegari S, McKinnon RA, Andrews S, de Barros Lopes MA. Atorvastatin-induced cell toxicity in yeast is linked to disruption of protein isoprenylation. FEMS Yeast Res. 2010;10(2):188–198. doi: 10.1111/j.1567-1364.2009.00593.x. [DOI] [PubMed] [Google Scholar]
- 35.Cicha I, Schneiderhan-Marra N, Yilmaz A, Garlichs CD, Goppelt-Struebe M. Monitoring the cellular effects of HMG-CoA reductase inhibitors in vitro and ex vivo. Arterioscler Thromb Vasc Biol. 2004;24(11):2046–2050. doi: 10.1161/01.ATV.0000145943.19099.a3. [DOI] [PubMed] [Google Scholar]
- 36.Laufs U, et al. Impact of HMG CoA reductase inhibition on small GTPases in the heart. Cardiovasc Res. 2002;53(4):911–920. doi: 10.1016/s0008-6363(01)00540-5. [DOI] [PubMed] [Google Scholar]
- 37.Rashid M, et al. Importance of Rac1 signaling pathway inhibition in the pleiotropic effects of HMG-CoA reductase inhibitors. Circ J. 2009;73(2):361–370. doi: 10.1253/circj.cj-08-0817. [DOI] [PubMed] [Google Scholar]
- 38.Sulston JE, Hodgkin JA. In: The Nematode Caernorhabditis elegans. Wood WB, editor. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1988. pp. 587–606. [Google Scholar]
- 39.Bigelow H, Doitsidou M, Sarin S, Hobert O. MAQGene: Software to facilitate C. elegans mutant genome sequence analysis. Nat Methods. 2009;6(8):549. doi: 10.1038/nmeth.f.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 41.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6(4):280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Lee S-J, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10(5):379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan KT, Luo S-C, Ho W-Z, Lee Y-H. Insulin/IGF-1 receptor signaling enhances biosynthetic activity and fat mobilization in the initial phase of starvation in adult male C. elegans. Cell Metab. 2011;14(3):390–402. doi: 10.1016/j.cmet.2011.06.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.