Significance
ParA-type partition systems self-organize and pattern the bacterial nucleoid to organize plasmids, chromosomes, and protein machinery spatially. To study how protein patterns generate cargo movement, we reconstituted and visualized the partition system of F plasmid using a DNA-carpeted flowcell as an artificial nucleoid surface. We found that the partition proteins could bridge plasmid to the DNA carpet dynamically and mediate plasmid motion. Our data favor a diffusion-ratchet mechanism inherently different from classical motor protein or actin/microtubule filament-based transport. We expect surface-mediated patterning to become increasingly recognized as a means of intracellular transport in all kingdoms of life.
Keywords: bacterial chromosome segregation, ParA ATPase, plasmid segregation, spatial pattern organization, chromosome dynamics
Abstract
Increasingly diverse types of cargo are being found to be segregated and positioned by ParA-type ATPases. Several minimalistic systems described in bacteria are self-organizing and are known to affect the transport of plasmids, protein machineries, and chromosomal loci. One well-studied model is the F plasmid partition system, SopABC. In vivo, SopA ATPase forms dynamic patterns on the nucleoid in the presence of the ATPase stimulator, SopB, which binds to the sopC site on the plasmid, demarcating it as the cargo. To understand the relationship between nucleoid patterning and plasmid transport, we established a cell-free system to study plasmid partition reactions in a DNA-carpeted flowcell. We observed depletion zones of the partition ATPase on the DNA carpet surrounding partition complexes. The findings favor a diffusion-ratchet model for plasmid motion whereby partition complexes create an ATPase concentration gradient and then climb up this gradient toward higher concentrations of the ATPase. Here, we report on the dynamic properties of the Sop system on a DNA-carpet substrate, which further support the proposed diffusion-ratchet mechanism.
Proper DNA segregation ensures the faithful inheritance of genomic information for all life forms. In bacteria, this fundamental process is poorly understood. Low-copy bacterial genomes, including plasmids and chromosomes, encode active partition (Par) systems to ensure stability. Par systems are minimalistic in that only three dedicated components are required: a partition site on the DNA, a partition site-binding protein, and a nucleoside triphosphatase (NTPase). Par systems have been classified according to the type of NTPase involved: Walker-type (generically called “ParA”), actin-like, or tubulin-like (reviewed in ref. 1). Reconstitution of purified Par components of R1 plasmid in a cell-free system unveiled the mechanism involving an actin-like ATPase, ParM, in which elongating filaments of the ATPase push plasmids to opposite cell poles (2). Tubulin-like GTPases also appear to function as a filament (3). However, all chromosome-based and most plasmid-based systems use ParAs, and mounting evidence shows that ParA-like ATPases also are responsible for transporting large protein machineries (reviewed in ref. 4). However, the underlying mechanism for reactions of this category remains unresolved.
The Sop system (stability of plasmid) of F plasmid is one of the first Par systems to be identified (5, 6) and is considered a paradigm for the study of ParA-mediated DNA segregation. The three plasmid-encoded system components are SopA (the ParA-type ATPase), SopB (or ParB in other systems; i.e., the partition site-binding protein), and sopC (or parS in other systems; i.e., the cis-acting partition site on the plasmid). The first task of a partition system is to identify its DNA cargo. SopB accomplishes this task by loading onto sopC and forming a partition complex, which has been visualized in vivo by fluorescence microscopy as punctuate foci (7, 8). The partition complex is believed to contain a large number of SopB dimers, some bound specifically to sopC and additional dimers bound near sopC (9–11).
SopB-stimulated ATPase activity of SopA is critical to the partition reaction and plasmid stability, but how ATP hydrolysis drives plasmid movement is unknown. SopA has weak ATPase activity that is mildly stimulated by SopB or nonspecific DNA (nsDNA) (12). However, when SopB and nsDNA are combined, synergistic stimulation is observed. These properties generally are shared by other ParAs (reviewed in ref. 1). In vitro, several ParAs also bind nsDNA, and this activity requires or is enhanced by ATP (reviewed in ref. 4). A conserved basic patch of C-terminal residues has been implicated as the nsDNA-binding interface (13, 14), and mutation of SopA at this interface damages the ATP-dependent nsDNA-binding activity in vitro and plasmid stability in vivo (15).
In vivo, nsDNA mainly takes the form of the nucleoid. Several fluorescent versions of plasmid and chromosomal ParAs display dynamic patterns on the nucleoid (reviewed in ref. 4). Both ATPase activity and the ability to interact with the cognate partition complex are essential for this dynamic patterning. With either capacity inactivated, dynamic patterning ceases. Overall, the evidence suggests that the ParB-induced patterning by ParAs on the nucleoid plays a key role in partition, but mechanistic insight is limited. ParA patterns on the nucleoid have been interpreted as filaments that pull the plasmid cargo (reviewed in ref. 1). We have proposed an alternative diffusion-ratchet model in which the partition complex stimulates the local release of nucleoid-bound ParA, generating a ParA gradient that provides the motive force for plasmid movement (16).
To gain further insight into Par-mediated cargo-transport mechanisms, we reconstituted the P1 and F plasmid partition reactions in a cell-free system, whereby an nsDNA-coated flowcell surface (the DNA carpet) functioned as an artificial nucleoid (17). Fluorescently labeled reaction components were visualized by total internal reflection fluorescence microscopy (TIRFM). For both F Sop and P1 Par, we found that partition complex clusters could form zones on the surrounding DNA carpet in which the partition ATPase was depleted. These depletion zones are a critical component of the diffusion-ratchet model (16). However, after formation of the ATPase depletion zone, most partition complex clusters dissociated from the DNA carpet without extensive lateral movement.
In vivo, the P1 Par system seems to transition between mobile and immobile phases to move and position the plasmid (18). The F Sop system, on the other hand, patterns the nucleoid via a dynamically oscillating behavior (19) that is similar to several other ParA-mediated partition systems (reviewed in ref. 4). In this study, we used our cell-free setup to focus on how SopA and SopB mediate the dynamic movement of F plasmid, as being more representative of this general class of reactions. We searched and did not find evidence for SopA filament formation on the DNA carpet. Instead, we observed many partition complex clusters exhibiting dynamic behaviors, including lateral movements on the DNA carpet without any detectable SopA filaments. Similar dynamics also have been observed in the P1 Par system (17). Our results demonstrate that the P1 and F systems use the same principles of the diffusion-ratchet mechanism to accomplish the same biological outcome—faithful inheritance of genomic information. However, subtle differences in their respective biochemistries likely underlie variations in the resulting patterns seen in vivo.
Results
ATP-Dependent SopA Binding to the DNA Carpet.
To study F plasmid partition in a cell-free system, we created a biomimetic of the nucleoid surface by carpeting the flowcell with sonicated DNA fragments (∼500 bp) at high density (∼1,000 fragments/μm2; Materials and Methods). TIRFM was used to visualize Sop proteins and plasmids interacting with the DNA carpet and with each other in real time.
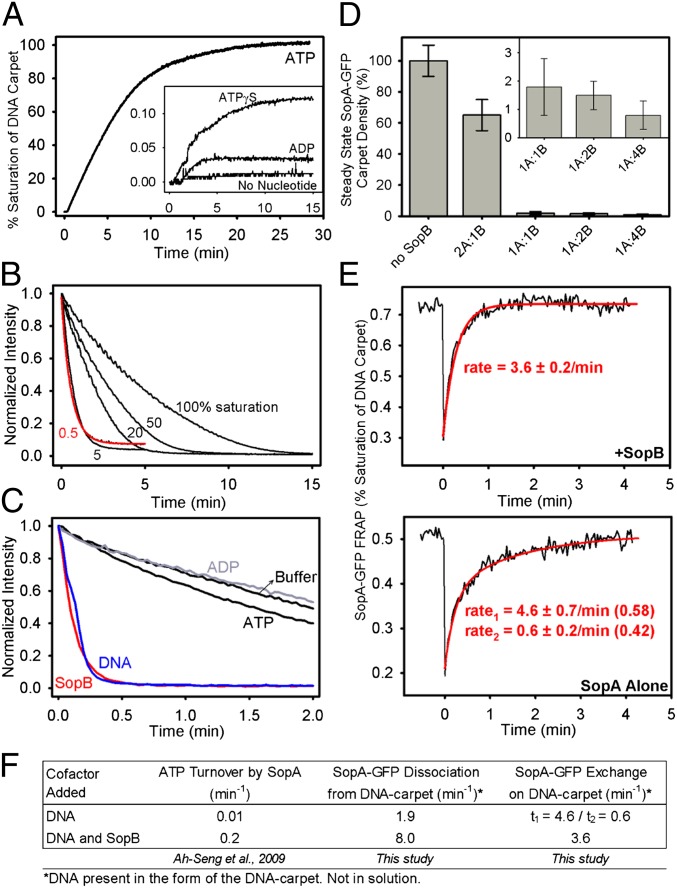
We first characterized the interaction between SopA and nsDNA by using a functional SopA protein fused at its C terminus to GFP (Fig. S1 A and B). When infused into the flowcell with ATP and Mg2+, SopA-GFP bound the DNA carpet with high affinity (Fig. 1A). As shown previously for native SopA (20), ATP was required for DNA binding; ATPγS or ADP could not replace ATP. Taking into account the fluorescence intensity of a single SopA-GFP monomer, we estimated its carpet density at saturation to be (6.1 ± 1.1) × 104 dimers/μm2 (Materials and Methods).
Fig. 1.
SopA-GFP interaction with the DNA carpet. (A) SopA binding to DNA is ATP specific. SopA-GFP (0.5 μM) was mixed with 1 mM of the indicated nucleotide upstream of the flowcell and was infused onto the DNA carpet at a rate of 20 μL/min. The fluorescence intensity of SopA-GFP that bound the DNA carpet (normalized to the saturated carpet intensity) was measured over time. (B) Kinetics of SopA disassembly from DNA. Using ATP, SopA-GFP was bound to the DNA carpet as in A. Once SopA-GFP binding reached the carpet densities indicated as a percentage of saturation, flow was switched to buffer without SopA (t = 0), and the decrease in fluorescence intensity was monitored over time. The dissociation curve measured at 0.5% saturation (red) was fit to a single exponential decay function to obtain an apparent dissociation rate of 1.9/min. (C) Cofactor effects on the kinetics of SopA disassembly from DNA. SopA-GFP was bound to the DNA carpet up to saturation. At t = 0, the flow was switched to wash buffer with or without the indicated cofactors (2 μM SopB, 100 μg/mL sonicated DNA, or 2 mM nucleotide). For B and C, the y axis is normalized to the SopA-GFP intensity before buffer switch. (D) SopB inhibits SopA binding to DNA. SopA-GFP (0.5 μM) was preincubated alone or with SopB at the molar ratios indicated for 30 min at 23 °C. Then ATP (1 mM) was added, and the sample was infused at 20 μL/min until SopA-GFP binding to the DNA carpet reached a plateau. Flow then was stopped, and steady-state SopA-GFP binding to the DNA carpet was measured. The y axis is the steady-state SopA-GFP intensity normalized to the saturated carpet intensity of SopA-GFP. Error bars represent the SD of at least three independent experiments. (E) FRAP analysis of SopA-GFP exchange on the DNA carpet. SopA-GFP (0.5 μM) was preincubated with or without stoichiometric amounts of SopB for 30 min at 23 °C. ATP (1 mM) was added, and the sample was infused into the flowcell at 20 μL/min. (Upper) In the presence of SopB, flow was stopped once SopA-GFP binding to the DNA carpet reached steady state. (Lower) Without SopB, flow was stopped once SopA-GFP reached a DNA-carpet density similar to that reached with SopB present. SopA-GFP on the DNA carpet then was photobleached, and recovery was measured over time. The data were fit to a single (+SopB) or double (SopA alone) exponential function. The y axis is SopA-GFP intensity normalized to the saturated carpet intensity of SopA-GFP. (F) Summary of rate data. Also see Fig. S1 and Movie S1.
We then looked at SopA-GFP release from the DNA carpet. SopA-GFP was flowed onto the carpet with ATP to varying densities. The solution then was switched to wash buffer (without SopA-GFP and ATP), and fluorescence intensity was monitored over time (Fig. 1B). With increasing initial SopA-GFP density on the DNA carpet, the apparent dissociation rate slowed. This effect was caused mainly by the rebinding of protein molecules that were released from the DNA carpet upstream of the observation area within the flowcell during washing. To eliminate the influences of rebinding, the flowcell was imaged immediately adjacent to the flow convergence point (SI Materials and Methods and Fig. S2) and at low SopA-GFP density (∼0.5% of saturation). We obtained an apparent dissociation rate of 1.9/min, which is similar to the rate of ∼1.5/min obtained previously for P1 ParA release from individual lambda DNA molecules (16).
Elimination of the rebinding artifact also was achieved by including competitor DNA in the wash buffer (Fig. 1C). DNA in the wash buffer blocks the dissociated SopA-GFP from rebinding the DNA carpet. When DNA was added to the wash buffer, the observed dissociation rate of SopA-GFP was 1.9/min regardless of where the measurement was done in the flowcell. The ATP-turnover rate of SopA in the presence of saturating DNA (0.01/min; ref. 21) is ∼200-fold slower than the rate of SopA release from DNA. Thus, DNA release is not obligatorily coupled to ATP hydrolysis. We conclude that an ATP-bound state of SopA can bind nsDNA dynamically before ATP hydrolysis. The dissociation rate of SopA-GFP from DNA is increased significantly by including SopB in the wash buffer (Fig. 1C). The rate of SopA dissociation in the presence of 2 μM SopB in the wash buffer was 8/min. Adding ADP or ATP to the wash buffer had no significant effect on SopA-GFP dissociation. The data suggest that SopB can accelerate SopA dissociation from DNA.
SopB Mediates SopA Interaction with the DNA Carpet.
To examine SopB effects on the steady-state DNA-binding activity of SopA, we preincubated SopA-GFP and ATP with varying concentrations of SopB. The SopA-GFP/SopB/ATP mixture was flowed onto the DNA carpet until the SopA-GFP density reached a steady state. When flow was stopped, the SopA-GFP density decreased to a new steady state (Fig. 1D). With no SopB, SopA-GFP (0.5 μM) quickly saturated the DNA carpet as above. With equimolar or greater amounts of SopB, the SopA-GFP density on the DNA carpet dropped to less than 2% of saturation (Fig. 1D, Inset). SopB (2 μM) reduced the steady-state binding of SopA to the DNA carpet by more than 100-fold, whereas the same concentration of SopB accelerated the dissociation of SopA from DNA by only fourfold (Fig. 1F). Thus, SopB-stimulated dissociation of SopA alone cannot explain the dramatic reduction in steady-state DNA binding by SopA in the presence of SopB. The data suggest that SopB also reduces the effective association rate of SopA-ATP and DNA.
To confirm this conclusion, we carried out fluorescence recovery after photobleaching (FRAP) analysis of SopA-GFP exchange on the DNA carpet with and without SopB (Fig. 1E). When ATP-bound SopA-GFP alone was infused, the recovery curve fit a double exponential function with recovery rates of 4.6 ± 0.7/min (58% of total fraction) and 0.6 ± 0.2/min (42% of total fraction), implying two distinct populations. In contrast, when 0.5 μM SopB was present, SopA-GFP exchange fit a single exponential function with a rate of 3.6 ± 0.2/min, implying that the slower-exchanging population was eliminated. However, once again, the minor increase in dissociation rate alone cannot account for the dramatic decrease in steady-state SopA binding to the DNA carpet in the presence of SopB. Therefore, SopB also must inhibit SopA binding to the DNA carpet from solution. Together, the findings suggest that a direct interaction with SopB converts SopA-ATP in solution to a non–DNA-binding state and/or prevents the generation of the DNA-binding form of SopA.
We also visualized SopB binding to the DNA carpet using SopB-Alexa 647 (mixed 1:9 with unlabeled SopB). SopB alone bound the carpet weakly (∼1,000 monomers/μm2 at 0.5 μM SopB in solution), with rapid exchange kinetics as measured by FRAP (Fig. S1). SopA-GFP did not affect the steady-state level of SopB binding or its exchange rate on the carpet significantly.
Thus, the impact of SopB on DNA-bound SopA must be exerted via a transient interaction and does not involve a stable SopA–SopB complex that lasts long enough to slow the DNA-bound SopB exchange rate. We have reported similar observations for the P1 ParA–ParB interaction on the DNA carpet (17).
No SopA-GFP Filaments Were Found on the DNA Carpet.
Several studies have shown that ParAs form filaments in vitro under varying conditions (reviewed in ref. 4). We searched for and did not find any evidence suggesting that SopA bound the DNA carpet as long filaments. First, SopA-GFP uniformly bound, exchanged, and dissociated from the DNA carpet without any pattern that suggested filamentation (Movie S1). SopA-GFP did form filament bundles on unpassivated glass or quartz surfaces, where nonspecific protein binding and denaturation is possible, as shown previously with unlabeled SopA (8). Second, the protein subunits within a long filament, if formed, would be relatively immobile compared with the subunits at the filament ends. Therefore, if SopA-GFP bound the DNA carpet as a filament, the majority of SopA-GFP molecules should be relatively static. From this viewpoint, we examined the motion of individual SopA-GFP molecules bound to the DNA carpet. By diluting SopA-GFP 10,000-fold with unlabeled SopA, we resolved single SopA-GFP molecules and studied their diffusion characteristics at varying SopA densities on the DNA carpet by single-particle tracking (Fig. 2A and Movie S2). At low SopA density (∼1% of saturation), the majority of SopA-GFP particles (91%, n = 183) were observed to diffuse randomly on the DNA carpet for distances >200 nm before releasing or photobleaching (Fig. 2B). Most particles diffused across the DNA carpet for several microns with an apparent diffusion coefficient of 0.85 ± 0.14 μm2⋅s−1 (Fig. 2C and Fig. S3A). These data confirm that SopA-GFP can hop intersegmentally from one DNA molecule to another in a fashion that seems to be independent of other SopA-GFP particles close by. This behavior was more apparent in the presence of flow, where particle movement was visibly biased by the flow direction (Fig. 2D and Fig. S3B).
Fig. 2.
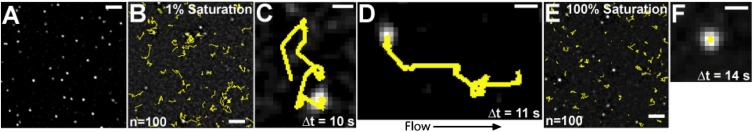
Single-particle tracking of SopA-GFP. (A) Total internal reflection fluorescence (TIRF) image of single SopA-GFP particles on the DNA carpet. SopA (500 nM) and SopA-GFP (50 pM) were mixed with 1 mM ATP and infused onto the DNA carpet. (Scale bar: 5 μm.) (B) SopA-GFP particle motion on the DNA carpet at low SopA density. At ∼1% saturation of the DNA carpet by SopA, flow was stopped, and the diffusion of SopA-GFP particles (n = 100) was tracked (yellow lines). (Scale bar: 5 μm.) (C) A typical SopA-GFP particle trajectory at 1% saturation. (Scale bar: 1 μm.) (D) An example of SopA-GFP particle motion influenced by flow (10 μL/min). (Scale bar: 1 μm.) (E) SopA-GFP particle motion on the DNA carpet at 100% saturation. SopA-GFP particle diffusion was tracked as in B. (Scale bar: 5 μm.) (F) A SopA-GFP particle trajectory with reduced mobility at 100% saturation. (Scale bar: 1 μm.) See also Fig. S3 and Movie S2.
The fraction of SopA molecules that exhibited restricted motion (the narrow peak at the center of the Gaussian distribution in Fig. S3C, Bottom) became significant only at very high densities of SopA on the DNA carpet. When carpet binding by SopA reached saturation, the percentage of SopA-GFP particles whose trajectories did not exceed 200 nm rose to 29% (n = 237) (Fig. 2 E and F and Fig. S3C). However, even at saturation, the diffusion properties of the majority of SopA molecules were not significantly different from those at lower SopA densities. The increase in the fraction of SopA-GFP molecules engaged in constrained diffusion may reflect excluded volume effects (molecular crowding), weak cooperative binding to DNA, or both. However, the SopA density on the DNA carpet that was necessary for this effect to be significant was at least two orders of magnitude greater than that expected on the nucleoid in vivo. If one assumes that 1,000 SopA dimers exist inside an Escherichia coli cell with one chromosome (22), ∼0.2% of the chromosomal DNA would be bound by SopA. Also, nucleoid-associated proteins and transcription factors dynamically occupy a significant fraction of the bacterial chromosome and likely prevent SopA from forming stable higher-order complexes on the nucleoid surface. Therefore, we conclude that at physiologically relevant SopA/DNA densities only a minor fraction of SopA molecules may engage in constrained diffusion, whereas the majority of SopA can bind independently and hop across DNA as dimers.
SopA Disassembly Is Required for Plasmid Movement on the DNA Carpet.
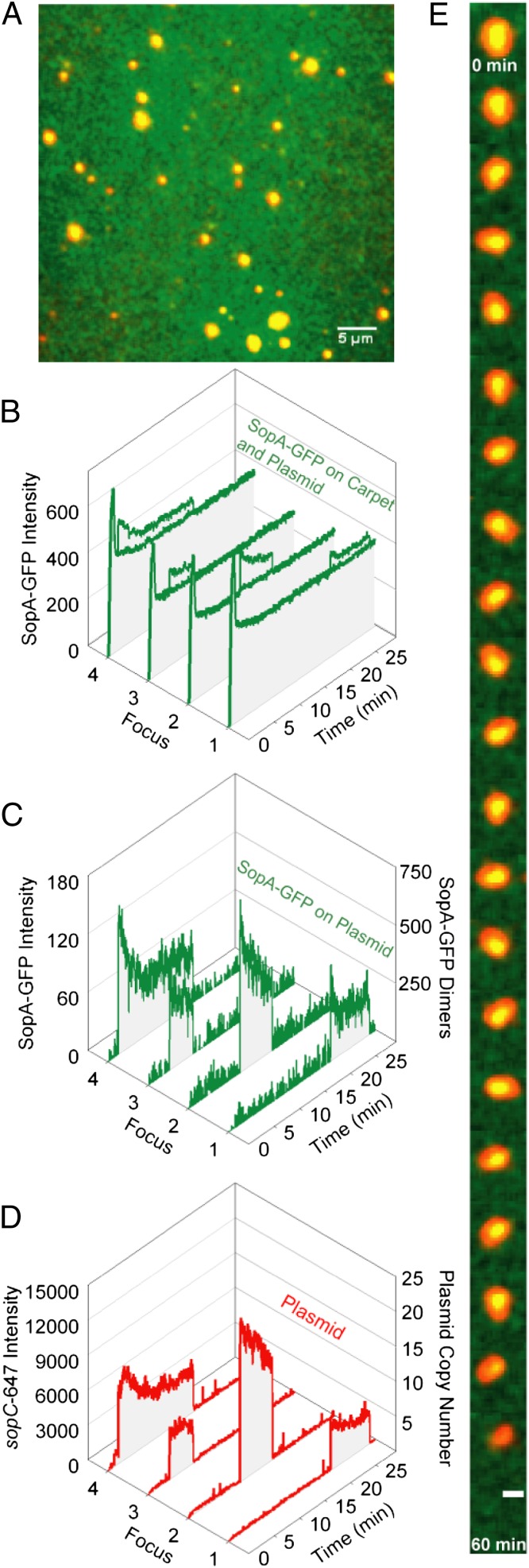
We have reported that in the reconstituted cell-free system of the F-plasmid partition reaction SopB-bound sopC plasmids were tethered transiently to the DNA carpet via DNA-bound SopA-ATP (17). Here we studied how Sop protein content governs the dynamics of partition complexes on the DNA carpet. A supercoiled pBR322::sopC plasmid (5 kb) labeled with Alexa647 (sopC-647) was preincubated with SopA-GFP, SopB, and ATP, and the mixture then was infused into the DNA-carpeted flowcell for observation. Preincubation before infusion results in the assembly of large partition complex clusters. After infusion, flow was stopped, and imaging was continued (Fig. 3A and Movie S3). SopA-GFP binding to the DNA carpet spiked transiently and decreased to a steady-state level within a few minutes (Fig. 3B). The plasmid clusters tethered to the DNA carpet with colocalized SopA-GFP. SopA-GFP then dissociated gradually before the plasmid clusters were released (Fig. 3C). Upon release from the DNA carpet, these clusters still contained a significant amount of SopA-GFP. Also, many of the larger clusters showed a decrease in plasmid fluorescence, concomitant with the decrease in SopA-GFP intensity, albeit at a slower rate. The finding indicates that some plasmid copies are released from the tethered cluster as SopA-GFP is released (Fig. 3D, foci 2 and 4). Interestingly, as SopA-GFP content decreased, most plasmid foci displayed tethered particle motion before release from the carpet, seen as dynamic movements around a central focal position (Fig. 3E and Movie S4). We infer that an initial static plasmid cluster is anchored to the DNA carpet with many tethers and that the number of tethers decreases as SopA-GFP disassembles from the complex, and the plasmid cluster wiggles on the few remaining tethers. Once all tethers are lost, the plasmid cluster is released. This progression confirms that SopB/sopC plasmid clusters are tethered to the DNA carpet via their associated SopA-GFP.
Fig. 3.
Sop system effects on plasmid-cluster dynamics. (A) Covisualization of SopA-GFP (green) and sopC-647 (red). SopA-GFP (0.5 μM), SopB (1 μM), and sopC-647 (0.1 nM) were preincubated together with 1 mM ATP for 15 min at 23 °C and were infused into the flowcell at 20 μL/min for 2 min before flow stoppage and movie acquisition. A freeze-frame TIRF image is shown. (Scale bar: 5 μm.) (B) SopA-GFP intensity was quantified at four different regions of the DNA carpet where plasmid-clusters bound and released within the time course indicated. (C) (Left) The average SopA-GFP intensity on the carpet was subtracted from the SopA-GFP intensities colocalized with the four plasmid foci (y axis). (Right) Intensity also was converted to an estimated number of SopA-GFP dimers (y axis). (D) (Left) The sopC-647 intensity was plotted over time to identify when the plasmid clusters bound and released the DNA carpet (y axis). (Right) Intensity also was converted to an estimated plasmid-copy number (y axis). (E) A time-lapse image series displays the typical decrease in SopA-GFP and the wiggling motion that is associated with large plasmid clusters before release. (Scale bar: 1 μm.) Also see Movies S3 and S4.
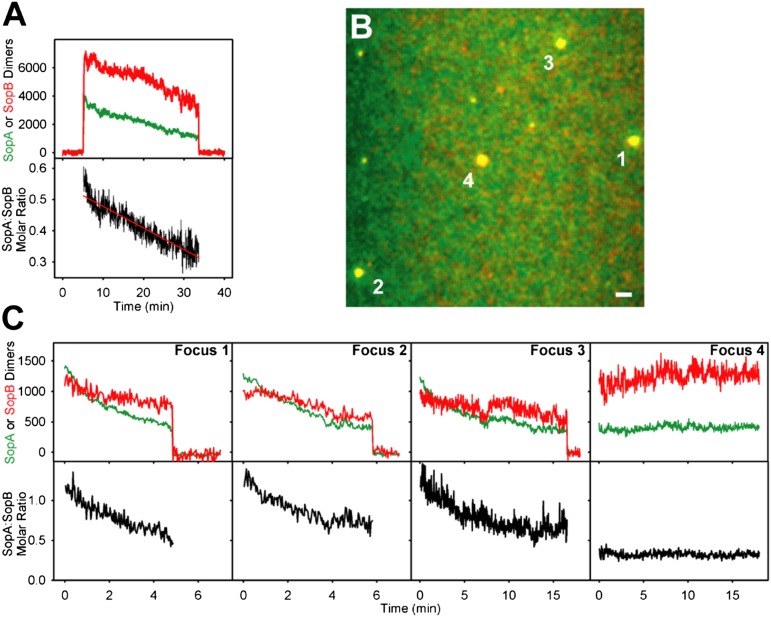
This inference is supported further by covisualization of SopA-GFP and SopB-Alexa647 (mixed 1:9 with unlabeled SopB) in the presence of unlabeled sopC plasmid. The Sop proteins, plasmid, and ATP were preincubated before infusion to form large plasmid clusters. SopA-GFP and SopB-Alexa647 formed large yellow foci that anchored transiently to the DNA carpet (Movie S5). The tethered foci subsequently faded to red and released. Quantification of the fluorescence intensities showed that both proteins disassembled from the complex, but SopA dissociated faster than SopB (Fig. 4A).
Fig. 4.
Effects of theSopA:SopB ratio on plasmid-cluster dynamics. (A) (Upper) Quantification of SopA and SopB dimers associated with plasmid-clusters. A region of the DNA carpet where a plasmid cluster bound and released was measured for SopA-GFP (green) and SopB-Alexa647 (red) intensities over time. Fluorescence on the DNA carpet was subtracted. The y axis is intensity converted to the number of SopA or SopB dimers (Materials and Methods). (Lower) Change in the SopA:SopB molar ratio is displayed for the duration in which the plasmid cluster was associated with the DNA carpet. (B) Freeze-frame image shows four plasmid clusters. (Scale bar: 2 μm.) (C) Effects of the SopA:SopB ratio on the dwell time of the clusters in B, quantified as in A. Also see Movies S5 and S6.
The details of the dynamics of individual plasmid clusters on the DNA carpet varied. Some clusters bound and released the carpet quickly; others stayed on longer. The SopA:SopB ratio of a plasmid cluster appeared to influence both the rate of SopA release and the dwell time of the cluster on the carpet. Those exhibiting the fastest decline in SopA:SopB ratio were the first to release from the carpet (Movie S6 and Fig. 4 B and C, foci 1 and 2). Foci that exhibited a slow decline in SopA:SopB ratio, or no decline at all, stayed on longer (Fig. 4 B and C, foci 3 and 4). The data suggest that excess SopB (over SopA) on the plasmid interacts with SopA on the carpet to bolster bridging complexes, and a prerequisite to plasmid movement is SopB-stimulated removal of SopA from the bridge at a rate that exceeds the protein-rebinding rate.
Some Plasmid Clusters Exhibited Lateral Motion.
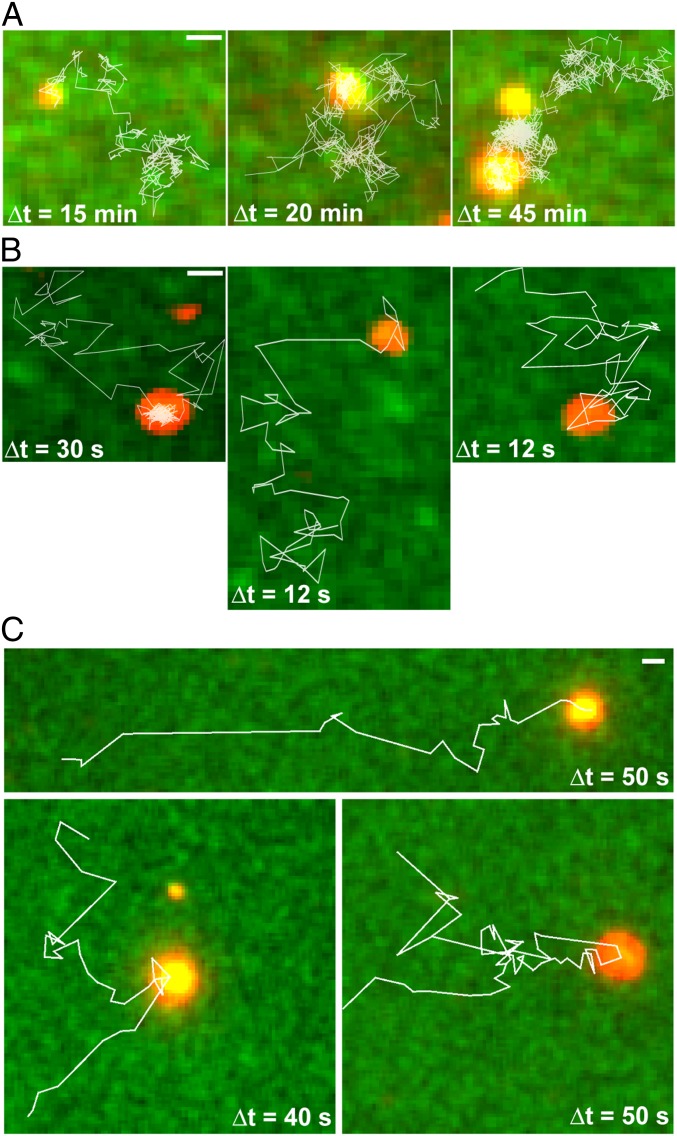
As SopA-GFP disassembled, some plasmid clusters moved laterally on the DNA carpet before dissociating fully (Fig. 5A and Movie S7). The motion was random in the absence of flow. With flow, some plasmid clusters rolled downstream while maintaining contact with the carpet (Fig. S4). The SopA:plasmid ratio of these traveling clusters was similar to that observed for other clusters at their time of carpet dissociation (Fig. 3C), suggesting that new anchor-points were being established at a rate similar to the rate at which old anchor-points were released.
Fig. 5.
Examples of F and P1 plasmid clusters moving laterally on the DNA carpet. (A) Trajectories (white lines) of F plasmid clusters on the DNA carpet up to the point of the first detachment. Images show the initial position of the plasmid clusters immediately before lateral movement. (B) Trajectories (white lines) of the F plasmid clusters during a brief revisit to the DNA carpet and moving laterally. (C) As in B, except the P1 Par system was observed. Split trajectories show when a P1 plasmid cluster splits into smaller complexes. SopA- or ParA-GFP is green, and sopC- or parS-plasmid is red. For all images, flow was stopped before movie acquisition. (Scale bar: 1 μm.) Also see Fig. S4 and Movies S7 and S8.
All plasmid clusters eventually dissociated into solution but occasionally returned to rebind the carpet transiently (Fig. 5B and Movie S7). These plasmid clusters paused intermittently on the DNA carpet, sometimes rolling laterally for several microns before falling back into solution. These revisiting clusters had significantly less SopA-GFP content than the clusters at the time of the initial dissociation from the DNA carpet. We believe this rolling mode of plasmid motion, although only transiently observed in our current setup, more closely reflects the in vivo dynamics of the Sop system (Discussion).
We also assembled the P1 Par system in our cell-free system by preincubating Alexa647-labeled parS plasmid with ParA-GFP, ParB, and ATP. As with F Sop, the P1 Par system generated large partition-complex clusters that released ParA-GFP over time, and some complexes began traveling laterally on the DNA carpet before full dissociation (Fig. 5C and Movie S8). In some instances, the plasmid clusters split into smaller complexes. These traveling clusters did not generate noticeable ATPase depletion zones on the DNA carpet as we have shown previously for more stably anchored partition complexes (17). We conclude that transient bridging allows the partition complexes to roll on the DNA carpet, but their dynamic movement and weak associations with the DNA carpet prevent the formation of an observable ParA/SopA depletion zone.
Discussion
The ParA family of ATPases can separate, transport, and position DNA and large protein machineries inside bacterial cells. ParAs that act on plasmids are key models for understanding how ATP-driven patterning of biological surfaces leads to cargo transport. ParAs bind nsDNA in vitro and colocalize with the nucleoid in vivo (reviewed in ref. 4), but how this interaction plays a role in the transport mechanism is a subject of considerable debate. Two models have been proposed. First, filament-pulling models propose that ParA polymerizes into a continuous filament that nonspecifically binds the nucleoid (23). When a filament end encounters the plasmid, ParB stimulates ParA disassembly but maintains contact with the depolymerizing end, which results in plasmid pulling. As an alternative, we previously proposed a diffusion-ratchet model (16) that does not invoke ParA filamentation. Here, and in a companion paper (17), we made use of a cell-free reaction system to try to distinguish between these two models. In this study, we examined Sop protein dynamics and the motion of F and P1 plasmid partition complexes on a DNA-carpeted flowcell surface; a biomimetic of the nucleoid.
No SopA Filaments Were Detected.
ParAs, including SopA, have been reported to form filament-like helices in vivo and filament-bundles in vitro, leading to models that resemble eukaryotic mitosis. However, the conditions for in vitro polymerization vary considerably among Par systems (reviewed in ref. 4). For SopA, in vitro polymerization was stimulated by SopB and inhibited by nsDNA (20). Because the majority of SopA colocalizes with the nucleoid (15, 19), where DNA concentration is very high, it is unlikely that SopA forms long, self-supporting filaments in vivo. Consistently, we found that SopA accumulated on the DNA carpet evenly as it was introduced with ATP into the flowcell, with no detectable filamentous structures. The fast exchange rate of carpet-bound SopA is inconsistent with long-lasting SopA filaments on the DNA carpet. The exchange rate also was faster than the steady-state rate of ATP hydrolysis by SopA, as has been observed for P1 ParA (17), indicating that DNA release is not obligatorily coupled to ATP hydrolysis. Subunits of a hypothetical SopA filament are expected to be relatively stationary during their lifetime within the filament. Instead, we found that the majority of individual SopA-GFP molecules hopped rapidly across DNA segments that made up the carpet. This finding agrees with previous in vitro studies showing the rapid disassembly of SopA filaments upon addition of nsDNA (20). We conclude that SopA does not form nucleoid-associated filaments that persist long enough for plasmid transport as subscribed by current filament-based models. However, the possibility of limited SopA polymerization on DNA still remains. Indeed, a fraction of carpet-bound SopA exchanged at a slower rate, possibly reflecting the presence of small polymers. However, this species was not observed in the presence of SopB.
DNA Bridging by SopA and SopB.
When SopB was infused together with SopA and ATP, SopB strongly inhibited SopA binding to the DNA carpet. One simple explanation is that SopB-stimulated ATP hydrolysis by SopA leads to its accelerated release from DNA. However, SopA exchange on the DNA carpet was only slightly faster when SopB was present. Together, the data indicate that SopB inhibits the SopA–DNA interaction mainly by preventing the accumulation of the DNA-binding form of SopA in solution without triggering ATP hydrolysis. Carpet-bound SopB exchanged quickly (t1/2 = 2 s) and with no significant influence by SopA, indicating that the SopA–SopB interaction on DNA is transient but commits DNA-bound SopA to ATP hydrolysis.
When plasmid DNA was preincubated with SopA and SopB in the presence of ATP, the proteins formed large plasmid clusters, which anchored to the DNA carpet when infused into the flowcell. When the sample contained SopA and SopB at ratios of 1:1 or lower, the steady-state SopA density on the carpet was ≤2% of saturation. Despite the low density, the plasmid was anchored to the carpet soon after infusion, indicating that SopB contributes to the formation of the DNA-bridging tether. Plasmid anchoring was followed by a decrease in plasmid-associated SopA, and eventually the plasmid detached.
The bridging activity of SopA and SopB studied here has features similar to the DNA-bridging complexes described for P1 and pSM19035 of Streptococcus pyogenes (14, 24). For all systems, DNA bridging required both proteins and ATP, and bridge disassembly was coupled to ParB-stimulated ATP hydrolysis. For P1, the complex was proposed to bridge plasmid and nucleoid and was referred to as the “nucleoid-adaptor complex” (NAC) (24). For pSM19035, the bridging was proposed to play a role in plasmid pairing as well as in forming an NAC-like complex (14). Likewise, the large plasmid clusters studied here likely are connected by the same protein-mediated contacts that tether plasmid to the DNA carpet. We note that the interplasmid contacts within a plasmid cluster generally persisted longer than the bridging interactions between a plasmid cluster and the carpet, suggesting that the high SopB content of the cluster contributes to the stability of plasmid-plasmid association.
After plasmid clusters anchored to the carpet, SopA content and, at a slower rate, SopB content decreased, typically within a few minutes, resulting in a reduction of the A/B ratio. We believe SopA release is primarily the result of a SopB-stimulated conformational change on SopA that eventually leads to ATP hydrolysis. The decrease in SopB likely reflects the release of plasmid copies from the cluster. We have shown that SopB stimulates the release of SopA not only from the plasmid but also from the surrounding DNA carpet (17). The development of SopA (or ParA) depletion zones by a partition complex is one of the central components of the diffusion-ratchet mechanism we have proposed (16).
Just before release from the DNA carpet, the plasmid clusters were held by a small number of anchor points and engaged in tethered particle motion. During this period, some complexes exhibited a “stepping” motion, whereby old anchor points were released while new anchor points were established. Some clusters stayed in this random-walk mode for some time before detaching completely from the carpet. This stepping motion is consistent with the diffusion-ratchet model, but it may reflect only one possible mode of plasmid motion in vivo.
Partition System Dynamics in Vivo.
Real-time in vivo cytology of Par systems has provided a foundation on which to build mechanistic models. Fluorescence microscopy of both the F Sop and pB171 Par systems found that the plasmid chases nucleoid-bound ParA at its movement front, leaving a region of the nucleoid devoid of ParA in the wake of plasmid movement (19, 23). Concurrently, ParA reassociates with the nucleoid away from the plasmid. When only one partition complex is present in the cell, the plasmid focus is relatively mobile. However, a decrease in mobility is observed when ParA appears to colocalize with the plasmid. Covisualization of P1 ParA and a plasmid carrying the P1 par locus clearly show two distinct populations of ParA: one colocalized with immobile plasmids and another dispersed on the nucleoid (18). Plasmid-associated P1 ParA foci disappeared from time to time, and this disappearance was coupled to plasmid movement. Chromosomal ParAs also have been observed as two discrete populations in vivo. Both ParA from Caulobacter crescentus and ParAI from Vibrio cholerae form foci that colocalize with their ParB-parS complexes during periods of relative immobility (25, 26). In both cases, ParA disappears from the mobile ParB-parS complex during segregation, and the nucleoid-bound population of ParA redistributes in response to ParB-parS motion. Our cell-free system recapitulated the immobile phase of the in vivo system dynamics as well as transition to the mobile phase. However, as discussed below, we have reproduced the mobile phase only partially.
Comparing F and P1 System Dynamics.
In vivo, P1 partition complexes cycle between immobile and mobile phases, whereas an F partition complex oscillates continuously from nucleoid pole to pole without extended pauses (19). Therefore, our finding that the Sop system also could anchor plasmid to the DNA carpet was unexpected. When measuring the ATPase content of partition complexes at the point of complex release from the DNA carpet, the SopA content (25 ± 8 dimers per plasmid copy) was significantly higher than the ParA content (6 ± 3 dimers per plasmid copy) (17). This result indicates that a smaller fraction of SopA than of P1 ParA is capable of bridging the F partition complex to the DNA carpet. Both systems generated ATPase depletion zones on the DNA carpet around anchored partition complexes (17). However, the zones were more robust and easier to detect for the P1 system. The “premature” release of F partition complexes from the DNA carpet could explain the weaker depletion zones. We believe that development of the robust SopA depletion zones necessary for plasmid movement in vivo requires spatial confinement of the partition complex (see below).
After carpet detachment, partition complex clusters were capable of interacting transiently with SopA-bound carpet, which we detected as short visits on the DNA carpet. These complexes had a low SopA content and often rolled across the DNA carpet for a few microns before dissociating into solution. Although rolling was observed only transiently with our current setup, it resembled Sop system dynamics in vivo. We found that some P1 partition complexes also revisited the DNA carpet after dissociation. However, the ATPase depletion zones did not develop around these traveling clusters, presumably because of the lack of persistent interaction with the DNA carpet, and the motion appeared random. Therefore, under the experimental conditions used, the behavior of the F Sop and P1 Par systems was similar. We conclude that both systems use the same diffusion-ratchet mechanism, but subtle biochemical differences between ParA-mediated systems could produce variations in the nucleoid-patterning dynamics observed in vivo. These differences may include the in vivo concentrations of Par proteins, the quantitative differences of a variety of kinetic parameters as we have observed (ref. 17 and this study), and possible crosstalk between the system and the in vivo environment.
Spatial Confinement.
We believe the plasmids did not roll persistently along the DNA carpet, as they do on the nucleoid in vivo, because our flowcell lacks a key requirement of the diffusion-ratchet mechanism, namely, spatial confinement. Inside bacterial cells, the gap between the nucleoid and inner membrane is narrow (27). As a result, cargo with a large excluded volume, such as a plasmid partition complex, cannot diffuse quickly away from or through the nucleoid. Spatial confinement of the partition complex would provide a persistent interaction with the nucleoid surface and an uninterrupted generation of the SopA depletion zone. This depletion would lead to the continuous and directionally biased Brownian motion of the partition complex as it searches for higher SopA concentrations on the nucleoid. The perpetual cycle of chasing and redistributing the ATPase would result in the oscillation of a single partition complex. Multiple partition complexes would repulse one another because of the ATPase depletion zone that develops on the nucleoid in between them. The 25-μm-thick flowcell we currently use does not provide the spatial confinement necessary for the continual motion observed in vivo. We currently are exploring several methods to provide sufficient confinement to our cell-free system because we believe it is one of the key parameters for persistent and directional transport of cargo.
If spatial confinement can sustain a SopA depletion zone in vivo, how does the Sop system maintain its ability to anchor the partition complex to the nucleoid and cluster plasmid copies? Interplasmid bridging may be a general prerequisite for partition to sort out the appropriate partners to be separated, not unlike sister chromatid cohesion and homolog pairing in mitosis and meiosis. In the context of plasmid partition, this sorting process may be needed when several plasmid copies occupy a single nucleoid. If too many plasmid copies formed independent partition complexes with accompanying ATPase depletion zones, these zones would merge, and the segregation mechanism that is based on the ATPase distribution gradient would break down. A corollary to this effect would be a weakened drive for the plasmid copies to move away from each other. It is attractive to speculate that this mechanism leads to interplasmid bridging, which reduces the number of cargo units to a level needed for efficient segregation and positioning.
Many biomolecular reactions rely on the internal dynamics of a single, large, molecular machine. In contrast, the system studied here does not assemble and operate as a large, single entity. Rather, the geometrical settings of the cell provide boundaries for the system components. The reaction steps that generate the overall system dynamics occur as parallel and repeated cycles of assembly and disassembly of many local complexes. When combined as a whole, the system yields a sensible biological outcome. We expect that this type of biomolecular-patterning and cargo-transport system exists in all kingdoms of life and shares many of the reaction principles identified here. We study a handful of examples that are composed of a relatively small number of components, including the MinD-MinE system that patterns the membrane to localize the bacterial divisome (28), the DNA-target search system used in transposition target immunity (29), and the P1 and F plasmid partition systems (17). We anticipate that what we learn from these relatively simple systems will lead to a better understanding of more complex pattern-mediated transport mechanisms.
Materials and Methods
Strains, Plasmids, Media, and Growth Conditions.
The construction details of strains and plasmids used in this study are given in SI Materials and Methods.
Protein Expression and Purification.
Hexahistidine-tagged SopA, SopA-GFP, and SopB proteins were purified by Ni-NTA affinity chromatography and subsequent ion exchange chromatography and/or gel filtration (SI Materials and Methods). Details on SopB labeling with Alexa Fluor 647 are given in SI Materials and Methods.
Flowcell Preparation.
Lipid bilayer and NeutrAvidin coating of the flowcell was performed as previously described (29). Sonicated salmon sperm DNA, biotinylated at both ends, was attached to the flowcell surface at high density as described (17) with minor modifications (SI Materials and Methods). The DNA fragments used to make the carpet were 500 ± 250 bp in length. Using an estimated DNA-binding length of 8 bp per SopA dimer (30), we calculated there are ∼60 dimers per DNA fragment at saturation. Therefore, the density of the DNA carpet is estimated to be 1,000 fragments/μm2.
TIRFM Setup.
The prism-type TIRFM setup using an Eclipse TE2000E microscope (Nikon) with a PlanApo 60× NA = 1.40 oil-immersed objective and magnifier setting at 1.5× was essentially as described (28), with minor modifications (SI Materials and Methods). FRAP experiments were carried out as described (28). The power of the 488-nm laser was sufficient for bleaching SopA-GFP and also bleached SopB-Alexa647 to a lesser degree.
Microfluidics and Sample Handling for TIRFM.
Unless stated otherwise, all experiments were performed in Sop buffer: 50 mM Hepes (pH 7.5), 100 mM KCl, 10% (vol/vol) glycerol, 5 mM MgCl2, 2 mM DTT, 0.1 mg/mL α-casein, 0.6 mg/mL ascorbic acid. Phosphoenolpyruvate (2 mM) (Sigma) and pyruvate kinase (10 μg/mL) (Sigma) were added also for ATP regeneration.
For buffer-switch experiments, a two-inlet flowcell was used. One inlet was connected to a syringe containing SopA-GFP, and the other inlet was connected to a syringe containing wash buffer (as specified). The Y-patterned flow channel was imaged at the point of flow convergence to minimize the effect of protein rebinding to the DNA carpet during measurements of the dissociation rate (SI Materials and Methods).
Partition-reaction components were preincubated before infusion and were loaded into a 300-μL loop connected to a polyether ether ketone Rheodyne injection valve. A syringe with Sop buffer was connected upstream of the valve to push the sample into the flowcell. Tubing downstream of the valve was connected to the flowcell inlet (dead volume = 20 μL). For most experiments, the sample was infused into the flowcell at 20 μL/min for 2 min, and flow was stopped for data acquisition.
Materials and conditions used for the P1 Par system experiments were as described in ref. 17.
Single-Particle Tracking.
SopA-GFP (50 pM) was incubated with 0.5 μM SopA and 1 mM ATP in Sop buffer at 23 °C for 15 min. The sample then was infused into the flowcell at 10 μL/min to the specified SopA densities on the carpet, based on the amount of SopA-GFP known to be required to saturate the DNA carpet. Before acquisition, background fluorescence was photobleached using 1.4 mW laser power for 10 s. Streaming movies were acquired at 10 Hz for 500 frames (50 s) with illumination and acquisition parameters as described (SI Materials and Methods). Data analysis details are given in SI Materials and Methods.
Estimating Protein Concentration and Plasmid Copy Number on the DNA Carpet.
The average fluorescence intensity of single labeled molecules was measured and used to calculate the number of fluorescent molecules within a region of interest. Details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Vassili Ivanov for help with the microscope setup, Yong-Woon Han for helpful suggestions regarding the study, and Barbara Funnell for the P1 Par proteins. This work was supported by the intramural research fund for the National Institute of Diabetes, and Digestive and Kidney Diseases, National Institutes of Health, US Department of Health and Human Services (K.M.). A.G.V. and L.C.H. were recipients of a Nancy Nossal Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302745110/-/DCSupplemental.
References
- 1.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141(6):927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 2.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315(5816):1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21(11):1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vecchiarelli AG, Mizuuchi K, Funnell BE. Surfing biological surfaces: Exploiting the nucleoid for partition and transport in bacteria. Mol Microbiol. 2012;86(3):513–523. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin SJ, Abeles AL. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J Mol Biol. 1983;169(2):353–372. doi: 10.1016/s0022-2836(83)80055-2. [DOI] [PubMed] [Google Scholar]
- 6.Ogura T, Hiraga S. Partition mechanism of F plasmid: Two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 7.Hirano M, et al. Autoregulation of the partition genes of the mini-F plasmid and the intracellular localization of their products in Escherichia coli. Mol Gen Genet. 1998;257(4):392–403. doi: 10.1007/s004380050663. [DOI] [PubMed] [Google Scholar]
- 8.Lim GE, Derman AI, Pogliano J. Bacterial DNA segregation by dynamic SopA polymers. Proc Natl Acad Sci USA. 2005;102(49):17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J Mol Biol. 2006;356(4):850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- 10.Lynch AS, Wang JC. SopB protein-mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc Natl Acad Sci USA. 1995;92(6):1896–1900. doi: 10.1073/pnas.92.6.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillet F, Sanchez A, Lane D, Anton Leberre V, Bouet JY. Centromere binding specificity in assembly of the F plasmid partition complex. Nucleic Acids Res. 2011;39(17):7477–7486. doi: 10.1093/nar/gkr457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe E, Wachi M, Yamasaki M, Nagai K. ATPase activity of SopA, a protein essential for active partitioning of F plasmid. Mol Gen Genet. 1992;234(3):346–352. doi: 10.1007/BF00538693. [DOI] [PubMed] [Google Scholar]
- 13.Hester CM, Lutkenhaus J. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc Natl Acad Sci USA. 2007;104(51):20326–20331. doi: 10.1073/pnas.0705196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soberón NE, Lioy VS, Pratto F, Volante A, Alonso JC. Molecular anatomy of the Streptococcus pyogenes pSM19035 partition and segrosome complexes. Nucleic Acids Res. 2011;39(7):2624–2637. doi: 10.1093/nar/gkq1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castaing JP, Bouet JY, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol Microbiol. 2008;70(4):1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- 16.Vecchiarelli AG, et al. ATP control of dynamic P1 ParA-DNA interactions: A key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78(1):78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang LC, et al. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013 doi: 10.1038/emboj.2013.34. 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatano T, Niki H. Partitioning of P1 plasmids by gradual distribution of the ATPase ParA. Mol Microbiol. 2010;78(5):1182–1198. doi: 10.1111/j.1365-2958.2010.07398.x. [DOI] [PubMed] [Google Scholar]
- 19.Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol Microbiol. 2007;64(5):1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: Regulation by DNA binding and SopB. Mol Microbiol. 2007;63(2):468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 21.Ah-Seng Y, Lopez F, Pasta F, Lane D, Bouet JY. Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein. J Biol Chem. 2009;284(44):30067–30075. doi: 10.1074/jbc.M109.044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouet JY, Rech J, Egloff S, Biek DP, Lane D. Probing plasmid partition with centromere-based incompatibility. Mol Microbiol. 2005;55(2):511–525. doi: 10.1111/j.1365-2958.2004.04396.x. [DOI] [PubMed] [Google Scholar]
- 23.Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106(46):19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Havey JC, Vecchiarelli AG, Funnell BE. ATP-regulated interactions between P1 ParA, ParB and non-specific DNA that are stabilized by the plasmid partition site, parS. Nucleic Acids Res. 2012;40(2):801–812. doi: 10.1093/nar/gkr747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20(23):3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ptacin JL, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12(8):791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mika JT, van den Bogaart G, Veenhoff L, Krasnikov V, Poolman B. Molecular sieving properties of the cytoplasm of Escherichia coli and consequences of osmotic stress. Mol Microbiol. 2010;77(1):200–207. doi: 10.1111/j.1365-2958.2010.07201.x. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci USA. 2010;107(18):8071–8078. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han YW, Mizuuchi K. Phage Mu transposition immunity: Protein pattern formation along DNA by a diffusion-ratchet mechanism. Mol Cell. 2010;39(1):48–58. doi: 10.1016/j.molcel.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui MP, et al. ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA. Proc Natl Acad Sci USA. 2010;107(10):4590–4595. doi: 10.1073/pnas.0913060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.