Abstract
Most in vivo studies of granulocytes draw conclusions about their trafficking based on examination of their steady-state tissue/blood levels, which result from a combination of tissue homing, survival, and egress, rather than direct examination of cellular trafficking. Herein, we developed a unique cell transfer system involving the adoptive transfer of a genetically labeled, bone-marrow–derived unique granulocyte population (eosinophils) into an elicited inflammatory site, the allergic lung. A dual polychromatic FACS-based biomarker-labeling system based on the IL4-eGFP transgene (4get) or Cd45.1 allele was used to track i.v. transferred eosinophils into the airway following allergen or TH2-associated stimuli in the lung in multiple mouse strains. The system was amenable to reverse tagging of recipients, thus allowing transfer of nonlabeled eosinophils and competitive tracking of multiple populations of eosinophils in vivo. The half-life of eosinophils in the blood was 3 h, and migration to the lung was dependent upon the dosage of transferred eosinophils, sensitive to pertussis toxin pretreatment, peaked at ∼24 h after adoptive transfer, and revealed a greater than 8-d eosinophil half-life in the lung. Eosinophil migration to the lung was dependent upon recipient IL-5 and IL-13 receptor α1 and donor eosinophil C-C chemokine receptor type 3 (CCR3) and interleukin 1 receptor-like 1 (ST2) in vivo. Taken together, this unique eosinophil transfer system provides an unprecedented opportunity to examine airway eosinophil migration without the need for extensive efforts to acquire donor source and time-consuming genetic crossing and has already been used to identify a long eosinophil half-life in the allergic lung and a definite role for ST2 in regulating eosinophil trafficking.
Keywords: asthma, allergy, eosinophilia, chemoattraction
Eosinophils are a circulating granulocyte population generated in the bone marrow and found in the blood and the gastrointestinal (GI) tissues under homeostatic conditions (1, 2). They have been shown to have a multifaceted role in regulating innate and adaptive immunity and host response against select pathogens (including parasites and viruses) as well as in eliciting inflammation, adaptive immunity, and tissue remodeling particularly in allergic disorders such as asthma (3–9). The level of eosinophilia positively correlates with asthma severity (10–12), and indeed several eosinophil-directed therapies have now been shown to improve asthma outcomes, particularly in patients with eosinophil-dominant allergic lung disease (13, 14).
Most studies of granulocyte trafficking in vivo rely on conclusions drawn from examination of granulocyte levels rather than direct examination of their homing. Whereas tissue eosinophil levels may partially reflect homing, they are influenced by a combination of factors including tissue influx, survival, and egress. These factors are particularly important as eosinophils do not develop in situ but rather migrate into tissues; thus, their ultimate tissue level is regulated by a balance between survival, cell death, and tissue egress. Adoptive transfer experiments offer the opportunity to directly examine eosinophil homing, particularly when labeled eosinophils are transferred and monitored in the target organ for a relatively short span of time (typically 24 h). The few studies that have used adoptively transferred eosinophils in vivo have been limited by the route of administration (e.g., direct intratracheal delivery as opposed to a more natural delivery) and by the use of cells that are derived from in vivo IL-5 overexpression models and are typically derived from the spleen rather than a more physiological source (4, 15–17).
Recently, an in vitro system for developing eosinophils from mouse bone marrow has become an emerging technique to generate large amounts of high-purity eosinophils (18, 19), bypassing the time-consuming eosinophil isolation procedure, which also alters eosinophil viability and introduces artificial alteration of other cellular properties due to ex vivo handling. Bone-marrow–derived eosinophils share properties of in vivo isolated eosinophils in terms of morphology (circular nuclei, eosinophilic granules) and unique surface marker expression [e.g., C-C chemokine receptor type 3 (CCR3) and sialic acid-binding Ig-like lectin F (Siglec F)] and are functionally competent as assessed by in vitro chemotaxis (18, 19). Herein, we aimed to develop a robust eosinophil adoptive transfer system, i.v. administering in vitro generated, bone-marrow–derived eosinophils to mice that were undergoing pulmonary allergen challenge. Furthermore, in vitro developed eosinophils offer the opportunity to study the consequences of genetic manipulation of eosinophils in vivo without the need to develop tissue- and/or cell-type–specific mutant mice. Accordingly, we used two genetically tagged strains of donor eosinophils, namely IL4-eGFP reporter mice (4get) (20) and the Cd45.1 congenic mice (21) (BALB/c and C57BL/6 backgrounds, respectively) to perform a series of eosinophil adoptive transfer experiments with bone-marrow–derived eosinophils to determine the feasibility of investigating eosinophil airway migration using genetically labeled donor eosinophils and recipient target tissue.
We demonstrated that in a model of intranasal allergen challenge cultured eosinophils migrate from the bloodstream to the airway in a pertussis-toxin–sensitive [i.e., G protein coupled receptors (GPCR), dependent] and allergen-dependent fashion. Proof-of-principle studies demonstrated a profound reduction of donor eosinophils, compared with levels in respective controls, when the donor eosinophils were transferred into Il13ra1−/− or Il5−/− recipients, or when Ccr3−/− donors were transferred into recipients. This approach was also used to determine eosinophil lifespan in vivo; blood and lung eosinophils had distinct half-lives of 3 h and 8 d, respectively. Competitive cotransfer of St2+/+ and St2−/− eosinophils revealed a dominant role for Interleukin 1 receptor-like 1 (Il1rl1/ST2) in regulating eosinophil responses. This unique eosinophil adoptive transfer system will be of significant value in revealing the fundamental mechanisms of airway eosinophil migration in the context of TH2 airway inflammation.
Results
In Vitro Generation of Robust Amounts of Bone-Marrow–Derived Eosinophils for Adoptive Transfer into Experimental Asthmatic Recipients.
By light microscopy, the day 14 cultured eosinophils have characteristic eosinophilic granules and circular polymorphonuclear morphology (Fig. S1A). Likewise, with the well-established allergen-induced experimental asthma model (22), the inflamed airway develops extensive eosinophilia, serving as the recipient’s target organ for donor eosinophil migration. A representative photomicrograph of an H&E-stained bronchoalveolar lavage fluid (BALF) cells from allergen-challenged mice demonstrates the presence of large amounts of eosinophils in the airway (Fig. S1B); in contrast, eosinophils are absent in the BALF from saline-challenged mice or from naïve mice (Fig. 1C). Compared with blood eosinophils, bone-marrow–derived eosinophils share common eosinophil surface expression profiles as reflected by surface CCR3, Siglec-F, and CD11b expression (Fig. S1C). To test whether bone-marrow–derived cultured eosinophils from 4get mice express robust levels of IL4-eGFP, we stained cultured eosinophils from 4get mice. 4get eosinophils express readily detectable level of the IL4-eGFP transgene after 14 d of culture (Fig. S1D). Eosinophil IL-4 expression, assessed by the FACS imaging, was intracellular (Fig. S1D, Right). Notably, airway eosinophils from allergen-challenged 4get mice were universally IL4-eGFP positive (Fig. S1E). In the C57BL/6 background, the Cd45.1 allele protein product can be readily distinguished from the wild-type Cd45.2 allele protein product (Fig. S1F). Therefore, these two donor eosinophil markers, IL4-eGFP and CD45.1, provide the foundation to track the eosinophil migration into target organs in BALB/c and C57BL/6 mice, respectively. Together with an eosinophil gating strategy that yields >99% purity in asthmatic BALF cells [Fig. S1G, FACS sorted by Siglec-F+CD11b+CD11c−side scatter (SSC)high], the FACS system allows both quantitative and qualitative assessments.
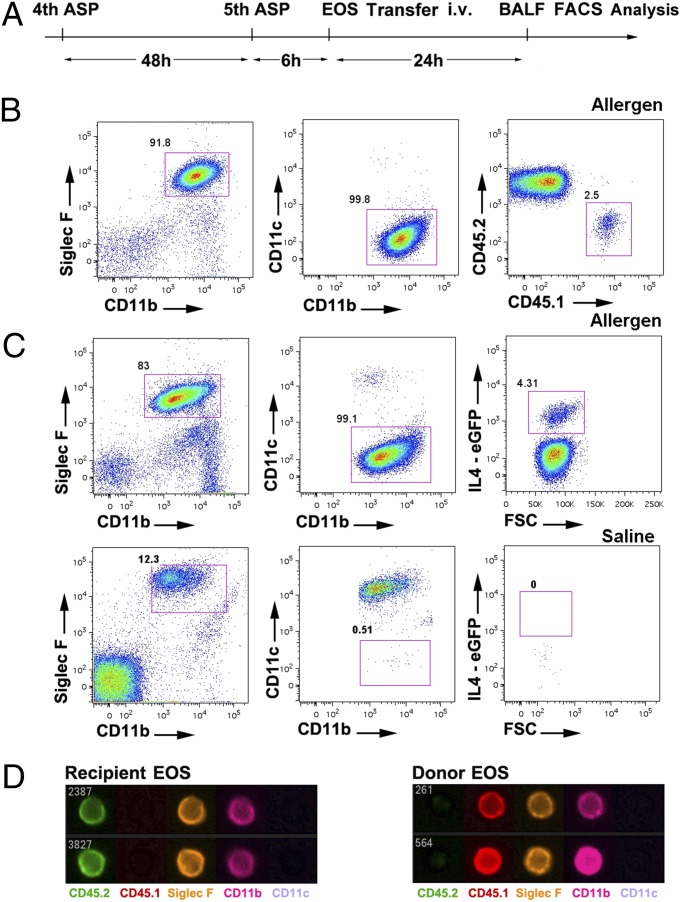
Fig. 1.
Identification of donor eosinophils in recipient airway with the dual-marker FACS system. (A) Schematic illustration of synchronizing the allergen (Aspergillus) challenge and eosinophil transfer. ASP, Aspergillus; EOS, eosinophil; i.v., Intravanously; BALF, bronchoalveolar lavage fluid; FACS, fluorescence-activated cell sorting. (B) Donor eosinophils (1.5 × 107, CD45.1 allele) were transferred i.v. 6 h after the fifth allergen challenge, and total BALF cells were harvested for FACS analysis 24 h later. After a serial gating strategy of SSChigh, Siglec F-CD11b double-positive, CD11c-negative events ensuring >99% eosinophil purity, CD45.1 donor eosinophils can be readily discriminated from CD45.2 recipient eosinophils despite sharing other characteristics. (C) IL4-eGFP-positive donor eosinophils (4get, 1.5 × 107) were transferred i.v. into the allergen-or saline-challenged BALB/c recipients. Whereas 4get donor eosinophils can be discriminated from recipient eosinophils in the GFP-positive gate after allergen challenge, saline-challenged BALB/c control mice exhibited no detectable airway eosinophilia. (D) Flow imaging micrograph showing the extracellular expression patterns of donor (CD45.1) and recipient (CD45.2) eosinophils from the BALF. All data are representative of at least three experiments.
Adoptively Transferred Eosinophils Home to the Airway Following Allergen Challenge by a Pertussis-Toxin–Inhibited Mechanism and Are Influenced by Endogenous IL-5.
We tested whether the cultured eosinophils introduced i.v. could migrate into the airway lumen. The schematic strategy for synchronizing the eosinophil culture and asthma model is illustrated in Fig. 1A. BALF cells were stained with an antibody panel of CD11c, Siglec-F, and CD11b, plus CD45.1 and CD45.2 for the CD45.1 system, or eGFP for the 4get system, which enabled identification of donor and recipient eosinophils. This eosinophil antibody panel and associated gating strategy (Fig. 1B) enabled acquisition of >99% eosinophils in the airway by morphological analysis of the FACS-sorted eosinophils (Fig. S1G). Indeed, both the CD45.1 and 4get systems demonstrated that donor eosinophils migrated into the airway lumen following allergen challenge but were not detectable in saline-challenged recipients (Fig. 1 B and C for CD45.1 and 4get systems, respectively). As an independent approach, FACS imaging of the antibody-stained BALF cells from experiments using the CD45.1 system verified the surface labeling of donor eosinophils (Fig. 1D). Of note, after airway migration, eosinophils from the donor and recipient share comparable expression levels of a panel of specific surface markers (Fig. S2).
Kinetic analysis with the 4get system revealed that donor eosinophils first appeared in the blood, turned over rapidly (Fig. 2A), and subsequently accumulated in the lung with a relatively low turnover rate (Fig. 2B). Kinetic analysis by enumeration of donor eosinophils in the airway of allergen-challenged mice (sustained challenge regimen of every other day) revealed that airway eosinophil levels peaked at 24 h and remained stable for 7 d. The overall donor eosinophil half-life in the lung was ∼8 d, which contrasts with the ∼3-h half-life of donor eosinophils in the blood. Notably, eosinophil homing to the lung was proportional to the quantity of donor cell input (Fig. 2C), whereas the spleen did not sequester significant numbers of donor eosinophils, as donor eosinophils in the spleen accounted for 2.5 ± 0.5% and 0.2 ± 0.1% (mean ± SD) of initial input (4.3 × 106) at 4-h and 24-h points, which is ∼8-fold and ∼80-fold lower than the residential spleen eosinophil population, respectively.
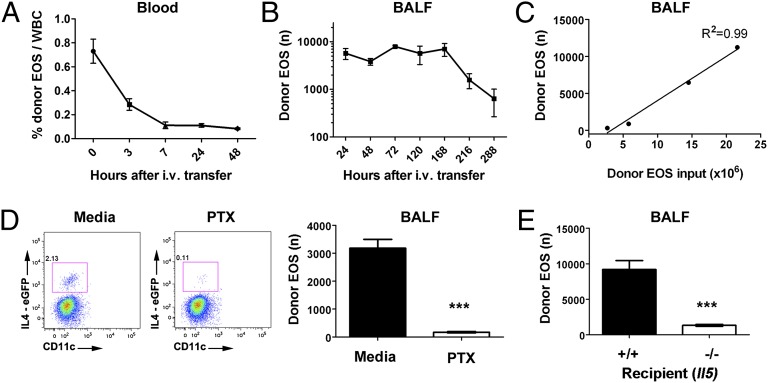
Fig. 2.
Kinetic analysis of donor eosinophils in the recipient circulation and airway. Donor 4get eosinophils (EOS, 6.5 × 106 i.v. input) were kinetically monitored in the blood (A) and BALF (B) of allergen-challenged recipients following the SSChigh, Siglec F+CD11b+CD11c− gating strategy by FACS over the indicated time period with the allergen challenge administrated every other day. (C) The linear relationship between i.v. donor eosinophil input and airway donor eosinophil recovery as illustrated with the 4get system by FACS. (D) 4get eosinophils (1.3 × 107) were pretreated with media or pertussis toxin (PTX, 100 ng/mL) for 2 h before the i.v. transfer, and airway donor eosinophil migration was tracked by FACS. Levels of donor eosinophils in the airway with or without PTX pretreatment. (E) 4get donor eosinophils were transferred into allergen-challenged Il5+/+ and −/− recipient mice and the number of airway donor eosinophils were enumerated (mean ± SEM, ***P < 0.001, two-tailed Student t test).
To examine whether the donor eosinophil migration was GPCR dependent, we treated cultured eosinophils with 100 ng/mL pertussis toxin for 2 h before the transfer. Indeed, the airway migration of pertussis-toxin–treated eosinophils was abolished (Fig. 2D). We also tested whether the donor eosinophil airway migration was influenced by IL-5, by transferring 4get eosinophils into allergen-challenged Il5+/+ and Il5−/− recipients. Notably, a 10-fold reduction in donor eosinophilia was observed in Il5−/− recipient mice (Fig. 2E).
Homing of Donor Eosinophils into the Recipient Airway Is Abolished in Il13ra1 Knockout Recipients (CD45.1 and 4get Systems).
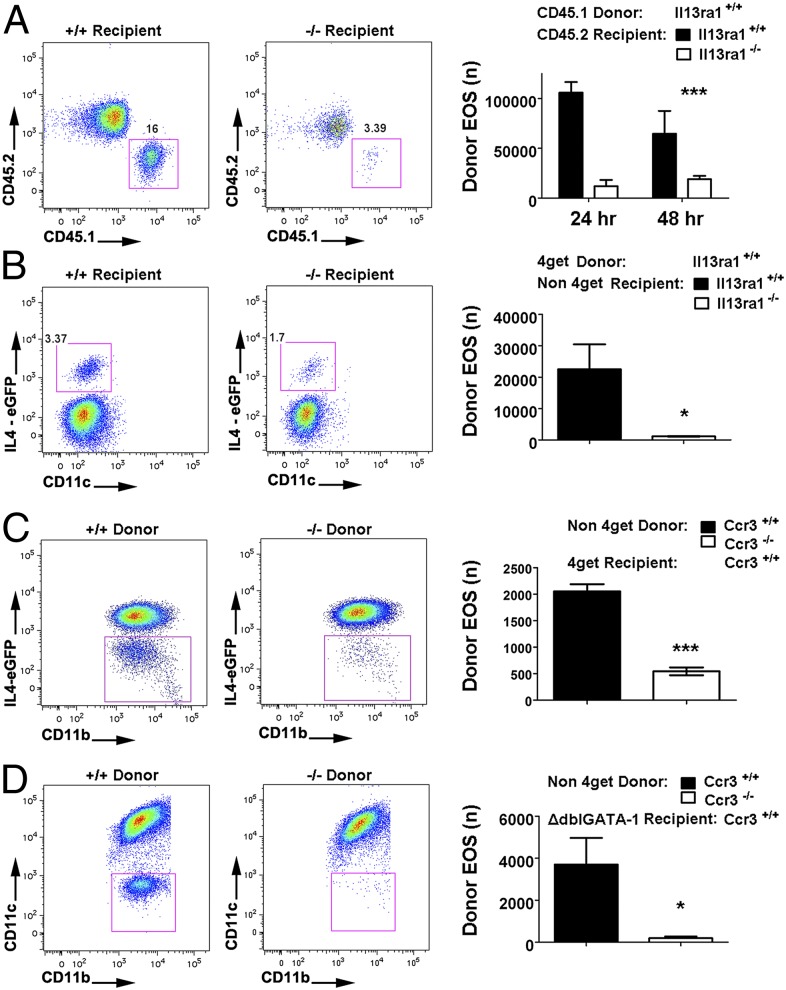
Il13ra1−/− mice are known to have reduced eosinophilia in the lung following allergen challenge (23). We therefore used this genetic system as the standard to demonstrate the experimental utility of the dual transfer systems and, more importantly, to rule out the possibility that the observed phenotype in Il13ra1−/− mice was due to IL-13RA1 expression on eosinophils. To address the effect of modifying the gene of interest (GOI) on the recipient side, we transferred CD45.1 and 4get (wild-type Il13ra1+/+) eosinophils into allergen-challenged Il13ra1+/+ and Il13ra1−/− mice in both the C57BL/6 and BALB/c backgrounds, respectively. Animals were killed 24 h after the donor eosinophil transfer. Notably, airway eosinophilia specific to the CD45.1 donor eosinophils was abolished in Il13ra1-deficient mice compared with wild-type control mice (Fig. 3A). Performing these same experiments with the 4get transfer system (BALB/c strain), we observed a profound reduction of eosinophil homing into the lung in Il13ra1-deficient mice following i.v. transfer of 4get donor eosinophils (Fig. 3B). Therefore, the dependence of allergen-induced airway eosinophilia on lung tissue IL-13RA1 was confirmed in both the C57BL/6 and BALB/c congenic backgrounds. Similarly, the dual-transfer system with CD45.1 and 4get, respectively, was validated.
Fig. 3.
Validation of the dual system with Il13ra1 and Ccr3 gene-targeted mice. (A) CD45.1 donor eosinophils (1.5 × 107) were i.v. transferred into allergen-challenged Il13ra1+/+ and −/− recipients of the C57BL/6 congenic background, and donor eosoinophil events in the BALF were identified by the above-mentioned gating strategy and quantified by FACS analysis. Representative FACS plots and quantification are shown (***P < 0.001, two-way ANOVA on genotype factor). (B) In the BALB/c congenic background, 4get donor eosinophils (7.5 × 106) were i.v. transferred into allergen-challenged Il13ra1+/+ and −/− recipients, and donor eosinophil events in the BALF were quantified by FACS analysis. (*P < 0.05, two-tailed Student t test) (C) Ccr3+/+ and −/− nonmarked eosinophils (1.8 × 107 cells) were i.v. transferred into allergen-challenged 4get recipients. Donor eosinophils are identified from IL4-eGFP-negative gate among the total airway eosinophils. (D) Ccr3+/+ and −/− nonmarked eosinophils (2.0 × 107) were transferred into allergen-challenged ΔdblGATA-1 mice. Following the eosinophil gating strategy, donor eosinophils can be readily quantified without interference of recipient/native eosinophils. (*P < 0.05, ***P < 0.001, two-tailed Student t test) All data are shown as mean ± SEM.
Eosinophil Adoptive Transfer with Reverse Genetic Tagging and Eosinophil Lineage Deficient (ΔdblGATA-1) Recipients.
When assessing an eosinophil GOI for its role in airway migration, nontagged GOI+/+ and GOI−/− eosinophils, instead of labeled eosinophils, can be transferred into recipient mice that bear a genetic tag. This reverse approach has the advantage that genetically engineered eosinophils could be directly studied without the need to develop double-engineered mice. Accordingly, we used Ccr3+/+ and Ccr3−/− mice as a source of donor eosinophils, and 4get mice as a recipient strain. As shown by the negative/donor gate below the major eGFP signal group, CCR3 disruption attenuated the airway donor eosinophil migration (Fig. 3C). Thus, it should be feasible to study eosinophil migration with unlabeled donor and labeled recipient eosinophils. These results broaden the applications of the transfer systems in that the GOI originated from both the donors and recipients can be assayed for alterations in eosinophil migration.
When studying eosinophil homing, involvement of host eosinophils may not be desired. Accordingly, we studied the feasibility of transferring Ccr3+/+ and Ccr3−/− eosinophils into an eosinophil lineage deficient (ΔdblGATA-1) mice (24). In the first approach, we transferred 4get eosinophils into the circulation of allergen-challenged ΔdblGATA-1 mice. Twenty-four hours later, BALF cells were evaluated for donor eosinophil levels. Notably, under these conditions, donor eosinophils were the predominant eosinophil population (representing >90% of the airway eosinophils, Fig. S3) in contrast to the wild-type recipients (representing 2–4% of the airway eosinophils, Fig. 1 B and C). Accordingly, we next transferred nontagged Ccr3+/+ and Ccr3−/− donor eosinophils into allergen-challenged ΔdblGATA-1 mice. Notably, the recipients receiving Ccr3+/+ donor eosinophils exhibited a pronounced donor eosinophilia in the allergic airway, whereas eosinophils were barely detected in the mice that received Ccr3−/− cells (Fig. 3D).
Competitive Transfer System to Simultaneously Assess the Homing Capacity of Two Distinct Populations of Eosinophils.
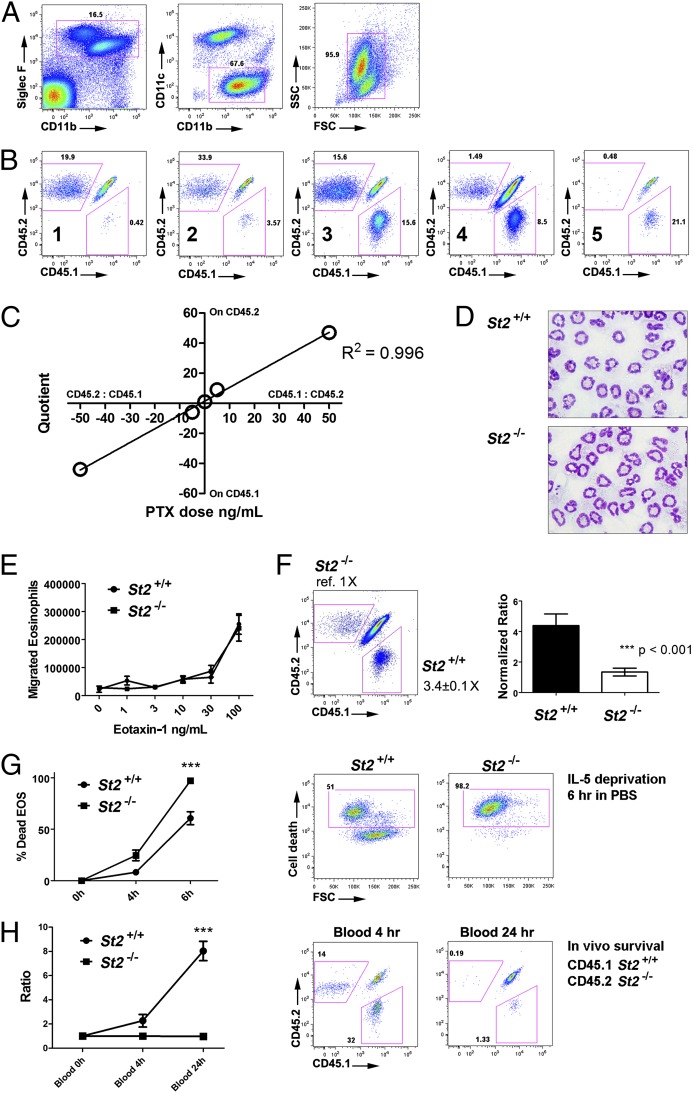
When investigating the role of a GOI in eosinophils, concurrently monitoring airway migration of two donor populations (GOI+/+ and GOI−/−) would be advantageous, as experimental variability would be internally controlled in each mouse. The leukocytes from the CD45.1–CD45.2 heterozygous mice have one copy of each allele (double positive), providing the opportunity to track the homing of CD45.1 and CD45.2 single-positive donor eosinophils by FACS. Herein, we evaluated the feasibility of simultaneously transferring an equal dose of CD45.1 and CD45.2 donor eosinophils into the circulation and monitoring their airway homing activity as driven by the intranasal administration of eotaxin-1 (4 μg) and IL-5 (1 μg). By the FACS gating indicated, the two single-positive donor eosinophil populations, as well as the double-positive recipient eosinophils, are readily distinguishable on the CD45.1/CD45.2 double plots (Fig. 4 A and B). To demonstrate that this cotransfer system is able to reflect the differential homing capacities of the two populations of donor eosinophils, we used a series of five suboptimal doses of pertussis toxin (PTX) of different combinations in 1:1 ratios (1.1 × 107 cells each): (i) CD45.2 untreated + CD45.1 treated with PTX (50 ng/mL); (ii) CD45.2 untreated + CD45.1 treated with PTX (5 ng/mL); (iii) CD45.1 untreated + CD45.2 untreated; (iv) CD45.1 untreated + CD45.2 treated with PTX (5 ng/mL); and (v) CD45.1 untreated + CD45.2 treated with PTX (50 ng/mL). FACS quantification of the two donor eosinophil populations in the five treatment groups indicated that the decrease in PTX-pretreated donor eosinophil homing as assessed by the ratio of the two donor eosinophil levels (media vs. PTX) was linearly correlated with the PTX doses (Fig. 4 B and C, negative value endowed to the CD45.1 eosinophil quantities and quotients to establish the linear regression model).
Fig. 4.
Competitive cotransfer model. (A) Equal dose of CD45.1 and CD45.2 D14 eosinophils were i.v. transferred into CD45.1–CD45.2 heterozygous mice that received an intranasal bolus of 4 μg eotaxin-1 and 1 μg IL-5 immediately before the transfer. After 24 h, BALF cells were stained and subjected to FACS. Eosinophils were gated sequentially for the presence of CD45.1-postive (donor 1), CD45.2-positive (donor 2), and double-positive (recipient) eosinophils. (B) Five different regimens were used to assess the linearity between suboptimal PTX treatment and airway migration as labeled: (1) CD45.2 untreated + CD45.1 treated with PTX (50 ng/mL); (2) CD45.2 untreated + CD45.1 treated with PTX (5 ng/mL); (3) CD45.1 untreated + CD45.2 untreated; (4) CD45.1 untreated + CD45.2 treated with PTX (5 ng/mL); and (5) CD45.1 untreated + CD45.2 treated with PTX (50 ng/mL). (C) The quotients of CD45.1/CD45.2 eosinophils airway percentages for naïve and treated donor eosinophils were used to set up the linear regression with PTX dosage. The quotient and dose related to CD45.1 treatment were assigned to negative values to establish a linear regression model. (D) Representative micrograph depicting the morphology of St2+/+ and −/− bone-marrow–derived eosinophils H&E stained before transfer. (E) Bone-marrow–derived St2+/+ and −/− eosinophils were subjected to in vitro transwell migration assay in the presence of IL-5, driven by escalating eotaxin-1 doses. (F) St2+/+ (CD45.1) and St2−/− (CD45.2) eosinophils were i.v. transferred into allergen-challenged CD45.1–CD45.2 heterozygous mice. Representative CD45.1–CD45.2 double plot of gated airway eosinophils reveals three major populations: the double-positive recipient eosinophils and the two single-positive donor eosinophils from distinct genotypes. Compared with St2−/− donors (normalizer), St2+/+ donors were present at 3.4 ± 0.1-fold (mean ± SEM) higher levels. Right: bar chart of St2+/+ vs. −/− migration percentage ratio derived from normalization to the median of St2−/− donor. (***P < 0.001); experiments were repeated three times independently with similar results. (G) St2+/+ and St2−/− eosinophils were washed, resuspended in PBS, and labeled with viability dye for a IL-5 deprivation assay in vitro. Death rates were quantified by FACS and representative plots are shown at 6 h. (***P < 0.001, representative of four experiments shown) (H) Equal amount of St2+/+ and St2−/− eosinophils were i.v. transferred into naïve nonallergen-challenged mice to assess survival in vivo, and the two donor eosinophil populations in the blood were measured by FACS. The ratio of St2+/+ and St2−/− eosinophil percentages (% of total eosinophils) to the median of St2−/− eosinophil percentages was graphed, with the representative FACS plots shown. (***P < 0.001, representative of two experiments shown). For survival studies measured at different time points across genotypes, two-way ANOVA was used to evaluate the overall genotype significance.
With this competitive transfer system, we next analyzed the airway migration of St2−/− bone-marrow–derived eosinophils; St2−/− mice have diminished allergen-induced eosinophilia, but it has not been determined whether this effect is directly mediated through eosinophils. Following bone marrow culture, St2−/− eosinophils developed normal morphology (Fig. 4D), comparable expression on a panel of specific markers (Fig. S4), adhesion molecules (Fig. S5), and similar adhesion to mouse endothelial cells in vitro (Fig. S6) before the transfer. Importantly, in vitro analysis of eosinophil migration did not reveal a migration difference between St2+/+ and St2−/− eosinophils following an escalating dose series of eotaxin-1 (Fig. 4E), as well as eotaxin-2 (Fig. S7), indicating an intact CCR3 axis. Notably, competitive transfer of St2+/+ and St2−/− eosinophils revealed a marked 3.4-fold attenuation of St2−/− eosinophil homing to the allergen-challenged lung (n = 8; Fig. 4F) compared with St2+/+ eosinophils. We next tested whether this deficit could be due to reduced survival of St2−/− eosinophils before reaching the airway. Indeed, under IL-5 deprivation conditions (in PBS) in vitro, a ∼2-fold higher death rate was observed in St2−/− eosinophils at multiple time points (Fig. 4G). Furthermore, we cotransferred an equal number of St2+/+ and St2−/− eosinophils to naïve recipients and examined the presence of both eosinophil genotypes in the blood after 4 and 24 h. A dominance of St2+/+ eosinophils was observed at both time points (Fig. 4H).
Discussion
Systems that allow adoptive transfer of leukocyte populations (e.g., lymphocytes and mast cells) have been critical for experimental analysis, but are not readily available for terminally differentiated cells such as granulocytes. Herein, we report a unique system that allows in vitro generated bone-marrow–derived eosinophils to be studied by adoptive transfer. The experimental system demonstrates that eosinophil airway migration is GPCR dependent, allergen induced, directly dependent on donor eosinophil input, and sensitive to recipient IL-13RA1 and IL-5 and donor eosinophil CCR3 and ST2 expression. The eosinophil transfer system introduced has unique merits and provides an opportunity to answer questions that have been difficult to explore via previously available approaches in the asthma/eosinophil field. For example, we have used this system to demonstrate a direct and key role for eosinophil ST2 in regulating eosinophil responses in vivo.
Notably, prior eosinophil studies did not distinguish donor and recipient eosinophils, with the only exception being the studies that addressed peritoneal homing under homestatic conditions using the 4get marker (16). We now report a scenario wherein adoptive transfer is able to answer questions that standard genetic models alone cannot readily answer. Using classic approaches, it would be arbitrary to attribute certain phenotypes to the expression of GOI on either eosinophils or migration target tissue without a comprehensive understanding of expression topology or efforts to make tissue-specific conditional knockout mice. In this study, we used Il13ra1−/− mice from two genetic backgrounds (BALB/c and C57BL/6) to demonstrate that the phenotype of abolished airway eosinophilia in Il13ra1−/− mice is observed across two mouse strains. The Il13ra1−/− recipient mice results demonstrate that the previously observed airway eosinophil migration deficit (23) was primarily due to the IL-13RA1 receptor expressed on recipient cells (likely respiratory epithelium) rather than to IL-13RA1 expressed on the eosinophils themselves (25, 26) or to an altered eosinophil developmental property driven by IL-13RA1. These findings not only confirm the Il13ra1−/− recipient phenotype but also substantiate the utility of these systems in that eosinophil migration phenotype detection can be readily performed using these two common genetic backgrounds in the allergy/eosinophil field, namely BALB/c and C57BL/6.
To demonstrate that our transfer system is also capable of detecting a phenotypic difference from a donor perspective, we used Ccr3−/− donor eosinophils (27) in conjunction with airway allergen-challenged 4get recipients to model the expected donor eosinophil migration deficit due to the disrupted eotaxin/CCR3 system. With nearly all of the eosinophils in allergen-challenged airway being IL4-GFP positive, we used the GFP-negative window (after eosinophil gating) to observe the migration of Ccr3+/+ and Ccr3−/− donor eosinophils without any labeling, observing the expected deficit in migration of Ccr3−/− donor eosinophils. This “reverse-transfer” approach, transferring unlabeled donor eosinophils into labeled recipients, can be applied to other GOIs to address their contributions to eosinophil migration without anatomical confounders of the target tissue.
Kinetic studies indicated that donor eosinophils had a much longer half-life in the recipient airway compared with the circulation (8 d vs. 3 h). This vast difference is likely due to the presence of eosinophil-promoting cytokines in the local lung environment, primarily IL-5. Even with the experimental asthma model, the distribution of IL-5 is biased toward the target tissue due to the fact that the cytokine-producing T helper cells are concentrated in the pathological site. Considering that there are a number of human diseases with eosinophilia as a primary characteristic and/or leading cause of pathogenesis (28), the system introduced here can also serve as a valuable tool to assess eosinophil half-life in multiple tissue types without the interference of concurrent eosinophilopoeisis in the recipient bone marrow.
We have also successfully demonstrated the feasibility of cotransferring two donor eosinophil populations and studying their respective airway homing capacities. Because the comparison parameter is a ratio of the two populations after airway migration, one of the major advantages of this design is that the two donor populations are mutually controlled for common variables in experimental asthma induction; this experimental design results in higher tolerance to experimental errors and hence higher experimental resolution. The demonstrated linearity of this model will allow assaying mild migration difference between wild-type (CD45.1) and mutant (CD45.2) eosinophils in the C57BL/6 background. Although we barely detect any phenotypic difference between the donor and recipient native eosinophils, such as migration and surface marker expression, a functional difference cannot be ruled out. The potential difference should not affect the experimental goals herein.
It is well recognized that the gene and protein expression levels of components of the IL-33/ST2 axis strongly correlate with both human and mouse asthmatic onset (29–31); however, despite well-documented ST2 expression on eosinophils (32), it is not known how ST2 regulates eosinophil migration and pulmonary homing per se. In the presence of IL-5, we did not observe a pronounced developmental defect in St2−/− eosinophils in vitro in terms of multiple eotaxin-driven migration or eosinophil-specific marker expression; nonetheless, we hypothesized that a survival difference driven by ST2 may reveal unique regulation specifically on eosinophils, rendering the St2−/− eosinophils quantitatively less competitive. The optimized competitive eosinophil transfer model introduced herein substantiated this suspected deficit as well as the utility of this approach. Indeed, we demonstrated the prosurvival function of ST2 on eosinophils both in vitro and in vivo, which may explain the airway migration phenotype, at least in part. We believe that this finding is a unique observation with respect to the role of ST2 on eosinophils and that this experiment exemplifies the merit of the transfer system in elucidating fine cellular mechanisms without extensive genetic engineering and crossing.
Collectively, we have established a unique, bone-marrow–derived eosinophil adoptive transfer system well suited to study the molecular mechanisms of eosinophil airway homing as a comprehensive function of eosinophil adhesion, chemotaxis, and survival (for summary and applications of the approaches performed herein, see Table 1). Compared with the existing eosinophil transfer models (4, 15–17), the strengths of this unique system include: (i) a readily available eosinophil source with high viability and quantity; (ii) the requirement for relatively small amounts of eosinophils to get a satisfactory window of detection; (iii) the capability of addressing both donor- and/or recipient-expressed genes without further crossing or chemical labeling; (iv) the utility of the dual detecting systems, enabling phenotyping studies to be performed in two common strains of mice, C57BL/6 and BALB/c; and (v) the ability to monitor airway migration of two populations of donor eosinophils simultaneously. Although the described system is currently focused on the asthmatic response, future studies will be directed toward other anatomical/pathological conditions, such as dextran sulfate sodium (DSS)-induced colitis and atopic dermatitis. The unique transfer system introduced herein has potential to facilitate future research in TH2-biased inflammation and basic eosinophil biology, allowing a more comprehensive understanding of the synchronized interaction of multiple eosinophil migration factors in the targeted tissues.
Table 1.
Summary of different variations of eosinophil adoptive transfer
| System | Stimuli | Background* | Donor | Recipient | GOI assessment | Advantage |
| 4get (IL4-eGFP) | Allergen† | BALB/c, 129 | 4get | GOI+/+ and −/− | Target tissue | Automatic tagging |
| Reverse 4get | Allergen | BALB/c, 129 | GOI+/+ and −/− | 4get | Eosinophil | Automatic tagging |
| ΔdblGATA1 | Allergen | BALB/c, 129, C57BL/6 | GOI+/+ and −/− | ΔdblGATA-1 | Eosinophil | Ablation of recipient eosinophils |
| CD45.1/CD45.2 | Allergen | C57BL/6 | CD45.2 or CD45.1 | CD45.1 or CD45.2 | Target tissue or Eosinophil | Donor/recipient interchangability |
| Competitive | Allergen | C57BL/6 | CD45.2(−/−) and CD45.1(+/+) | CD45.1–CD45.2+/− | Eosinophil | Concurrent mutual control |
| Intranasal chemokine | Eotaxin-1/IL-5 | Any | Any | Any | Eosinophil | “In vivo Transwell” simple setup |
*Or any additional strains, in which the 4get and CD45.1 allele congenic mice are available.
†Aspergillus fumigatus
Materials and Methods
Mice.
C57BL/6J wild-type (CD45.2) mice were obtained from Charles River, and BALB/c mice were obtained from Taconic Farms. The initial breeders of BALB/c IL4-eGFP transgenic (4get) (20) and C57BL/6 CD45.1 mice (21) were obtained from Yui-Hsi Wang and Simon Hogan (Cincinnati Children’s Hospital Medical Center, Cincinnati), respectively, and are commercially available from multiple vendors including Jackson Laboratory and Charles River on multiple backgrounds. St2−/− mice are a generous gift obtained from Andrew McKenzie’s laboratory (Cambridge University, Cambridge, United Kingdom). Experimental animals were housed in specific pathogen-free conditions at Cincinnati Children’s Hospital Medical Center under Institutional Animal Care and Use Committee-approved protocols.
Induction of Experimental Asthma in Synchronization with Eosinophil Adoptive Transfer.
Allergic lung inflammation was established with intranasal challenges of Aspergillus fumigatus in BALB/c and C57BL/6 mice (22). Briefly, a total of five intranasal challenges with Aspergillus extract (Greer) dissolved in 70 µL of sterile saline were administered on an every-other-day regimen at the dose of 100 µg total protein (BCA assay, Pierce; 23225). The eosinophil culture and asthma induction were synchronized so that the fifth Aspergillus challenge was performed on day 14 of the eosinophil culture. Six hours after the fifth Aspergillus challenge, a bolus of cultured eosinophils was transferred i.v. in PBS by tail vein injection. After 24 h of incubation, animals were subsequently euthanized with CO2, and BALF was collected by lavage for polychromatic FACS analysis.
Statistical Analysis.
Statistical significance was analyzed using a two-tailed Student t test in all instances unless noted otherwise in the figure legends. Numerical data were graphed as mean ± SEM. Linear regression was established with the Pearson method.
Supplementary Material
Acknowledgments
The authors thank Dr. Yui-Hsi Wang and Dr. Ariel Munitz for constructive scientific discussions and professional experimental suggestions; and Shawna Hottinger for editorial assistance. This work was supported by the National Institutes of Health Grants R37 A1045898 and R01 AI083450, the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation, the Food Allergy Research & Education (FARE) Foundation, and the Buckeye Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220572110/-/DCSupplemental.
References
- 1.Barrett NA, Austen KF. Innate cells and T helper 2 cell immunity in airway inflammation. Immunity. 2009;31(3):425–437. doi: 10.1016/j.immuni.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 3.Walsh ER, Stokes K, August A. The role of eosinophils in allergic airway inflammation. Discov Med. 2010;9(47):357–362. [PubMed] [Google Scholar]
- 4.Shen HH, et al. A causative relationship exists between eosinophils and the development of allergic pulmonary pathologies in the mouse. J Immunol. 2003;170(6):3296–3305. doi: 10.4049/jimmunol.170.6.3296. [DOI] [PubMed] [Google Scholar]
- 5.Jacobsen EA, Zellner KR, Colbert D, Lee NA, Lee JJ. Eosinophils regulate dendritic cells and Th2 pulmonary immune responses following allergen provocation. J Immunol. 2011;187(11):6059–6068. doi: 10.4049/jimmunol.1102299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Górski P, et al. Eotaxin but not MCP-3 induces eosinophil influx into nasal fluid in allergic patients. Allergy. 2002;57(6):519–528. doi: 10.1034/j.1398-9995.2002.03555.x. [DOI] [PubMed] [Google Scholar]
- 7.Broide DH, Finkelman F, Bochner BS, Rothenberg ME. Advances in mechanisms of asthma, allergy, and immunology in 2010. J Allergy Clin Immunol. 2011;127(3):689–695. doi: 10.1016/j.jaci.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 8.Song DJ, et al. Anti-Siglec-F antibody reduces allergen-induced eosinophilic inflammation and airway remodeling. J Immunol. 2009;183(8):5333–5341. doi: 10.4049/jimmunol.0801421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aceves SS, Broide DH. Airway fibrosis and angiogenesis due to eosinophil trafficking in chronic asthma. Curr Mol Med. 2008;8(5):350–358. doi: 10.2174/156652408785161023. [DOI] [PubMed] [Google Scholar]
- 10.Li SH, Luo YL, Lai WY. [Expression of eosinophil major basic protein mRNA in bronchial asthma] Nan Fang Yi Ke Da Xue Xue Bao. 2006;26(9):1330–1333. Chinese. [PubMed] [Google Scholar]
- 11.Saeed W, Badar A, Hussain MM, Aslam M. Eosinophils and eosinophil products in asthma. J Ayub Med Coll Abbottabad. 2002;14(4):49–55. [PubMed] [Google Scholar]
- 12.Boyce JA, Austen KF. No audible wheezing: Nuggets and conundrums from mouse asthma models. J Exp Med. 2005;201(12):1869–1873. doi: 10.1084/jem.20050584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes PJ. New drugs for asthma. Nat Rev Drug Discov. 2004;3(10):831–844. doi: 10.1038/nrd1524. [DOI] [PubMed] [Google Scholar]
- 14.Corren J. Anti-interleukin-5 antibody therapy in asthma and allergies. Curr Opin Allergy Clin Immunol. 2011;11(6):565–570. doi: 10.1097/ACI.0b013e32834c3d30. [DOI] [PubMed] [Google Scholar]
- 15.Pero RS, et al. Galphai2-mediated signaling events in the endothelium are involved in controlling leukocyte extravasation. Proc Natl Acad Sci USA. 2007;104(11):4371–4376. doi: 10.1073/pnas.0700185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohnmacht C, Pullner A, van Rooijen N, Voehringer D. Analysis of eosinophil turnover in vivo reveals their active recruitment to and prolonged survival in the peritoneal cavity. J Immunol. 2007;179(7):4766–4774. doi: 10.4049/jimmunol.179.7.4766. [DOI] [PubMed] [Google Scholar]
- 17.Walsh ER, et al. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J Exp Med. 2008;205(6):1285–1292. doi: 10.1084/jem.20071836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyer KD, et al. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol. 2008;181(6):4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyer KD, Percopo CM, Rosenberg HF. 2009. Generation of eosinophils from unselected bone marrow progenitors: Wild-type, TLR- and eosinophil-deficient mice. Open Immunol J 2:163–167. [DOI] [PMC free article] [PubMed]
- 20.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15(2):303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 21.Yakura H, Shen FW, Bourcet E, Boyse EA. On the function of Ly-5 in the regulation of antigen-driven B cell differentiation. Comparison and contrast with Lyb-2. J Exp Med. 1983;157(4):1077–1088. doi: 10.1084/jem.157.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra A, Weaver TE, Beck DC, Rothenberg ME. Interleukin-5-mediated allergic airway inflammation inhibits the human surfactant protein C promoter in transgenic mice. J Biol Chem. 2001;276(11):8453–8459. doi: 10.1074/jbc.M009481200. [DOI] [PubMed] [Google Scholar]
- 23.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci USA. 2008;105(20):7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195(11):1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myrtek D, et al. Expression of interleukin-13 receptor alpha 1-subunit on peripheral blood eosinophils is regulated by cytokines. Immunology. 2004;112(4):597–604. doi: 10.1046/j.1365-2567.2004.01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dulkys Y, Buschermöhle T, Escher SE, Kapp A, Elsner J. T-helper 2 cytokines attenuate senescent eosinophil activation by the CXCR4 ligand stromal-derived factor-1alpha (CXCL12) Clin Exp Allergy. 2004;34(10):1610–1620. doi: 10.1111/j.1365-2222.2004.02063.x. [DOI] [PubMed] [Google Scholar]
- 27.Fulkerson PC, et al. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc Natl Acad Sci USA. 2006;103(44):16418–16423. doi: 10.1073/pnas.0607863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valent P, et al. 2012. Contemporary consensus proposal on criteria and classification of eosinophilic disorders and related syndromes. J Allergy Clin Immunol 130(3):607–612.
- 29.Stolarski B, Kurowska-Stolarska M, Kewin P, Xu D, Liew FY. IL-33 exacerbates eosinophil-mediated airway inflammation. J Immunol. 2010;185(6):3472–3480. doi: 10.4049/jimmunol.1000730. [DOI] [PubMed] [Google Scholar]
- 30.Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: The new kid in the IL-1 family. Nat Rev Immunol. 2010;10(2):103–110. doi: 10.1038/nri2692. [DOI] [PubMed] [Google Scholar]
- 31.Akhabir L, Sandford A. Genetics of interleukin 1 receptor-like 1 in immune and inflammatory diseases. Curr Genomics. 2010;11(8):591–606. doi: 10.2174/138920210793360907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pecaric-Petkovic T, Didichenko SA, Kaempfer S, Spiegl N, Dahinden CA. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood. 2009;113(7):1526–1534. doi: 10.1182/blood-2008-05-157818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.