Abstract
Cancer is a complex disease with a range of genetic and biochemical markers within and among tumors, but a general tumor characteristic is extracellular acidity, which is associated with tumor growth and development. Acidosis could be a universal marker for cancer imaging and the delivery of therapeutic molecules, but its promise as a cancer biomarker has not been fully realized in the clinic. We have discovered a unique approach for the targeting of acidic tissue using the pH-sensitive folding and transmembrane insertion of pH (low) insertion peptide (pHLIP). The essence of the molecular mechanism has been elucidated, but the principles of design need to be understood for optimal clinical applications. Here, we report on a library of 16 rationally designed pHLIP variants. We show how the tuning of the biophysical properties of peptide–lipid bilayer interactions alters tumor targeting, distribution in organs, and blood clearance. Lead compounds for PET/single photon emission computed tomography and fluorescence imaging/MRI were identified, and targeting specificity was shown by use of noninserting variants. Finally, we present our current understanding of the main principles of pHLIP design.
Keywords: membrane-associated folding, tumor acidity, thermodynamics, kinetics, diagnostics
The promise of exploiting tumor acidosis as a cancer biomarker has not been fully realized in clinical practice. The problem has been to find a practical way to target acidity. While studying membrane protein folding, we discovered a peptide called pH (low) insertion peptide (pHLIP) that reversibly folds and inserts across membranes in response to pH changes, and this discovery has led to a unique way to target acidic tissue. Our biophysical studies have revealed the molecular mechanism of pHLIP action (1–3), and we have shown that pHLIP can target acidic tissue and selectively translocate polar, cell-impermeable molecules across cell membranes in response to low extracellular pH (1, 4–9). pHLIP conjugated with fluorescent dyes, PET [64Cu-DOTA (1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid) and 18F], single photon emission computed tomography (SPECT; 99Tc) probes, and gold nanoparticles targets acidic tumors (1, 4, 5, 10, 11). All prior studies in vivo were carried out with the WT-pHLIP sequence and showed that a good contrast and tumor-to-blood ratio can be achieved only more than 24 h after pHLIP injection, when it has accumulated in the tumor and largely cleared from the blood (5, 6). However, for the use of pHLIP-based radioactive imaging agents in the clinic, a more rapid background signal reduction is absolutely essential. To address this important need, to tune tumor-targeting properties, and to broaden our understanding of the molecular mechanism of pHLIP action, we designed a set of 16 pHLIP variants based on chemical and physical principles. Comprehensive biophysical studies performed with nonlabeled peptides were correlated with investigations of tumor targeting and organ distributions of fluorescent versions of the pHLIP variants. As a result, a set of design criteria has been established and pHLIP candidates for imaging and therapeutic applications, including lead compounds for PET/SPECT imaging and fluorescence imaging/MRI, have been identified.
Results
Design of pHLIP Variants.
We have reported the basic molecular mechanism of the interaction of WT-pHLIP with lipid bilayers (refs. 1, 2, 12). In these studies, we found three states of the peptide: state I, in solution as an unstructured monomer at neutral pH when no lipid membrane is present; state II, bound at the surface of a lipid bilayer as a largely unstructured monomer at neutral pH; and state III, inserted across the bilayer as a monomeric helix at acidic pH. To broaden our understanding of the main principles of pHLIP interactions with membranes and to select the best sequences for clinical use, we used our knowledge and designed 16 variants of the WT-pHLIP [variant (Var) 0] sequence:
Var0-WT: ACEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT
Var1: ACEDQNPYWARYADWLFTTPLLLLDLALLVDG
Var2: ACEDQNPYWRAYADLFTPLTLLDLLALWDG
Var3: ACDDQNPWRAYLDLLFPTDTLLLDLLW
Var4: ACEEQNPWRAYLELLFPTETLLLELLW
Var5: ACDDQNPWARYLDWLFPTDTLLLDL
Var6: CDNNNPWRAYLDLLFPTDTLLLDW
Var7: ACEEQNPWARYLEWLFPTETLLLEL
Var8: CEEQQPWAQYLELLFPTETLLLEW
Var9: CEEQQPWRAYLELLFPTETLLLEW
Var10: ACEDQNPWARYADWLFPTTLLLLD
Var11: ACEEQNPWARYAEWLFPTTLLLLE
Var12: ACEDQNPWARYADLLFPTTLAW
Var13: ACEEQNPWARYAELLFPTTLAW
Var14: Ac-TEDADVLLALDLLLLPTTFLWDAYRAWYPNQECA-Am
Var15: CDDDDDNPNYWARYANWLFTTPLLLLNGALLVEAEET
Var16: CDDDDDNPNYWARYAPWLFTTPLLLLPGALLVEAEET
Biophysical Studies.
We studied the interaction of the pHLIP variants with lipid bilayers using the fluorescence and CD spectroscopic techniques previously used (2). The intrinsic tryptophan fluorescence and CD spectra of nonlabeled variants were measured in solution at pH 8 in the absence and presence of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) liposomes and in the presence of liposomes at low pH (pH 4–5), and they show that each peptide interacts with a lipid bilayer in a pH-dependent manner and that the peptides form transmembrane (TM) helices in state III (Fig. S1 and Table S1).
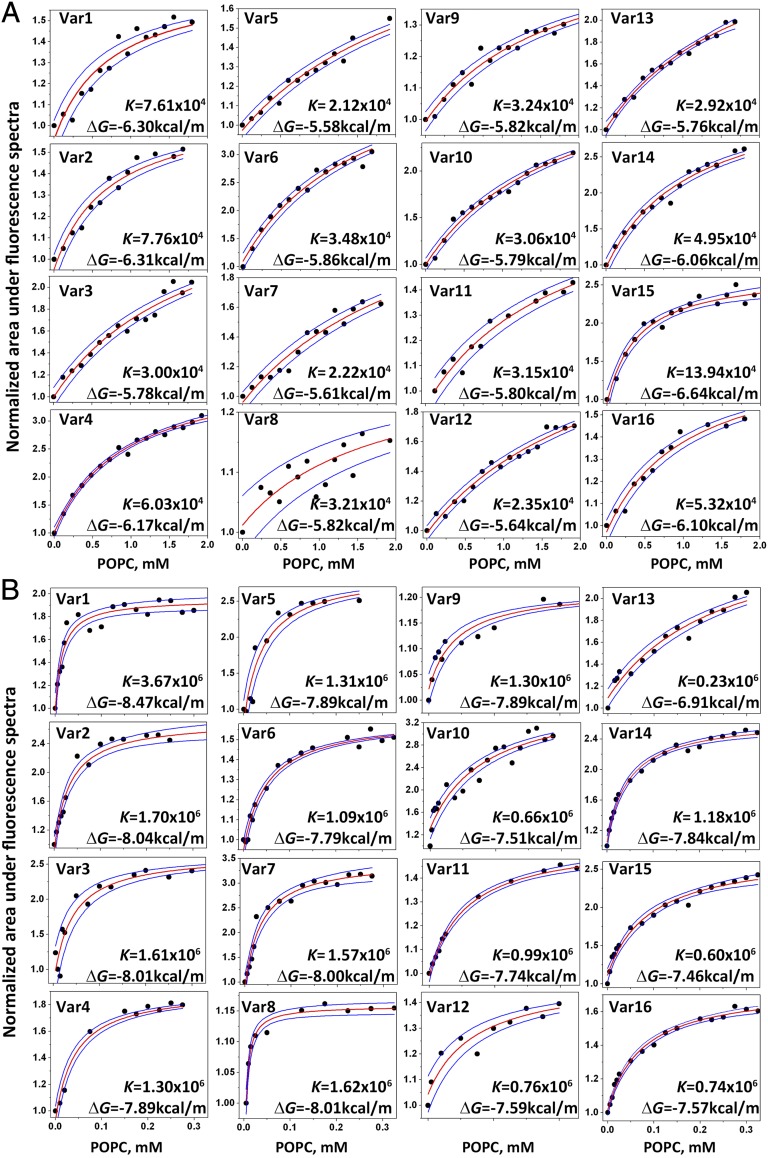
The partitioning of pHLIP variants into bilayers at pH 8 and pH 4–5 was assessed in titration experiments by measuring changes of the intrinsic peptide fluorescence (Fig. 1). On average, the partitioning of the variants into the membrane at low pH is about 50-fold higher than at high pH. As expected, the truncated variants (Var5–13) have lower affinities for the membrane at neutral pH compared with WT and other variants, because a number of hydrophobic residues were removed from the sequences. In general, variants with Glu residues have slightly higher affinities compared with the same variants with Asp residues (because Glu is more hydrophobic due to the additional methylene group). The highest affinity was observed for Var15, which forms a helical structure at neutral pH. At low pH, the lowest affinity for the lipid bilayer was observed for the most truncated versions Var10–13 and variants (Var15 and Var16) with no protonatable residues in the TM, which reflects the reduced stability of the TM helix. The differences between the Gibbs free energies (ΔGs) of the interactions of variants with the membrane at low and high pHs (in kilocalories per mole), ΔΔG = ΔGpH4 − ΔGpH8, are as follows:
Var1 Var2 Var3 Var4 Var5 Var6 Var7 Var8
2.17 0.28 2.23 1.72 2.31 1.93 2.39 2.19
Var9 Var10 Var11 Var12 Var13 Var14 Var15 Var16
2.07 1.79 1.94 1.95 1.15 1.78 0.82 1.47
Fig. 1.
Titration of pHLIP variant binding to liposomes at high and low pHs. The titration of the pHLIP variants with increasing concentrations of POPC liposomes at pH 8 (A) and pH 4–5 (B) is followed using intrinsic peptide fluorescence changes. For comparison the mole-fraction partition coefficients for Var0 (WT-pHLIP) at pH 8 and pH 5 are 4.87 × 104 and 1.54 × 106, respectively, and the corresponding ΔGs are 6.04 and 7.98 kcal/mol, respectively. The red and blue lines are fitting curves and 95% confidence intervals, respectively.
The largest differences are observed for Var3, Var5, and Var7. The smallest differences are found for Var2 and Var15 due to the strong interactions of these peptides with the bilayer in state II at pH 8.
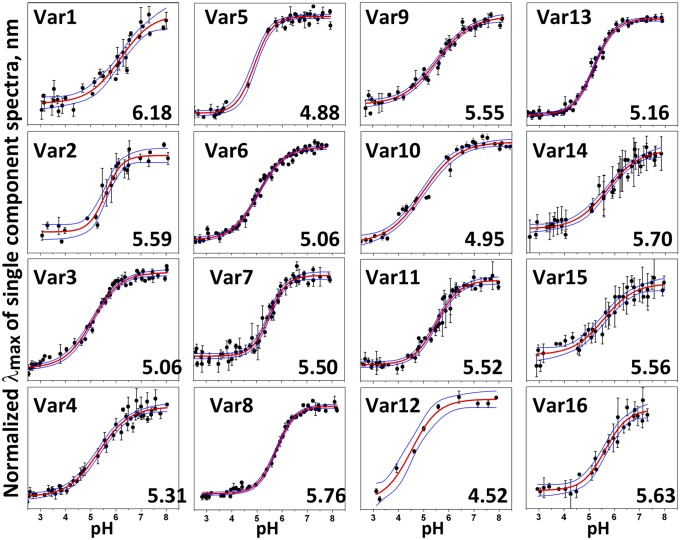
The pKa of pHLIP variant insertion was measured by following the shift of the position of the intrinsic peptide emission maximum as the pH is changed from pH 8 to pH 2 (Fig. 2). The shift of pKa to lower pHs correlates with the truncation of the pHLIP sequence: The pKa of Var10, with a single Asp residue in its TM, is the lowest (pKa = 4.5). All variants containing Glu residues (Var7–9, Var11, and Var13) have higher pKa values compared with the related sequences with Asp residues (Var5–6, Var10, and Var12).
Fig. 2.
pH-dependent bilayer insertion of variants. Peptide intrinsic fluorescence changes are used to follow the insertion as a function of pH (transition from state II to state III as the pH is lowered). The red and blue lines are fitting curves and 95% confidence intervals, respectively.
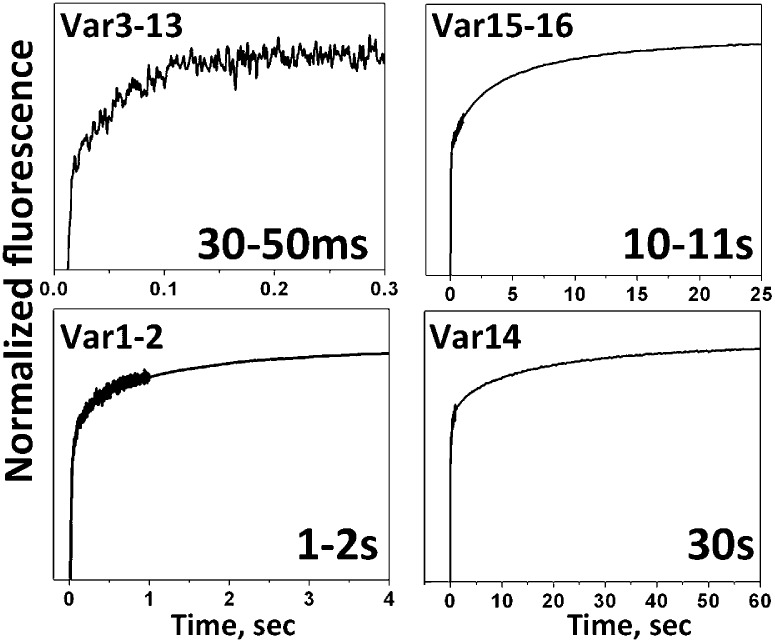
To this point, we have considered the equilibrium energies of progressive binding and insertion events; however, for some applications, it is also important that sequence variation will alter the kinetics of insertion. We have found in recent work that the kinetics of insertion correlate with the number of protonatable groups at the inserting end or the presence of polar cargo (13). The characteristic membrane insertion time for WT-pHLIP is about 30 s, and slightly faster kinetics were observed for Var14, which is the reverse sequence of WT. Var14 has the same number of protonatable residues at its N terminus as WT has at its C terminus, except for the free C terminus itself. The data are consistent with the view that the acetylated N terminus of Var14 inserts across the bilayer and the amidated C terminus stays outside, reversing the direction of insertion. The characteristic time of Var1–2 insertion, which has fewer protonatable residues at its C terminus than WT, was about 1–2 s, which correlates well with our recent findings (13). Further truncation of the peptide-inserting end, which reduces the number of protonatable residues, resulted in faster peptide insertion, completed within first 30–100 ms for Var3–13. There are no protonatable residues in the TM in Var15–16, although they have the same number of Asp/Glu residues as WT at the inserting end; as a result, the time of insertion was reduced to 10–11 s relative to WT. The kinetics data are shown in Fig. 3.
Fig. 3.
Kinetics of insertion. Representative kinetic curves for the insertion of variant peptides into the lipid bilayer are shown. The characteristic time of insertion of Var0 (WT-pHLIP) is about 40 s.
Tumor Targeting and Distribution of Alexa750-pHLIPs in Organs.
We used a mouse xenograft model to assess the tumor targeting and organ distribution of the pHLIP variants. Tumors were created by s.c. injection of HeLa-GFP cancer cells in the right flanks of each mouse, and they were grown to about 5 mm in diameter. To perform biodistribution studies, pHLIP variants were conjugated with Alexa750. We demonstrated that the kinetics of insertion of Alexa750-WT-pHLIP into lipid bilayers are similar to the kinetics of insertion of nonlabeled peptide (Fig. S2). Fluorescently labeled pHLIPs were given as single i.v. injections, and the major organs and tumors were collected at 4 and 24 h after fluorescent pHLIP administration. The GFP and near infrared (NIR) signals were monitored in tumors and organs after killing the mice. The mean intensities of NIR fluorescence normalized to the signal in kidneys at 4 h are shown in Fig. S3. The mean intensities of NIR fluorescence of all organs and the number of mice used in the study for each variant are given in Tables S2 and S3.
The pHLIP variants that show higher affinities for membranes at high pH have higher short-term liver uptake (Var1, Var2, Var4, and Var14–16); however, the liver is cleared at 24 h, except for Var1. Var3 has high tumor targeting compared with kidney and compared with other pHLIPs as well. Whole-body and organ imaging confirmed that the truncated variants Var5–13 have much faster blood clearance than WT; at 4 h, the signal has accumulated in the tumor, although being washed out of the organs. Tumor targeting was minimal for the shortest pHLIP variants Var12–13. The tumor/muscle, tumor/kidney, and tumor/liver ratios are given in Fig. 4 for all pHLIP variants calculated at 4 and 24 h after injection. Var0, Var3–4, and Var14–16 have the highest tumor/muscle ratio at 24 h. The same pHLIP variants have the highest tumor/kidney ratios, whereas only Var3 shows significantly less accumulation in the liver. Among the truncated pHLIP variants, Var7 demonstrates the highest tumor targeting.
Fig. 4.
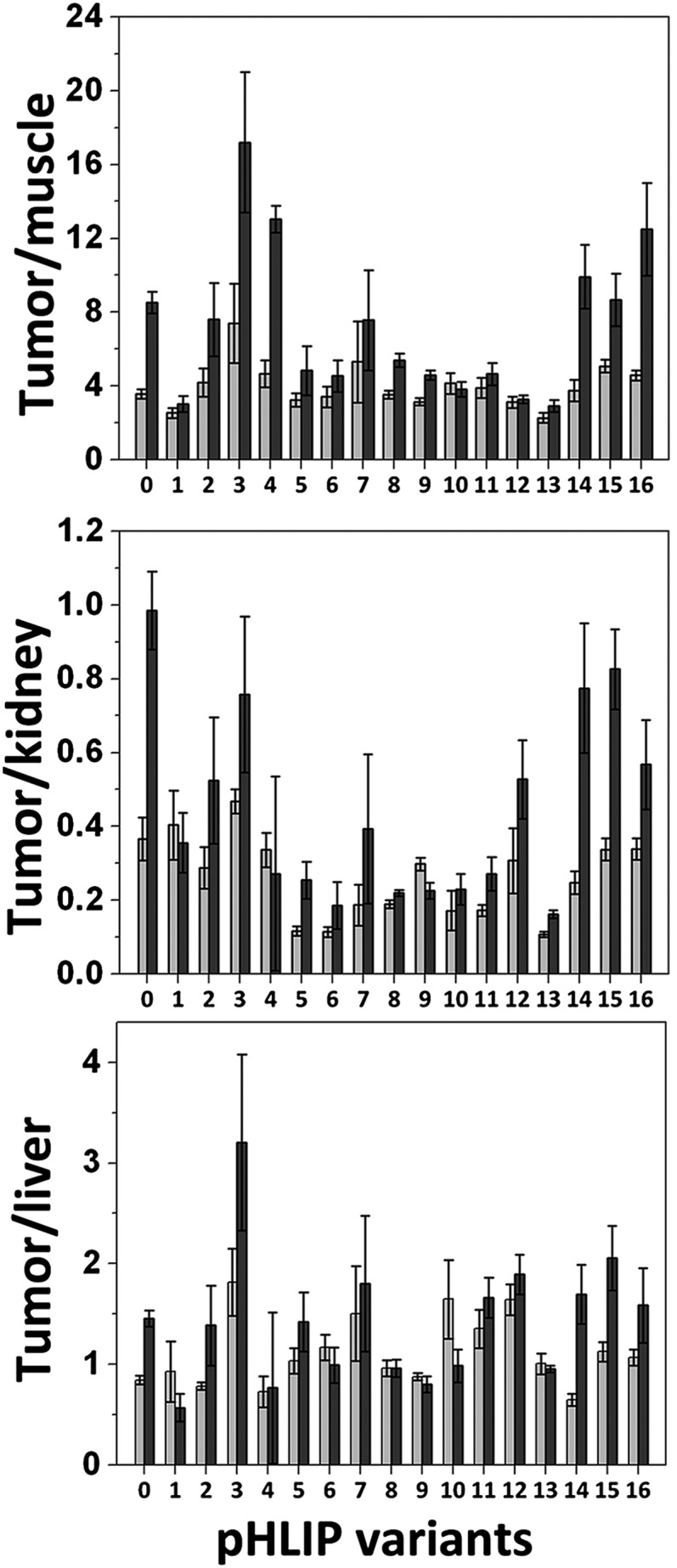
Tumor-to-organ fluorescence ratios. Tumor-to-muscle, tumor-to-kidney, and tumor-to-liver ratios calculated at 4- and 24-h time points are shown for all pHLIP variants.
This body of work allows us to propose different pHLIP variants for different applications. We have selected Var3 and Var7 as the best candidates for future studies for fluorescence imaging/MRI and PET/SPECT imaging, respectively, guided by the fact that the imaging time window for the stable fluorescence and MRI agents can be much wider than for the short t1/2 of PET and SPECT agents. Var3 had the highest tumor/organ ratios, especially at 24 h after construct administration. Var7 had a lower tumor/organ ratio but showed much faster blood clearance. The cytotoxicity of the selected pHLIP variants was evaluated on human cervix adenocarcinoma (HeLa) and human lung carcinoma (A549) cells, as well as on human cervical (HCvEpC), mammary (HMEpC), and bronchial (HBEpC) epithelial cells. Peptides at concentrations of up to 8 μM were evaluated at up to 72 h of incubation with cells, and no toxicity was observed.
pH-Insensitive K-pHLIPs.
As a control, we have previously used K-pHLIP-WT, where the two key D (Asp) residues in the TM part of the peptide were replaced by K (Lys) residues (1). K-pHLIP cannot interact with a membrane in a pH-dependent manner over the range of neutral and low pHs we are studying; therefore, it should not target acidic tumors. Because of the potential clinical applications of Var3 and Var7, we synthesized and tested K-versions of each:
K-Var3: ACDDQNPWRAYLKLLFPTKTLLLKLLW
K-Var7: ACEEQNPWARYLKWLFPTKTLLLKL
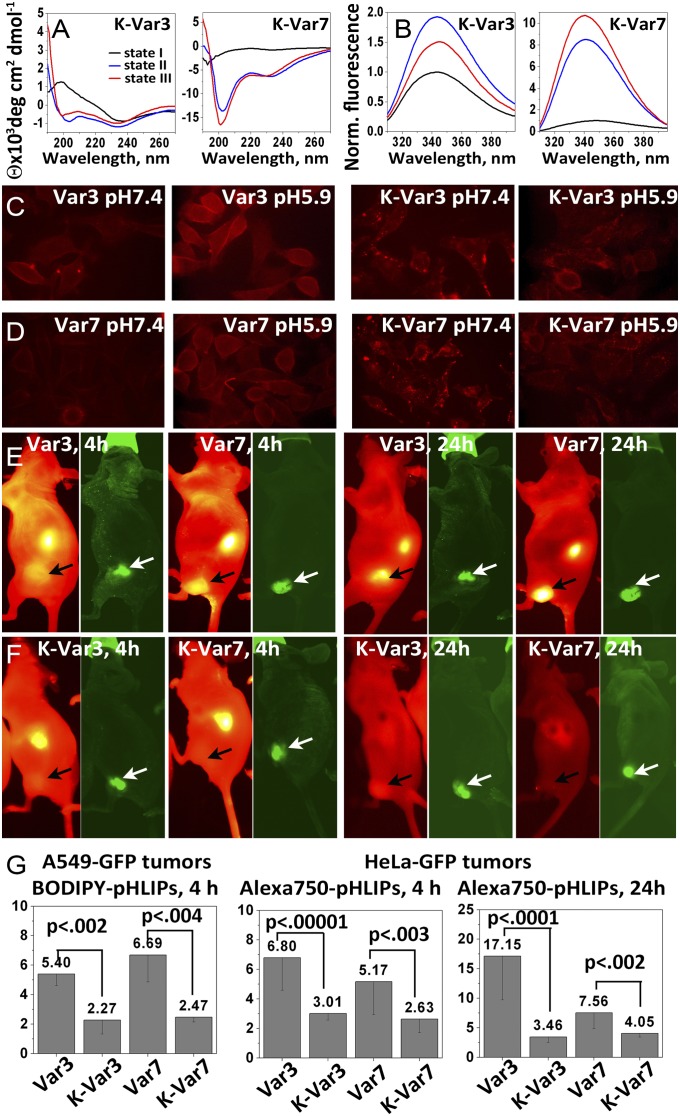
Each of these peptides failed to interact with lipid bilayers in a pH-dependent fashion (Fig. 5 A and B). No shift in the position of the peptide fluorescence maximum was seen for either of the K-pHLIPs in the presence of lipid as a result of pH decrease, and the CD spectra do not change and do not exhibit the minima at 208 and 225 nm that are characteristic signals reporting α-helical conformations. The CD spectra reveal a negative band at 234 nm and a positive band at 221 nm, suggestive of an exciton split doublet (2, 14) possibly arising from peptide aggregation.
Fig. 5.
Comparisons with control K-pHLIPs. The CD (A) and tryptophan fluorescence (B) spectra of K-Var3 and K-Var7 show that these variants lose the ability to interact with a lipid bilayer in a pH-dependent manner. (C and D) Cellular uptake at pH 7.4 and pH 5.9 of peptides labeled with rhodamine is shown. The images were obtained under an inverted optical microscope with a 60× objective. The cellular uptake of peptides labeled with BODIPY is shown at both pHs in Fig. S4. Whole-body NIR fluorescence images of mice obtained at 4 and 24 h after i.v. administration of Var3 and K-Var3 (E) and Var7 and K-Var7 (F) conjugated with Alexa750 are shown. The tumors are indicated by arrows. Images obtained at different time points are presented with maximum contrast; the contrast-to-noise ratios are as follows: Var3, 4 h = 103; Var3, 24 h = 186; Var7, 4 h = 85; Var7, 24 h = 360; K-Var3, 4 h = 87; K-Var3, 24 h = 54; K-Var7, 4 h = 22; and K-Var7, 24 h = 16. (G) A549-GFP tumor-to-muscle ratios calculated for 4 h for BODIPY-pHLIPs and BODIPY-K-pHLIPs and HeLa-GFP tumor-to-muscle ratios calculated for 4- and 24-h time points for Alexa750-pHLIPs and Alexa750-K-pHLIPs are shown. The mean fluorescence of HeLa-GFP and A549-GFP tumors and organs at 4 and 24 h after administration of Alexa750-K-pHLIPs and at 4 h after administration of BODIPY-pHLIPs and BODIPY-K-pHLIPs are given in the Tables S4 and S5, respectively.
Var3 and Var7, as well as K-Var3 and K-Var7, were conjugated with two different fluorescent dyes, zwitterionic tetramethylrhodamine (Rho) and noncharged boron-dypyrromethene (BODIPY), to ensure that distribution and localization of fluorescent peptides is independent of the choice of dye. The fluorescence images demonstrating cellular uptake of Rho- and BODIPY-labeled Var3, Var7, K-Var3, and K-Var7 are shown in Fig. 5 C and D and Fig. S4, respectively. The fluorescence signal of cells treated with fluorescent Var3 and Var7 at pH 7.4 was weaker compared with that of cells treated with peptides at pH 5.9. The cellular uptake of Var3 was 1.12-fold higher at low pH compared with neutral pH, and uptake of Var7 was 1.34-fold higher at low pH. Our biophysical data show that the affinity of Var3 for a membrane at pH 8 is higher than the affinity of Var7 at the same pH; therefore, we see slightly higher cellular uptake of Var3 at pH 7.4 compared with Var7 and a less pronounced difference in cellular uptake between neutral and low pHs for Var3. For the cases of K-Var3 and K-Var7, the fluorescence signal was very similar when cells were treated at either high or low pH; moreover, the fluorescence signal at pH 7.4 was slightly higher than at pH 5.9. Also, the distributions of fluorescent signals for D (Asp)- and E (Glu)-variants and K (Lys)-variants were different. K-Var3 and K-Var7 were distributed as cytoplasmic dots. In contrast to pHLIP variants, K-pHLIPs are positively charged and partially aggregated, and, most probably, they might be taken up by endocytosis and trapped in endosomes in a pH-independent manner, whereas cellular uptake of Var3 and Var7 is pH-dependent.
Biodistribution and tumor targeting of Alexa750-K-pHLIPs were also investigated using xenograft models in mice. In contrast to Var3 and Var7, K-pHLIPs did not show significant tumor targeting (Fig. 5 E and F and Table S4). To demonstrate that tumor targeting by Var3 and Var7 and the lack of tumor targeting by K-Var3 and K-Var7 are independent of the choice of tumor model and imaging fluorescent probe, we used BODIPY-labeled Var3, Var7, K-Var3, and K-Var7, which were tested on tumors created by s.c. injection of A549-GFP human lung carcinoma cells. The organs were collected at 4 h and analyzed (Table S5). The tumor-to-muscle ratio at 4 h for Var3 and Var7 labeled with BODIPY and Alexa750 was 6.0 ± 0.9, whereas for K-Var3 and K-Var7, it was 2.6 ± 0.3 (Fig. 5G). Thus, the replacement of the Asp/Glu residues by Lys in the TMs of Var3 and Var7 leads to the loss of both pH-dependent interactions with membranes and targeting of acidic tumors, supporting a central role for the carboxyl groups in targeting. However, we note that although K-pHLIPs may be considered as controls for in vitro studies, they should be used with caution in vivo, because the positive charges of K-pHLIPs might promote interactions with receptors and endocytotic uptake, which might vary for different tumor types.
Discussion
An acidic extracellular milieu favors tumor growth, invasion, and development (15–20), and the pHLIP peptides can exploit tumor acidity as a useful biomarker. Based on the results of our previous and current investigations, design principles can be formulated to set directions for different clinical uses:
The apparent pK of insertion of a pHLIP into a membrane depends on a combination of the pKs of protonation of all protonatable groups. Each pK of protonation of a carboxyl group depends on its environment, and it is increased as the surrounding dielectric constant is lowered. Thus, carboxyl pKs will be gradually raised with peptide propagation deeper into the membrane. The pKa of peptide insertion determines the proportion of available peptide molecules inserted into the plasma membrane of cancer cells at various pHs. Currently, we have pHLIP peptides with pKs varying over the range from 4.5 to 6.5 for insertion into POPC lipid bilayers. Various pHLIP variants might be used to probe the pHs of more or less acidic tumors. The insertion of pHLIP responds to the pH at the surfaces of cancer cells rather than that of the surrounding fluid, because the TM potential gradient causes protons to accumulate at the surfaces of cells and the pH is lower than in the bulk extracellular environment or in the blood. Current methods of measuring pH in vivo report the average value of the extracellular pH in tumors, which can vary from 6.0 to 7.2, but the pHs at the surfaces of cancer cells are likely to be lower. Moreover, the cellular pH gradient observed in tumors will favor cell entry of compounds with pKs in range of 4.5–6.5 (21, 22).
pHLIP is a relatively hydrophobic peptide; therefore, significant initial association with a lipid bilayer or plasma membrane occurs at neutral pH. The strength of this interaction (state II) is determined by the balance between hydrophobic and polar/charged residues in the pHLIP sequence. Stronger partitioning of the peptides onto bilayers at neutral pH increases the time of circulation in the blood, because the pHLIPs bind to the surface of blood cells. A long circulation time is a disadvantage for PET/SPECT imaging, because short t1/2 time probes are used and tumor targeting and blood clearance need to be very fast (preferably within 4 h). However, long circulation times of pHLIP variants in the blood may be well suited for fluorescence or MR imaging, as well as for the delivery of therapeutic agents. We find that all pHLIP variants that have higher affinities for membranes at neutral pH show high tumor/organ ratios at 24 h postinjection.
An important parameter is the difference between the ΔΔG of peptide binding to membranes at low pH vs. high pH. This parameter reflects the difference in the affinity of the peptide to the membrane at low and neutral pHs, as well as the strength of formation of the TM helix at low pH. A larger ΔΔG will ensure a greater differentiation between the inserted and noninserted peptides as a function of pH.
For applications in vivo, the kinetics of peptide insertion across the lipid bilayer are important for rapid equilibration with tissues and clearance from the blood. Based on insertion kinetics, we can group all pHLIP peptides into the following: (i) Peptides with protonatable residues both in the TM and in the inserting end show the slowest kinetics of insertion (minutes); (ii) peptides that are truncated at the inserting end and have few or no protonatable residues at the inserting end partition in bilayer much faster (seconds); (iii) peptides that have protonatable residues at the inserting end but not in the TM show intermediate times of insertion (∼20 s); and (iv) peptides with only one protonatable residue in the TM have the fastest kinetics of insertion, which coincide with the time of formation of the helical structure (∼100 ms).
Exit kinetics should be different in cells and liposomes, because peptides that have protonatable residues translocated across a cell membrane move them to the neutral pH of the cytoplasm. To exit the cell membrane, these groups must be protonated, which is much less likely in the cytoplasm; thus, these peptides become anchored in a cell. Such an “anchor” can significantly (by orders of magnitude) reduce the rate of peptide exit, and the peptide could stay in the plasma membrane for weeks. Such an effect would explain our observation that mouse tumors are stably labeled with fluorescent WT-pHLIP.
We have selected two pHLIP variants for further translational development (in addition to WT-pHLIP): Var3 and Var7. Var3 demonstrates high tumor-to-organ ratios at 24 h (similar to WT) and could be well suited for optical imaging or MRI 1 d after administration. Var3 might also be suitable for fluorescence-guided surgery, marking a tumor with fluorescently labeled pHLIP and performing surgery on the next day, when the contrast index is the highest. Var7 is one of the truncated versions of pHLIP, which could be appropriate for PET/SPECT imaging, because it demonstrates fast tumor targeting and blood clearance.
Materials and Methods
The details of experimental protocols can be found in SI Materials and Methods.
Materials.
pHLIP variants were prepared by solid-phase peptide synthesis at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Variants were conjugated with malemides of the dyes: Alexa750, Rho, or BODIPY. The purity of products was assessed by analytical HPLC, and peak identity was confirmed by surface-enhanced laser desorption/ionization TOF MS. Large unilamellar POPC (Avanti Polar Lipids, Inc.) vesicles were prepared by extrusion.
Steady-State Fluorescence and CD Measurements.
Peptide fluorescence spectra were recorded with the spectral widths of excitation and emission slits set at 4 and 2 nm, respectively, using excitation wavelengths of 295 or 280 nm. The concentrations of the peptides and POPC liposomes were 7 μM and 1.5 mM, respectively.
Oriented CD Measurements.
Oriented CD was measured from the supported bilayers deposited on a stack of quartz slides with special polish for far UV measurements as described previously (13).
Titration Experiments.
The intrinsic fluorescence spectra of samples containing 5 μM peptides and varying concentrations of lipids were measured using 280-nm excitation at 25 °C, with the emission polarizer set at 90°. The titration data were fitted by the peptide-membrane partition model to calculate the mole-fraction partition coefficient.
pH Dependence.
The pH-dependent partitioning of the peptides into lipid bilayers was investigated by the shift of the position of the peptide intrinsic fluorescence spectral maximum for the pHLIP variants induced by a drop of pH from pH 8 to pH 3 by means of the addition of HCl in the presence of POPC liposomes. Peptide (3 μM) was incubated overnight with 100 nm of POPC liposomes (2 mM), and the pH was decreased by the addition of aliquots of 4, 2, 1, and 0.1 M HCl. The data were analyzed using the Henderson–Hasselbalch equation.
Stopped-Flow Fluorescence Measurements.
Stopped-flow fluorescence measurements were carried out on an SFM-300 mixing apparatus (Biologic, Inc.) connected to a spectrometer. All solutions were degassed for several minutes under vacuum before loading into the syringes of the stopped-flow apparatus to minimize air bubbles. pHLIP variants (7 μM) were preincubated with POPC (1.5 mM) at pH 8.0 to reach binding equilibrium, and insertion was induced by fast mixing (5-ms dead time) of equal volumes of pHLIP-POPC variants at pH 8.0 and appropriately diluted HCl to obtain a drop of pH from 8 pH to the desired value.
Cell Lines.
Human cervix adenocarcinoma (HeLa and HeLa-GFP cells) and lung carcinoma (A549 and A549-GFP cells), HCvEpCs, HMEpCs, and HBEpCs were used in studies.
Fluorescence Microscopy.
HeLa-GFP and A549-GFP cells were incubated with 2 μM Rho- or BODIPY-labeled Var3, K-Var3, Var7, or K-Var7 peptide at pH 5.9 or pH 7.4. After 30 min of incubation, cells were washed four to five times and fluorescent images were acquired.
Quantification of Cellular Uptake.
For quantification of cellular uptake of BODIPY-labeled Var3 and Var7 at pH 7.4 and pH 5.9, we used A549-GFP cells in suspension. Fluorescently labeled peptides (2 μM) were incubated with cells in DMEM media at pH 7.4 and pH 5.9 for 30 min at 37 °C and 5% (vol/vol) CO2 and 95% (vol/vol) air. Cells were then pelleted by centrifugation and resuspended, and fluorescent signals were measured and analyzed using the ImagePro Plus program (Media Cybernetics, Inc.).
Cytotoxicity Assay.
The increasing amounts of pHLIP variants (1, 2, 4, and 8 μM) were added to HCvEpCs, HMEpCs, or HMEpCs, or growth medium was replaced with the medium without FBS containing increasing amounts of pHLIP. After 3 h of incubation, an equal volume of the medium containing 20% FBS was added in the case of HeLa and A549 cells. After 24, 48, and 72 h of incubation, a colorimetric reagent (CellTiter 96 AQueous One Solution Assay; Promega) was added for 1 h, followed by measuring absorbance at 490 nm to assess cell viability.
Animal Studies.
In total, 266 athymic female nude mice were used in the experiments. Mouse tumors were established by s.c. injection of HeLa-GFP or A549-GFP cells (106 cells per 0.1 mL per flank) in the right flank of each mouse. When tumors reached 5–6 mm in diameter, tail vein injections of 100 μL of Alexa750-pHLIPs (40 μM) were performed. Animals were imaged at 4 and 24 h postinjection on an FX Kodak in vivo image station under gas anesthesia, with supplemental heat provided to maintain core body temperature. Animals were euthanized at 4 or 24 h, and a necropsy was performed immediately after euthanization. Tumors and major organs were collected for further imaging. All animal studies were conducted according to the University of Rhode Island Institutional Animal Care and Use Committee animal protocol (AN07-01-015), in compliance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Animals.
Supplementary Material
Acknowledgments
We thank Prof. Jason Lewis, Memorial Sloan–Kettering Cancer Center, for reading the manuscript and for helpful discussions. This work was supported by National Institutes of Health (NIH) Grant CA138468 (to Y.K.R.) and Grants CA133890 and GM073857 (to O.A.A., D.M.E., and Y.K.R.). MS was performed at the Rhode Island Institutional Development Award (IDeA) Network of Biomedical Research Excellence core facility funded by National Center for Research Resources/NIH Grant P20RR016457.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303708110/-/DCSupplemental.
References
- 1.Andreev OA, et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci USA. 2007;104(19):7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: In solution, attached to, and inserted across lipid bilayers. Biophys J. 2007;93(7):2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera FN, et al. Roles of carboxyl groups in the transmembrane insertion of peptides. J Mol Biol. 2011;413(2):359–371. doi: 10.1016/j.jmb.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reshetnyak YK, et al. Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol Imaging Biol. 2011;13(6):1146–1156. doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vāvere AL, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69(10):4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macholl S, et al. In vivo pH imaging with (99m)Tc-pHLIP. Imaging. 2012;14(6):725–734. doi: 10.1007/s11307-012-0549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci USA. 2006;103(17):6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107(47):20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wijesinghe D, Engelman DM, Andreev OA, Reshetnyak YK. Tuning a polar molecule for selective cytoplasmic delivery by a pH (Low) insertion peptide. Biochemistry. 2011;50(47):10215–10222. doi: 10.1021/bi2009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daumar P, et al. Efficient (18)F-labeling of large 37-amino-acid pHLIP peptide analogues and their biological evaluation. Bioconjug Chem. 2012;23(8):1557–1566. doi: 10.1021/bc3000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao L, et al. pHLIP peptide targets nanogold particles to tumors. Proc Natl Acad Sci USA. 2012;110(2):465–470. doi: 10.1073/pnas.1219665110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreev OA, et al. pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path. Proc Natl Acad Sci USA. 2010;107(9):4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karabadzhak AG, et al. Modulation of the pHLIP transmembrane helix insertion pathway. Biophys J. 2012;102(8):1846–1855. doi: 10.1016/j.bpj.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy RS, et al. Peptide hairpins with strand segments containing alpha- and beta-amino acid residues: Cross-strand aromatic interactions of facing Phe residues. Biopolymers. 2005;80(6):787–799. doi: 10.1002/bip.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiche J, Brahimi-Horn MC, Pouysségur J. Tumour hypoxia induces a metabolic shift causing acidosis: A common feature in cancer. J Cell Mol Med. 2010;14(4):771–794. doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grillon E, et al. The spatial organization of proton and lactate transport in a rat brain tumor. PLoS ONE. 2011;6(2):e17416. doi: 10.1371/journal.pone.0017416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehncrona S. Brain acidosis. Ann Emerg Med. 1985;14(8):770–776. doi: 10.1016/s0196-0644(85)80055-x. [DOI] [PubMed] [Google Scholar]
- 19.Swietach P, Vaughan-Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev. 2007;26(2):299–310. doi: 10.1007/s10555-007-9064-0. [DOI] [PubMed] [Google Scholar]
- 20.Ihnatko R, et al. Extracellular acidosis elevates carbonic anhydrase IX in human glioblastoma cells via transcriptional modulation that does not depend on hypoxia. Int J Oncol. 2006;29(4):1025–1033. [PubMed] [Google Scholar]
- 21.Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: Potential exploitation for the treatment of cancer. Cancer Res. 1996;56(6):1194–1198. [PubMed] [Google Scholar]
- 22.Kozin SV, Shkarin P, Gerweck LE. The cell transmembrane pH gradient in tumors enhances cytotoxicity of specific weak acid chemotherapeutics. Cancer Res. 2001;61(12):4740–4743. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.