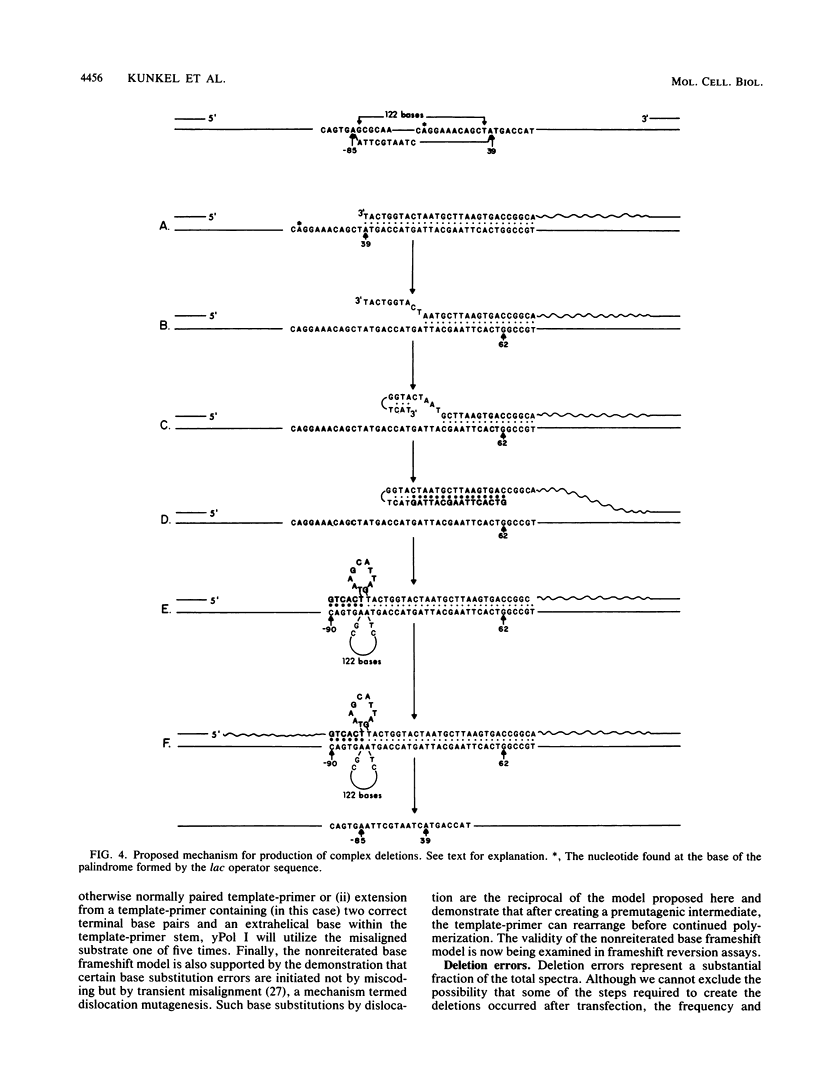
Abstract
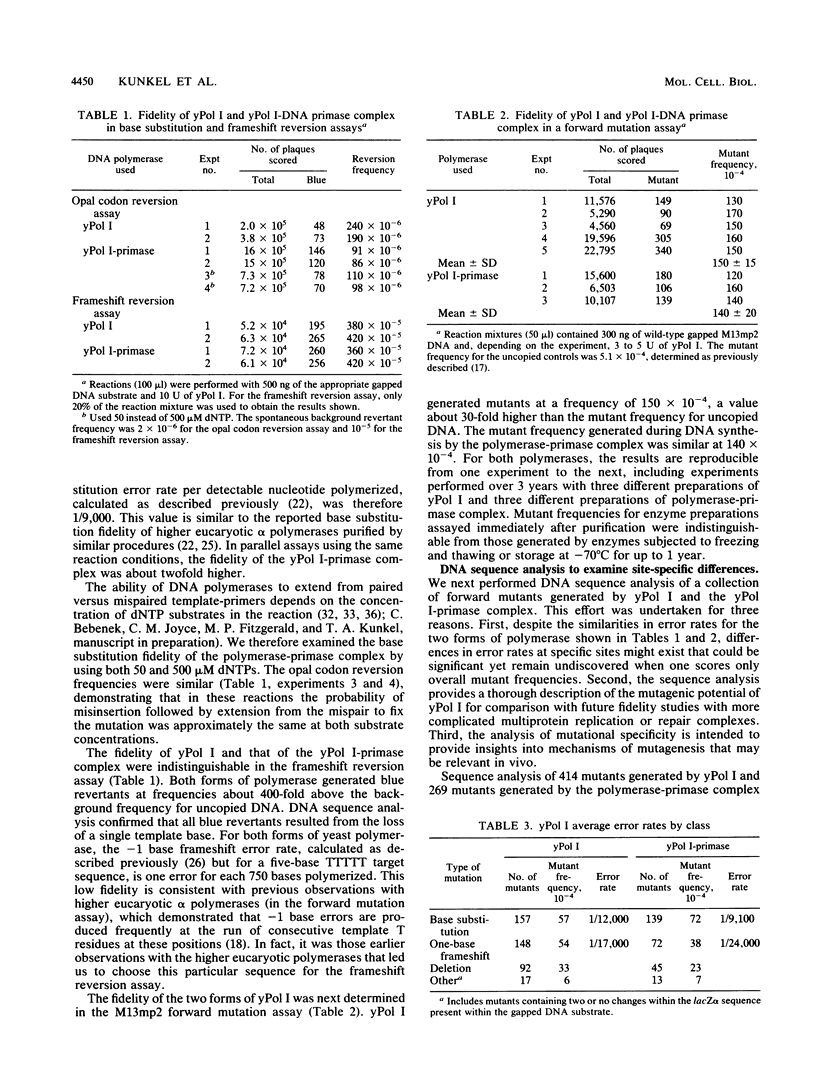
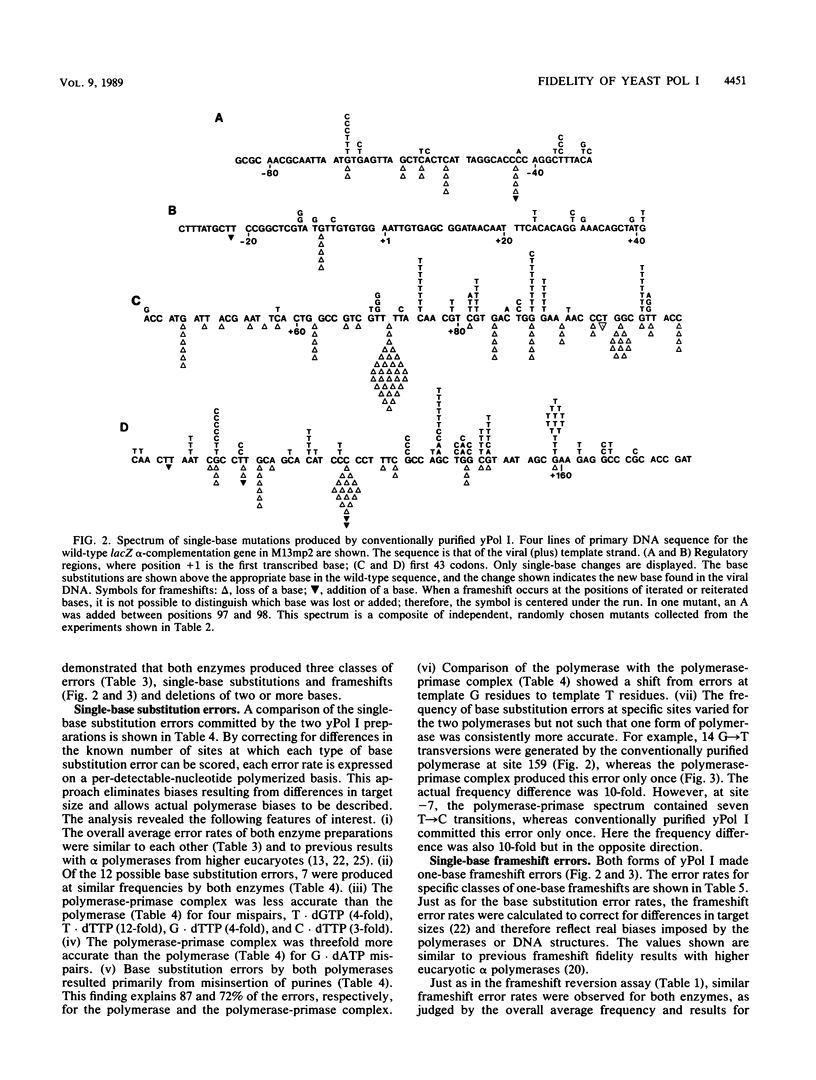
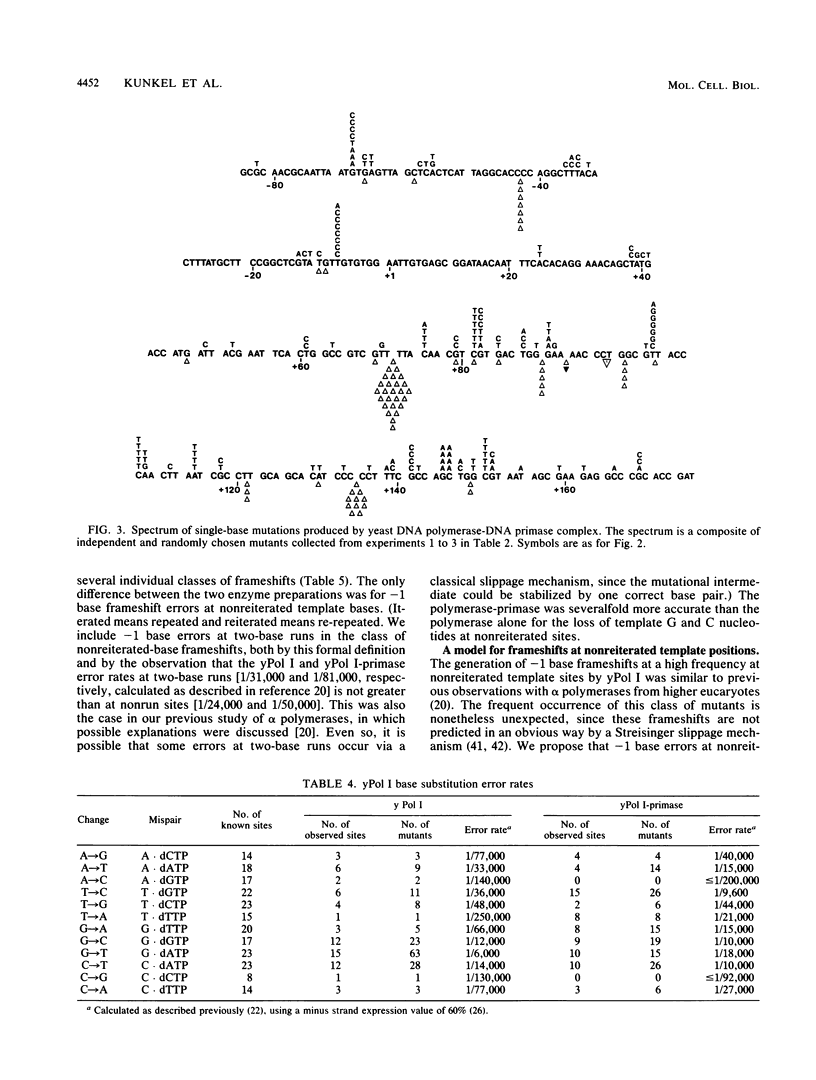
We have determined the fidelity of DNA synthesis by DNA polymerase I (yPol I) from Saccharomyces cerevisiae. To determine whether subunits other than the polymerase catalytic subunit influence fidelity, we measured the accuracy of yPol I purified by conventional procedures, which yields DNA polymerase with a partially proteolyzed catalytic subunit and no associated primase activity, and that of yPol I purified by immunoaffinity chromatography, which yields polymerase having a single high-molecular-weight species of the catalytic subunit, as well as three additional polypeptides and DNA primase activity. In assays that score polymerase errors within the lacZ alpha-complementation gene in M13mp2 DNA, yPol I and the yPol I-primase complex produced single-base substitutions, single-base frameshifts, and larger deletions. For specific errors and template positions, the two forms of polymerase exhibited differences in fidelity that could be as large as 10-fold. Nevertheless, results for the overall error frequency and the spectrum of errors suggest that the yPol I-DNA primase complex is not highly accurate and that, just as for the polymerase alone, its fidelity is not sufficient to account for a low spontaneous mutation rate in vivo. The specificity data also suggest models to explain -1 base frameshifts in nonrepeated sequences and certain complex deletions by a direct repeat mechanism involving aberrant loop-back synthesis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams W. T., Skopek T. R. Statistical test for the comparison of samples from mutational spectra. J Mol Biol. 1987 Apr 5;194(3):391–396. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- Badaracco G., Bianchi M., Valsasnini P., Magni G., Plevani P. Initiation, elongation and pausing of in vitro DNA synthesis catalyzed by immunopurified yeast DNA primase: DNA polymerase complex. EMBO J. 1985 May;4(5):1313–1317. doi: 10.1002/j.1460-2075.1985.tb03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battula N., Loeb L. A. On the fidelity of DNA replication. Lack of exodeoxyribonuclease activity and error-correcting function in avian myeloblastosis virus DNA polymerase. J Biol Chem. 1976 Feb 25;251(4):982–986. [PubMed] [Google Scholar]
- Boosalis M. S., Petruska J., Goodman M. F. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987 Oct 25;262(30):14689–14696. [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L. Eukaryotic DNA replication. Annu Rev Biochem. 1986;55:733–771. doi: 10.1146/annurev.bi.55.070186.003505. [DOI] [PubMed] [Google Scholar]
- Chang L. M. DNA polymerases from bakers' yeast. J Biol Chem. 1977 Mar 25;252(6):1873–1880. [PubMed] [Google Scholar]
- Cotterill S. M., Reyland M. E., Loeb L. A., Lehman I. R. A cryptic proofreading 3'----5' exonuclease associated with the polymerase subunit of the DNA polymerase-primase from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5635–5639. doi: 10.1073/pnas.84.16.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. A., Korn D. Enzymological characterization of KB cell DNA polymerase-alpha. III. The polymerization reaction with single-stranded DNA. J Biol Chem. 1979 Nov 10;254(21):11040–11046. [PubMed] [Google Scholar]
- Giroux C. N., Mis J. R., Pierce M. K., Kohalmi S. E., Kunz B. A. DNA sequence analysis of spontaneous mutations in the SUP4-o gene of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Feb;8(2):978–981. doi: 10.1128/mcb.8.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse F., Krauss G., Knill-Jones J. W., Fersht A. R. Accuracy of DNA polymerase-alpha in copying natural DNA. EMBO J. 1983;2(9):1515–1519. doi: 10.1002/j.1460-2075.1983.tb01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., Lehman I. R. Eukaryotic DNA polymerase-primase: structure, mechanism and function. Biochim Biophys Acta. 1988 Jul 13;950(2):87–101. doi: 10.1016/0167-4781(88)90001-2. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Alexander P. S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986 Jan 5;261(1):160–166. [PubMed] [Google Scholar]
- Kunkel T. A. Exonucleolytic proofreading. Cell. 1988 Jun 17;53(6):837–840. doi: 10.1016/s0092-8674(88)90189-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986 Oct 15;261(29):13581–13587. [PubMed] [Google Scholar]
- Kunkel T. A., Gopinathan K. P., Dube D. K., Snow E. T., Loeb L. A. Rearrangements of DNA mediated by terminal transferase. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1867–1871. doi: 10.1073/pnas.83.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A. Fidelity of mammalian DNA polymerases. Science. 1981 Aug 14;213(4509):765–767. doi: 10.1126/science.6454965. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J Biol Chem. 1988 Mar 25;263(9):4450–4459. [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988 Oct 15;263(29):14784–14789. [PubMed] [Google Scholar]
- Kunkel T. A. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J Biol Chem. 1985 May 10;260(9):5787–5796. [PubMed] [Google Scholar]
- Kunkel T. A. The mutational specificity of DNA polymerases-alpha and -gamma during in vitro DNA synthesis. J Biol Chem. 1985 Oct 15;260(23):12866–12874. [PubMed] [Google Scholar]
- Lawrence C. W. Mutagenesis in Saccharomyces cerevisiae. Adv Genet. 1982;21:173–254. doi: 10.1016/s0065-2660(08)60299-0. [DOI] [PubMed] [Google Scholar]
- Lee G. S., Savage E. A., Ritzel R. G., von Borstel R. C. The base-alteration spectrum of spontaneous and ultraviolet radiation-induced forward mutations in the URA3 locus of Saccharomyces cerevisiae. Mol Gen Genet. 1988 Nov;214(3):396–404. doi: 10.1007/BF00330472. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Francesconi S., Foiani M., Badaracco G., Plevani P. Yeast DNA polymerase--DNA primase complex; cloning of PRI 1, a single essential gene related to DNA primase activity. EMBO J. 1987 Mar;6(3):737–742. doi: 10.1002/j.1460-2075.1987.tb04815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini G., Mazza C., Scacheri E., Plevani P. Genetic mapping of the Saccharomyces cerevisiae DNA polymerase I gene and characterization of a pol1 temperature-sensitive mutant altered in DNA primase-polymerase complex stability. Mol Gen Genet. 1988 Jun;212(3):459–465. doi: 10.1007/BF00330850. [DOI] [PubMed] [Google Scholar]
- Papanicolaou C., Ripley L. S. Polymerase-specific differences in the DNA intermediates of frameshift mutagenesis. In vitro synthesis errors of Escherichia coli DNA polymerase I and its large fragment derivative. J Mol Biol. 1989 May 20;207(2):335–353. doi: 10.1016/0022-2836(89)90258-1. [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Loeb L. A. Differential extension of 3' mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J Biol Chem. 1989 Feb 15;264(5):2898–2905. [PubMed] [Google Scholar]
- Petruska J., Goodman M. F., Boosalis M. S., Sowers L. C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevani P., Foiani M., Valsasnini P., Badaracco G., Cheriathundam E., Chang L. M. Polypeptide structure of DNA primase from a yeast DNA polymerase-primase complex. J Biol Chem. 1985 Jun 10;260(11):7102–7107. [PubMed] [Google Scholar]
- Preston B. D., Poiesz B. J., Loeb L. A. Fidelity of HIV-1 reverse transcriptase. Science. 1988 Nov 25;242(4882):1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- Reyland M. E., Lehman I. R., Loeb L. A. Specificity of proofreading by the 3'----5' exonuclease of the DNA polymerase-primase of Drosophila melanogaster. J Biol Chem. 1988 May 15;263(14):6518–6524. [PubMed] [Google Scholar]
- Reyland M. E., Loeb L. A. On the fidelity of DNA replication. Isolation of high fidelity DNA polymerase-primase complexes by immunoaffinity chromatography. J Biol Chem. 1987 Aug 5;262(22):10824–10830. [PubMed] [Google Scholar]
- Roberts J. D., Kunkel T. A. Fidelity of a human cell DNA replication complex. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7064–7068. doi: 10.1073/pnas.85.19.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. D., Preston B. D., Johnston L. A., Soni A., Loeb L. A., Kunkel T. A. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989 Feb;9(2):469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson F. E., Sugino A. Purification of a DNA primase activity from the yeast Saccharomyces cerevisiae. Primase can be separated from DNA polymerase I. J Biol Chem. 1985 Jul 5;260(13):8173–8181. [PubMed] [Google Scholar]
- Wintersberger U., Blutsch H. DNA-dependent DNA polymerase from yeast mitochondria. Dependence of enzyme activity on conditions of cell growth, and properties of the highly purified polymerase. Eur J Biochem. 1976 Sep;68(1):199–207. doi: 10.1111/j.1432-1033.1976.tb10779.x. [DOI] [PubMed] [Google Scholar]