Abstract
Efforts to design an effective antibody-based vaccine against HIV-1 would benefit from understanding how germ-line B-cell receptors (BCRs) recognize the HIV-1 gp120/gp41 envelope spike. Potent VRC01-like (PVL) HIV-1 antibodies derived from the VH1-2*02 germ-line allele target the conserved CD4 binding site on gp120. A bottleneck for design of immunogens capable of eliciting PVL antibodies is that VH1-2*02 germ-line BCR interactions with gp120 are uncharacterized. Here, we report the structure of a VH1-2*02 germ-line antibody alone and a germ-line heavy-chain/mature light-chain chimeric antibody complexed with HIV-1 gp120. VH1-2*02 residues make extensive contacts with the gp120 outer domain, including all PVL signature and CD4 mimicry interactions, but not critical CDRH3 contacts with the gp120 inner domain and bridging sheet that are responsible for the improved potency of NIH45-46 over closely related clonal variants, such as VRC01. Our results provide insight into initial recognition of HIV-1 by VH1-2*02 germ-line BCRs and may facilitate the design of immunogens tailored to engage and stimulate broad and potent CD4 binding site antibodies.
Keywords: crystallography, structural biology, virology
Over 30 y after the emergence of HIV-1, there is no effective vaccine, and AIDS remains an important threat to global public health. After infection by HIV-1, the host immune response is unable to clear the virus because of a variety of factors, including rapid viral mutation and establishment of latent reservoirs (1). The only target of neutralizing Abs is the trimeric envelope (Env) spike complex (2, 3), but HIV-1 can usually evade antispike Abs because of rapid mutation of its two spike glycoproteins, gp120 and gp41, and structural features that allow the spike to hide conserved epitopes. These structural features include a dense layer of carbohydrates that mask its surface, conformational flexibility, and variable loops emanating from gp120 (4, 5). The oligomeric structure of the spike further conceals conserved regions at subunit interfaces or in narrow inaccessible pockets (6–8), and the low density of spikes on the surface of HIV virions impedes bivalent binding of Abs (9, 10).
Nevertheless, broadly neutralizing Abs (bNAbs) that can neutralize many HIV-1 strains have been identified in HIV-1–infected individuals (11, 12). The potential of the first such bNAbs, typified by 4E10, 2F5, 2G12, and b12, to prevent HIV infection in macaques (13–16) and delay viral rebound after highly active antiretroviral therapy (HAART) secession (17) made them particularly interesting for vaccine design efforts. Recently, highly potent bNAbs that have unprecedented potency and breadth for HIV neutralization were isolated from infected individuals (18–22). Of particular interest are Abs that target the relatively conserved binding site on gp120 for CD4, the host receptor for HIV-1 (20, 23–25). Crystal structures of several such Abs bound to gp120 (23–25) revealed molecular details of their neutralization mechanisms and facilitated structure-based rational design to improve their potency and breadth (24). The therapeutic potential of the new bNAbs is significant; for the first time, a mixture of passively delivered bNAbs was shown to suppress and control HIV-1 viremia in humanized mice (26). Thus, eliciting similar bNAbs by vaccination would be highly desirable.
Of importance for potential vaccine efforts, potent anti-CD4 binding site (CD4bs) Abs protect against HIV-1 infection in animal models (27, 28). These Abs were isolated from several different individuals (19, 20), and many of the more potent Abs are derived from a common germ-line gene segment allele (VH1-2*02) that is present in up to 95% of the population (19, 20, 23, 29). These observations suggest that similar CD4bs bNAbs could be elicited in other individuals. Available structural information, together with sequence and neutralization data for CD4bs Abs, allowed us to rationalize the structural basis of the VH1-2*02 germ-line gene usage, determine which Abs bind gp120 similarly, and identify the critical sequence features that permit this binding (29). Based on inspection of Ab variable domain sequences, we found that VH1-2*02-derived Abs completely conserve Arg71HC, Trp50HC, Asn58HC, and Trp100BHC (Trp102HC in NIH45-46 numbering) within the heavy chain. Within the light chain, Glu96LC and a complementarity-determining region (CDR) L3 length of exactly 5 amino acids are conserved (29). We proposed a nomenclature to describe the class of Abs including this set of sequence characteristics: potent VRC01-like (PVL) Abs, reflecting the first antibody of this class to be isolated (19). The required signature residues rationalize the VH1-2*02 germ-line gene origins of PVL Abs (29).
The initial recognition of HIV-1 by the VH1-2*02 B-cell receptor (BCR) might be a limiting factor for eliciting protective PVL Abs (30). The details of recognition of antigen by a germ-line BCR are not fully understood, but presumably, the interaction is sufficiently strong in certain individuals to yield a clonal expansion of the B cells carrying a VH1-2*02 BCR. The binding interaction is then strengthened by somatic hypermutation and clonal selection, ultimately leading to a PVL Ab. Although the rare emergence of B cells that produce bNAbs remains poorly understood, with structural information about the VH1-2*02 interaction, it may be possible to design immunogens capable of initiating clonal expansion from this germ-line allele, leading to an increased chance of maturation to a PVL bNAb. Here, we investigate the structural basis of recognition by a putative VH1-2*02 germ-line Ab of HIV-1 gp120 through analyses of the crystal structures of a chimeric VH1-2*02 germ-line/mature light-chain Ab bound to gp120 and the unbound germ-line Ab. Structural comparisons show that the heavy-chain PVL signature residues make the same contacts to the gp120 outer domain in the germ-line and mature NIH45-46 Abs but that critical contacts with the gp120 inner domain and bridging sheet are not formed by the germ-line Ab. These results suggest a pathway by which PVL Abs mature to achieve broad and potent neutralization and provide insights to guide vaccine immunogen design to eliciting PVL Abs.
Results
Construction of Germ-Line Precursor Antibody.
We constructed a putative VH1-2*02 germ-line sequence based on the sequence of NIH45-46, a more potent clonal variant of VRC01 that was isolated from the same donor (20). We used the ImMunoGeneTics database (IMGT) (31) to predict the V-D-J and V-J assignments for the heavy and light chains (IGHV1-2*02–IGHD2-15*01–IGHJ2*01/IGKV3-11*01–IGKJ2*01). Mature NIH45-46 (NIH45-46mature) differs from the resulting putative germ-line Ab (NIH45-46GL) by a large number of somatic mutations in both the heavy and light chains (44 of 126 and 27 of 103 residue differences, respectively) (Fig. 1A and Fig. S1).
Fig. 1.
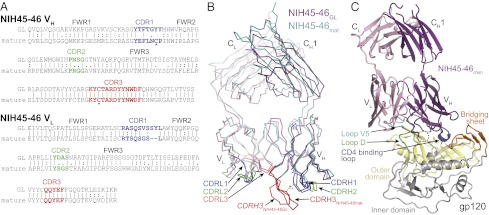
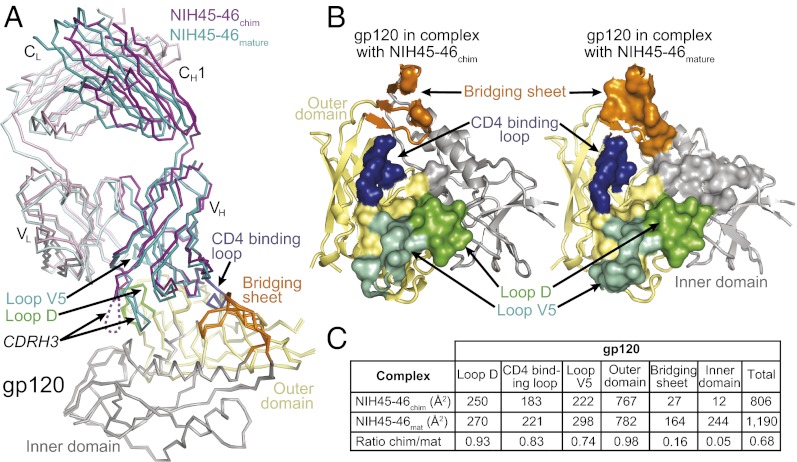
Crystal structures of NIH45-46GL Fab and NIH45-46chim/gp120 complex. (A) Alignments of VH and VL sequences of mature NIH45-46 (NIH45-46mature) and its putative germ-line progenitor (NIH45-46GL). Residues forming the CDR loops are colored blue (CDR1), green (CDR2), and red (CDR3). Two cysteines forming a disulfide bond between CDRH1 and CDRH3 of NIH45-46mature are marked with an asterisk. The CDR3 regions were taken from NIH45-46mature, because the germ-line configuration is unknown. (B) Superimposition of NIH45-46GL Fab and NIH45-46mature Fab shown as wire representations with NIH45-46GL Fab HC/LC in magenta/light pink and NIH45-46mature Fab HC/LC in teal/light teal. The disulfide bond formed between C32CDRH1 and C98CDRH3 of NIH45-46mature but not NIH45-46GL is marked with an asterisk. The CDR loops are colored as in A. (C) Ribbon diagram of crystal structure of NIH45-46chim (purple, germ-line heavy chain; pink, mature light chain) bound to gp120 core (yellow, inner domain; gray, outer domain). The disordered tip of CDRH3 is shown as a dashed line. The gp120 core construct is derived from 93TH053 gp120, but it lacks three variable loops (V1-V2 and V3) and contains N- and C-terminal truncations (25). Subdomains of gp120 are colored as follows: orange, bridging sheet; blue, CD4 binding loop; green, loop D; cyan, loop V5.
Because of the difficulty of accurately predicting the residues present in the CDR3 loops of a germ-line progenitor, the mature CDRH3 and CDRL3 sequences were used in our NIH45-46GL construct. We also included a four-residue “insertion” (residues Ala99a-Arg99b-Asp99c-Tyr99d) within the CDRH3 of NIH45-46 in our putative germ-line precursor sequence. These residues, which account for the increased potency of NIH45-46 compared with VRC01 (20, 24), could have resulted from N-region addition during V-D-J recombination of the germ-line gene segments or through somatic hypermutation. However, sequencing of VRC01-like Ab genes from the VRC01/NIH45-46 donor suggested the existence of these residues early in the development of VH1-2*02-derived bNAbs (32), implying that the NIH45-46 germ-line BCR contained the “insertion”.
Crystal Structure of Germ-Line NIH45-46 Fab.
To examine the structural determinants of differences between mature PVL Abs and their VH1-2*02-derived germ-line precursor, we crystallized and solved the structure of the NIH45-46GL Fab (Fig. 1B and Table S1). Compared with NIH45-46mature, NIH45-46GL Fab showed no major displacements of CDRs or framework regions (RMSD = 1.40 Å for 212 Cα atoms), with the exception of CDRH3 (third CDR in the heavy chain) (Fig. 1B). A noncanonical disulfide bond joining CDRH1 and CDRH3 in NIH45-46mature is not present in NIH45-46GL, which lacks Cys32CDRH1 (Fig. S2C). The conformation of CDRH3 of NIH45-46GL is, therefore, not constrained by a disulfide bond, and this increased flexibility could facilitate the different packing in the NIH45-46GL Fab crystals (Fig. S2D). Although affinity maturation did not produce significant structural rearrangements of the combining site with the exception of CDRH3, the large number of somatic mutations introduced in CDRs and framework regions of both heavy and light chain resulted in a significantly resurfaced combining site in NIH45-46mature compared with the putative germ-line precursor (Fig. S2E).
Crystal Structure of Chimeric Germ-Line NIH45-46 Fab in Complex with gp120.
Consistent with previous reports of no detectable HIV-1 antigen binding by bNAb germ-line precursors (20, 25, 29, 33, 34), NIH45-46GL failed to interact with clade A/E 93TH057 gp120 coupled to a sensor chip in surface plasmon resonance (SPR) experiments (Fig. S3A) and did not neutralize HIV-1 pseudoviruses in an in vitro neutralization assay (Fig. S3B). However, a chimeric Ab consisting of the germ-line heavy chain and the mature NIH45-46 light chain (NIH45-46chim) showed an intermediate level of binding in SPR experiments and weak neutralization in three of seven HIV-1 strains that were potently neutralized by NIH45-46mature (Fig. S3).
Detectable binding between the NIH45-46chim construct and gp120 allowed us to investigate the VH1-2*02 interaction with Env, and because the majority of gp120 contacts in PVL Abs is due to interactions with the heavy chain (23–25, 29), the structural information gathered using the chimera would be useful in understanding the native VH1-2*02 germ-line Ab interaction. To visualize the interaction between a germ-line variable heavy (VH) domain and gp120, we solved the crystal structure of NIH45-46chim Fab bound to 93TH057 gp120 at 3.25 Å resolution (Fig. 1C and Table S1). As used for previous crystallographic studies (20, 23–25), the gp120 was a core construct with truncations (N/C termini and loops V1-V2 and V3). We superimposed the gp120 cores from NIH45-46chim/gp120 and NIH45-46mature/gp120 complex structures (Fig. 2A) to compare the recognition of gp120 by the germ-line and NIH45-46mature Ab heavy chains (24). The variable domains of NIH45-46chim and NIH45-46mature aligned closely (RMSD = 0.68 Å; 209 Cαs), showing that the angle at which the Ab approaches gp120 is not altered by affinity maturation. Also, similar to the NIH45-46mature/93TH057 complex structure (Protein Data Bank ID code 3U7Y), NIH45-46chim contacts the outer domain of gp120, including the CD4 binding loop, loop D, and loop V5.
Fig. 2.
Comparison of the binding interfaces of NIH45-46chim/gp120 and NIH45-46mature/gp120. (A) Superimposition of complex crystal structures of NIH45-46chim/gp120 (purple, Ab heavy chain; light pink, Ab light chain) and NIH45-46mature/gp120 (teal, Ab heavy chain; light teal, Ab light chain). The crystal structures were superimposed on their gp120s (domain coloring as in Fig. 1C). The protein backbones are shown as wire diagrams, and the disordered tip of CDRH3 is shown as a dashed line. (B) Comparison of binding interfaces in NIH45-46chim/gp120 and NIH45-46mature/gp120 complexes. Surface area buried by Fabs on gp120 because of complex formation was calculated using a 1.4-Å probe. Residues at each contact interface are highlighted on the ribbon diagrams of gp120 complexed with NIH45-46chim Fab (Left) or NIH45-46mature (Right) as surfaces enclosing the contact residues. Coloring as in A. (C) Table listing BSAs (Å2) and ratios between surface areas buried on gp120 domains by NIH45-46chim and NIH45-46mature. The total column is the sum of outer domain, bridging sheet, and inner domain.
To quantify differences in the binding interfaces between the NIH45-46chim and NIH45-46mature complexes with gp120, we calculated their buried surface areas (BSAs) upon binding (Fig. 2 B and C and Fig. S4). Like NIH45-46mature, NIH45-46chim primarily contacts gp120 through its heavy chain (84% and 85% of the BSA for NIH45-46chim and NIH45-46mature, respectively), including gp120 contacts with all CDRH loops and residues in heavy-chain framework regions (FWRs) 2 and 3 (Fig. S4). The BSA on gp120 in the NIH45-46chim complex is ∼68% of the surface area buried in the interface with NIH45-46mature (Fig. 2 B and C). However, almost all of the additional contacts made by NIH45-46mature involve the inner domain and bridging sheet of gp120, whereas an equal surface area of the outer domain is contacted in both complexes (Fig. 2 B and C). NIH45-46mature makes these additional interactions using CDRH3 and somatically mutated residues in CDRH1 and FWR3 (Fig. S4). Therefore, although the location of the contact surfaces is similar in both complexes (except for the gp120 inner domain), affinity maturation increased the surface area buried on both the antibody and gp120 by 66% and 47%, respectively, which improved the strength and specificity of binding.
NIH45-46 Germ-Line Heavy Chain Makes PVL Signature and CD4 Binding Loop Contacts.
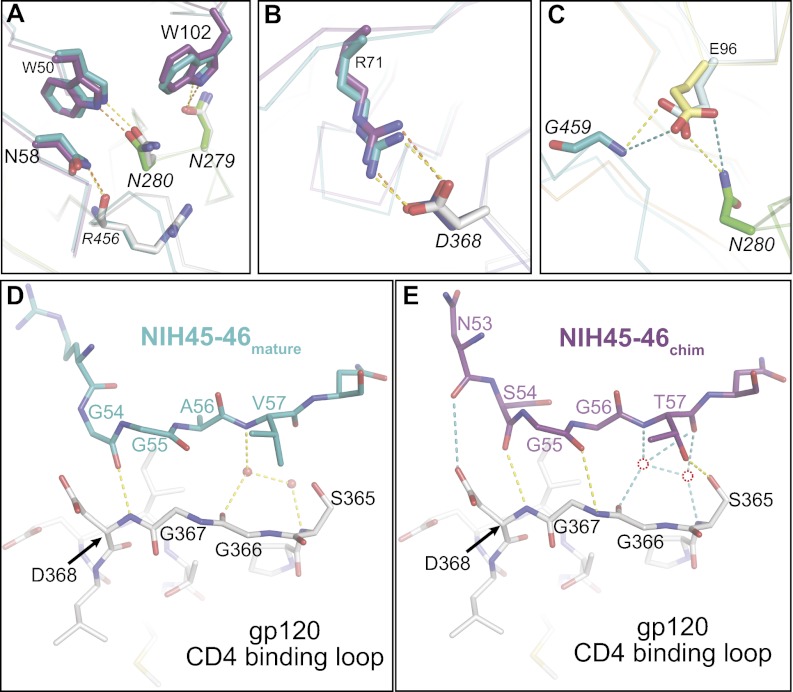
The VH1-2*02 germ-line allele and PVL Abs derived from it share a set of distinct residues [Trp50HC, Asn58HC, Arg71HC, and Trp102HC (Trp100BHC in VRC01 numbering)] predicted to engage in conserved interactions with the CD4bs on gp120 (29); thus, we were able to examine the conformations and potential interactions of these residues in the NIH45-46chim/gp120 complex structure. We found that the germ-line VH of NIH45-46chim is positioned to make all of the conserved PVL signature VH/gp120 contacts (Fig. 3 A and B). In particular, Trp50HC (FWR2) and Asn58HC (FWR3) are within hydrogen bonding distance of Asn280gp120 and Arg456gp120, respectively, and Trp102HC (Trp100BHC in VRC01 numbering; CDRH3) (29) hydrogen bonds with Asn279gp120 (Fig. 3A). Lastly, the salt bridge between Arg71HC (FWR3) and Asp368gp120, which mimics the Arg59CD4–Asp368gp120 interaction (6), is also made by the germ-line VH of NIH45-46chim (Fig. 3B). Preservation of the PVL signature residue interactions in the germ-line VH domain confirms the prediction that these residues are critical for establishing binding to gp120 (29).
Fig. 3.
Comparison of PVL signature and CD4 mimicry contacts in the NIH45-46chim/gp120 complex. (A–C) NIH45-46chim establishes PVL signature interactions with gp120 as observed in the NIH45-46mature/gp120 complex. Superimposition of complex crystal structures of NIH45-46chim/gp120 (purple, heavy chain; light pink, light chain; green, gp120 loop D; blue, gp120 CD4 binding loop; teal, gp120 loop V5), NIH45-46mature/gp120 (teal, heavy chain; light teal, light chain; gray, gp120), and NIH45-46GL (orange, heavy chain; yellow, light chain). Protein backbones are shown as wire diagrams, and interacting residues are shown as sticks (red, oxygen; blue, nitrogen). (A) Trp50HC-Asn280gp120, Asn58HC-Arg456gp120, Trp102HC-Asn279gp120 (in VRC01 numbering, Trp100BHC-Asn279gp120) (29), Glu96LC-Asn280gp120, and Glu96LC-Gly459gp120. (B) Arg71HC-Asp368gp120. Orange and yellow dashed lines show potential hydrogen bonds in NIH45-46chim/gp120 and NIH45-46mature/gp120 complexes, respectively. (C) Superimposition of NIH45-46GL Fab on the NIH45-46chim/gp120 complex. The PVL-characteristic Glu96LC–Asn280gp120 and Glu96LC–Gly459gp120 interactions can be established by the germ-line light chain and are shown as teal dashed lines. (D and E) Contacts between the gp120 CD4 binding loop (gray) and the heavy chain of (D) NIH45-46mature (teal) or (E) NIH45-46chim (purple). The protein atoms are shown as sticks (red, oxygens; blue, nitrogens); ordered water molecules in the NIH45-46mature/gp120 complex are shown as red spheres, and their corresponding locations in the NIH45-46chim/gp120 complex (not observed at 3.25 Å resolution) are shown as red circles. Probable hydrogen bonds (distance < 3.5 Å, A-H–D angle > 90°) are shown as yellow dashed lines. Possible hydrogen bonds (distance < 4 Å or rely on water molecules not observed) are shown as teal dashed lines.
The two PVL signature residues of the variable light chain (VL; Trp67 and Glu96) were acquired by somatic hypermutation and thus, could not contribute to the binding of the germ-line Ab to gp120. To investigate limiting factors that restrict germ-line VL/gp120 interaction, we superimposed the structure of NIH45-46GL onto the NIH45-46chim/gp120 complex (Fig. S5A). Gln96LC GL (CDRL3) is positioned within hydrogen bonding distance of Gly459gp120 and Asn280gp120, similar to Glu96LC NIH45-46 (Fig. 3C). The conserved Trp67LC that interacts with the Asn276gp120-attached N-glycan (29) in mature PVL Abs is a serine in the germ-line Ab. Combined with the increased length of CDRL1 caused by a two-residue insertion in NIH45-46GL (Fig. 1A and Fig. S1), VL GL may not be compatible with interacting with the Asn276gp120-attached N-glycan (Fig. S5B).
PVL Abs mimic CD4 by engaging the CD4 binding loop on gp120 with backbone atoms in the C″ strand of VH (23–25). NIH45-46mature makes direct and water-mediated hydrogen bonds using main-chain atoms of Gly54HC and Val57HC to contact Ser365gp120, Gly366gp120, and Asp368gp120 (Fig. 3D), mimicking the contacts between Leu44CD4 and Lys46CD4 with the CD4 binding loop (24). In addition to a direct main-chain hydrogen bond between Ser54HC and Asp368gp120, the germ-line VH of NIH45-46chim makes two other hydrogen bonds: between Gly55HC and Asp367gp120 and between the side chains of Thr57HC (Val57 in NIH45-46mature) and Ser365gp120 (Fig. 3E). Although ordered water molecules were not resolved in the 3.25 Å NIH45-46chim/gp120 complex structure, Thr57HC NIH45-46 chim could participate in a hydrogen bonding network analogous to and possibly more extensive than the network established by Val57HC NIH45-46 and Gly366gp120/Asp368gp120 in the NIH45-46mature/gp120 structure that included ordered water molecules (Fig. 3E). In summary, our structural analyses showed that NIH45-46GL relies on C″ strand contacts with the CD4 binding loop of gp120 as well as the PVL signature contacts with the CD4 binding loop, the V5 loop, and loop D to bind to gp120.
Loss of gp120 Inner Domain Contacts by the Germ-Line NIH45-46 Heavy Chain.
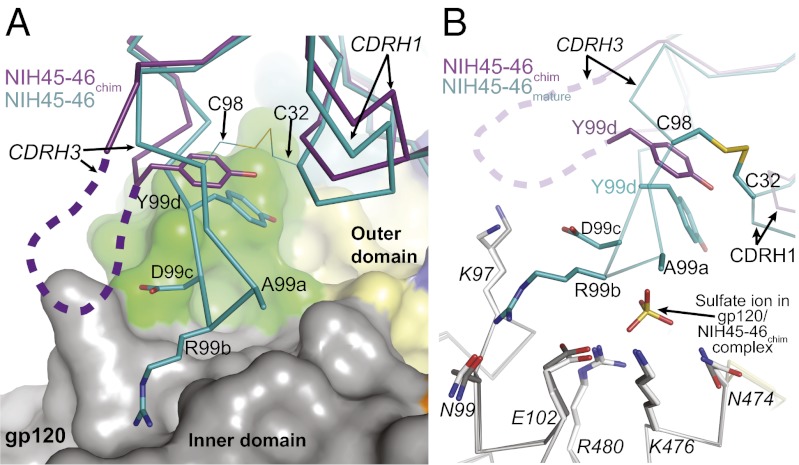
Although many contacts made by VH NIH45-46 mature with gp120 are also observed in the NIH45-46chim/gp120 structure, NIH45-46chim does not appreciably interact with the inner domain of gp120. By contrast, NIH45-46mature uses a four-residue “insertion” in CDRH3 (Ala99a-Arg99b-Asp99c-Tyr99d), which is critical for its increased neutralization potency compared with VRC01, to contact the gp120 inner domain (24). The loss of gp120 inner domain contacts by NIH45-46chim results from disordered residues forming the tip of CDRH3NIH45-46 chim (heavy-chain residues 99–99c), including three of four “insertion” residues (Fig. 1C). In addition, the base of CDRH3NIH45-46chim is turned away from gp120 by ∼5.5 Å (Fig. 4A), possibly because of the lack of the CDRH1–CDRH3 disulfide bond in the germ-line VH domain and the bulky side chain of Tyr33HC in NIH45-46chim occupying space created by the smaller Pro33HC residue in NIH45-46mature (Fig. S6A). The Cys32CDRH1–Cys98CDRH3 disulfide bond helps fix the conformation of CDRH3NIH45-46, potentially facilitating extensive interactions with the inner domain of gp120. The “insertion” in CDRH3NIH45-46 contacts inner domain residues, including Lys97gp120, Asn99gp120, Glu102gp120, Asn474gp120, Lys476gp120, and Arg480gp120, whereas NIH45-46chim makes none of these inner domain contacts because of its displaced CDRH3 (Fig. 4B and Fig. S6B). In the NIH45-46chim/gp120 structure, inner domain residues Asn474gp120, Lys476gp120, and Arg480gp120 coordinate a sulfate ion not found in the NIH45-46mature/gp120 complex, verifying that CDRH3NIH45-46chim does not interact with gp120 (Fig. 4B and Fig. S6B). The fourth CDRH3 “insertion” residue, Tyr99dNIH45-46 chim, is ordered, but because of the displacement of CDRH3, it cannot interact with gp120 loop D residues Ala281gp120 and Lys282gp120 as in the gp120 complex with NIH45-46mature (Fig. S6C). The inner domain and loop D contacts made by NIH45-46mature contribute most of the additional BSA on gp120 and therefore, may be partially responsible for the improved binding affinity and neutralization potency of NIH45-46mature over NIH45-46GL.
Fig. 4.
Displacement of CDRH3 in NIH45-46chim/gp120 structure results in loss of gp120 inner domain contacts. (A) Superimposition of the NIH45-46chim/gp120 and NIH45-46mature/gp120 complexes on gp120 shows that the base of CDRH3NIH45-46chim (Cys98) is displaced away from gp120 by ∼5.5 Å relative to the NIH45-46mature/gp120 complex. The four residues forming the tip of CDRH3 are disordered in our NIH45-46chim/gp120 complex structure and could extend even farther away from gp120 (purple, NIH45-46chim heavy chain; teal, NIH45-46mature heavy chain; gp120 domain coloring as in Fig. 1C). (B) Detailed view of gp120 inner domain contacts made by CDRH3NIH45-46 mature (teal) but not CDRH3NIH45-46 chim (purple). Protein backbones are shown as wire representations, and amino acid side chains are shown as sticks. The disordered part of CDRH3NIH45-46 chim is shown as a dashed line. A sulfate ion coordinated by Asn474gp120, Lys476gp120, and Arg480gp120 in the NIH45-46chim–bound structure (gp120 residues in gray) but not the NIH45-46mature–bound structure (gp120 residues in white) is shown as sticks. Sulfur, oxygen, and nitrogen atoms are colored yellow, red, and blue, respectively.
Discussion
The recently isolated group of PVL Abs directed against the CD4bs on HIV-1 gp120 comprises the most potent and broadly neutralizing set of anti–HIV-1 Abs characterized to date, making it a promising target for HIV-1 vaccine development (20, 23–25, 29). Despite being derived from different HIV-1-infected donors in response to different eliciting viruses (20), PVL Abs share a set of conserved heavy-chain residues that are crucial for gp120 recognition and present in the VH1-2*02 germ line from which they are derived (29). Detailed structural information about how germ-line precursor Abs might engage gp120 would aid immunogen design by identifying specific features that foster the clonal selection of these Abs (30). However, structures of a PVL germ-line precursor bound to gp120 or any anti–HIV-1 germ-line Ab bound to an HIV-1 antigen have been unavailable. Indeed, although such structures would facilitate design of immunogens capable of eliciting neutralizing Abs, structural characterizations of antigen recognition by germ-line Abs against other viruses are rare: other examples include germ-line Ab recognition of influenza (35) and human cytomegalovirus (36) antigens. Here, we report a crystal structure of a gp120 complexed with a Fab containing a putative germ-line heavy chain: in this case, a chimeric Fab consisting of the germ-line heavy chain and mature light chain of the PVL Ab NIH45-46 (NIH45-46chim). Comparisons with the structure of mature NIH45-46 (NIH45-46mature) bound to the same gp120 (24) are relevant to understanding how clonal selection progresses from initial recognition of a germ-line Ab to rounds of somatic hypermutation that improve binding, which is critical information for design of immunogens to elicit PVL Abs. Although we cannot be certain that this germ-line heavy chain is identical to the heavy chain of the initial BCR in patient 45 from whom NIH45-46 was isolated (20), it is representative of the various germ-line regions in the general population that will need to be targeted by a vaccine.
The orientation by which the 93TH057 gp120 is recognized by NIH45-46chim and NIH45-46mature is similar. Thus, the general mechanism by which the VH domain of a PVL Ab mimics CD4 (25) is present in the initial BCR/gp120 recognition event. Importantly, we found that extensive contacts are established by VH1-2*02 residues (including most of the PVL signature residues) of the germ-line Ab with the gp120 outer domain. Indeed, the size of the germ-line VH domain interface with the gp120 outer domain was similar to the interface formed with the mature VH domain. However, additional contacts with the gp120 inner domain and bridging sheet caused nearly a 50% increase in the BSA on gp120 by NIH45-46mature compared with NIH45-46chim. This additional contact area likely contributes to the greatly increased binding affinity and neutralization potency of NIH45-46mature. Thus, specific outer domain contacts, including PVL signature residue contacts and main-chain contacts between CDRH2 and the CD4 binding loop, were present in the VH domain germ-line precursor–gp120 complex, whereas contacts with the gp120 inner domain and bridging sheet required changes induced by somatic hypermutation and therefore, were largely absent in the NIH45-46chim/gp120 complex structure. Of direct relevance to immunogen design efforts, this result suggests that modified outer domain-only gp120 constructs might be capable of triggering a germ-line BCR for a VH1-2*02-derived Ab but that affinity maturation to a PVL Ab would likely require subsequent boosting with Env constructs, including native residues in the gp120 inner domain and bridging sheet. Thus, modified outer domain immunogens having an increased affinity for the VH1-2*02-derived germ line could only serve to trigger the initial immune response; more complete gp120 immunogens containing the inner domain and the bridging sheet would be required to drive affinity maturation.
The finding that the chimeric Ab relied almost exclusively on VH1-2*02 germ-line contacts with the gp120 outer domain is also important for understanding the structural correlates of broad and potent neutralization by PVL Abs. Previous structural studies defined the initial CD4 attachment site on the gp120 outer domain as a desirable target, concluding that increased contact with the CD4bs on the gp120 inner domain and bridging sheet (defined as outside the initial CD4 attachment site) correlated with decreased breadth and potency (23, 25). Therefore, CD4/bNAb contact regions on the gp120 inner domain/bridging sheet were deliberately modified to create a resurfaced gp120 core, which was used as bait for isolating bNAbs (19, 23) and as a candidate HIV immunogen (37). However, structural comparison between two closely related bNAbs, NIH45-46 and VRC01 (86% identity in VH; 96% in VL), subsequently revealed that increased contact area with regions outside of the gp120 outer domain does not correlate with decreased neutralization potency and breadth, because NIH45-46 showed more extensive contacts (relative to VRC01) with the inner domain and bridging sheet of gp120 (24) but is more potent (20). The present study, comparing germ-line and mature antibody recognition of gp120, more dramatically illustrates this principle, because NIH45-46chim shows much weaker binding and neutralization activity than NIH45-46mature but interacts almost exclusively with the gp120 outer domain.
The germ-line version of the VH1-2*02 VH domain used for our complex structure included a four-residue “insertion” within the CDRH3 of NIH45-46 responsible for the increased potency of NIH45-46 compared with VRC01 (20, 24). Although the insertion made extensive contacts with the gp120 inner domain in the NIH45-46mature/gp120 structure, this region of CDRH3 was partially disordered in the NIH45-46chim/gp120 complex. The lack of contacts with CDRH3NIH45-46chim may be, in part, because of the missing CDRH1–CDRH3 disulfide bond and/or a bulkier residue (Tyr33NIH45-46GL compared with Pro33NIH45-46 mature) displacing CDRH3 away from gp120. The absence of this disulfide bond leaves an unpaired cysteine residue at the base of CDRH3, which is energetically unfavored and may lead to aggregation. It is possible that the CDRH3 sequence in the germ-line Ab from the NIH45-46/VRC01 donor that was the precursor to NIH45-46 contained different residues that would have been replaced or deleted during maturation and are not reflected in the CDRH3 sequence used in our construct. It is also possible that the higher flexibility of the germ-line Ab lacking the disulfide linkage would broaden the number of similar targets that can be recognized while having a minimal effect on binding. The CDRH1–CDRH3 disulfide bond may play an important role in maturation by restricting the allowable CDRH3 conformations, perhaps providing more rigidity (decrease in entropy) that permits a collection of relatively weak interactions to form a functional binding surface. Interestingly, the heavy chains of some shark Abs [type II immunoglobulin new antigen receptors (IgNAR)] contain an analogous CDRH1–CDRH3 disulfide bond that is thought to stabilize select conformations of their extended CDRH3 loop (38). Furthermore, increasing the rigidity of CDRs during affinity maturation was also described for broadly neutralizing antiinfluenza antibodies (39). Even if the CDRH1–CDRH3 disulfide bond is not essential for the neutralization potency of NIH45-46mature, immunogens could be designed to encourage its formation by supporting the introduction of a CDRH1 cysteine (Tyr32Cys; i.e., an immunogen that would not rely on contacts with Tyr32HC) or favor the CDRH3 conformation resulting from the disulfide bond.
Key questions to consider for efforts to design immunogens capable of eliciting PVL Abs include why all HIV-1–infected individuals do not develop these Abs and why previous Env-based vaccines failed to elicit protective PVL Abs (reviewed in ref. 30). The prevalence of a suitable VH1-2*02 allele in the general population is high (29) and cannot account for the rarity of PVL Abs. It was shown that B cells expressing BCRs corresponding to the germ-line precursor of the cross-reactive 2F5 antibody were eliminated through immunological tolerance (40), but elimination of PVL precursors seems unlikely, because most PVL Abs do not show obvious cross-reactivity (20). However, PVL Abs undergo much more extensive somatic hypermutation than typical human IgGs (19, 20, 23, 41), with 96 nucleotide mutations in the VH of NIH45-46 (20) compared with an average of 18 mutations in human IgG memory B cells (42). The high level of somatic hypermutation required for neutralization breadth and potency (19, 20) is likely caused by the evolution of gp120 over the course of several years. The apparent requirement for extensive Ab hypermutation complicates immunogen design, because successive administration of different immunogens may be necessary to drive proper affinity maturation on the path from a VH1-2*02 germ-line predecessor to a highly potent and broadly neutralizing HIV-1 antibody. Our results illustrate how a BCR might recognize gp120 at the starting point of affinity maturation.
Materials and Methods
Construction of the Germ-Line Version of NIH45-46.
NIH45-46GL (20) was constructed by reverting all somatic mutations in the V(D)J regions of both heavy and light chains. The respective germ-line genes were identified by BLAST of the antibody sequences in the IMGT database (43). CDR3s were left in the original mutated forms, because it is impossible to reliably determine their germ-line configurations.
Protein Expression and Purification.
Protein expression and purification are described in detail in SI Materials and Methods. NIH45-46, NIH45-46chim (germ-line heavy chain/mature light chain), and NIH45-46GL IgGs were expressed, purified, and cleaved with papain to generate Fabs as previously described (24). A 93TH057 gp120 core was produced as described previously (24). For crystallization, purified NIH45-46chim Fab and 93TH057 gp120 were incubated at a 1:1 molar ratio and treated with Endoglycosidase H followed by size exclusion chromatography purification.
Crystallization and Structure Determination.
Crystallization, data collection, structure determinations, and analyses are described in SI Materials and Methods. Briefly, crystals of Fab NIH45-46GL (space group P212121, a = 56.0 Å, b = 70.1 Å, c = 225.1 Å; two molecules per asymmetric unit) were obtained in 30% (wt/vol) PEG 3350, 0.2 M (NH4)2SO4, and 0.1 M Bis⋅Tris, pH 5.5, at 20 °C. Crystals of NIH45-46chim–93TH057 gp120 (space group P212121, a = 60.7 Å, b = 66.1 Å, c = 206.7 Å; one molecule per asymmetric unit) were obtained in 5% (vol/vol) isopropanol, 16% (wt/vol) PEG 10,000, 0.1 M Bis⋅Tris, pH 6.5, and 80 mM ammonium sulfate at 20 °C. The Fab NIH45-46GL and NIH45-46chim–93TH057 gp120 complex structures were solved by molecular replacement and refined to 1.65 (Rwork = 17.4%; Rfree = 20.3%) and 3.25 Å resolution (Rwork = 22.7%; Rfree = 26.7%), respectively. The Fab NIH45-46GL and NIH45-46chim–93TH057 gp120 complex structures had 98.6%, 1.4%, and 0.0% or 98.5%, 1.4%, and 0.1% of the residues in the favored, allowed, and disallowed regions, respectively, of the Ramachandran plot.
SPR Measurements.
Binding of 93TH057 gp120 core to NIH45-46mature, NIH45-46chim, and NIH45-46GL IgGs was compared using a Biacore T200 instrument (GE Healthcare). Purified gp120 core was immobilized at a coupling density of 2,000 resonance units on a CM5 sensor chip (Biacore) in 10 mM acetate, pH 5.0, using primary amine coupling chemistry (Biacore). One flow cell on the sensor chip was mock-coupled using buffer to serve as a blank. Experiments were performed at 25 °C in 20 mM Hepes, pH 7.0, 150 mM NaCl, and 0.005% (vol/vol) P20 surfactant, and the sensor chips were regenerated using 10 mM glycine, pH 2.5. Abs were injected at a concentration of 62.5 nM (NIH45-46mature) or 1 μM (NIH45-46chim and NIH45-46GL) at a flow rate of 90 μL/min.
In Vitro Neutralization Assays.
We used a pseudovirus neutralization assay (44) to compare the neutralization potencies of mature, chimeric and germ-line NIH45-46 IgGs. Briefly, neutralization was assessed by measuring the reduction in luciferase reporter gene expression in the presence of a potential inhibitor after a single round of pseudovirus infection in TZM-bl cells. Antibodies were preincubated with 250 infectious viral units in a three- or fourfold dilution series for 1 h at 37 °C before adding 10,000 TZM-bl cells per well for a 2-d incubation. Cells were then lysed, and luciferase expression was measured using BrightGlo (Promega) and a Victor3 luminometer (PerkinElmer). Nonlinear regression analysis using the program Prism (GraphPad) was used to calculate the concentrations at which half-maximal inhibition was observed (IC50 values) as described (45). Samples were screened in duplicate.
Supplementary Material
Acknowledgments
M.C.N. and P.J.B. are Howard Hughes Medical Institute Investigators. R.D. is incumbent of Tauro Career Development Chair in Biomedical Research. This research was supported by the International AIDS Vaccine Initiative and the Bill and Melinda Gates Foundation [Collaboration for AIDS Vaccine Discovery Grants 38619s (to M.C.N.) and 38660 (to P.J.B.) and Comprehensive Antibody-Vaccine Immune Monitoring Consortium Grant 1032144], NIAID [Grant P01AI100148 (to P.J.B. and M.C.N.; contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID or NIH)], the Enoch Foundation (R.D.) and the American Cancer Society (Grant PF-13-076-01-MPC to L.S.). The Molecular Observatory at Caltech is supported by the Gordon and Betty Moore Foundation. Operations at the Stanford Synchrotron Radiation Lightsource are supported by the US Department of Energy and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4JDV (NIH45-46GL Fab) and 4JDT (NIH45-46chim/gp120 complex)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303682110/-/DCSupplemental.
References
- 1.Pierson T, McArthur J, Siliciano RF. Reservoirs for HIV-1: Mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roux KH, Taylor KA. AIDS virus envelope spike structure. Curr Opin Struct Biol. 2007;17(2):244–252. doi: 10.1016/j.sbi.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 5.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 6.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wyatt R, et al. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71(12):9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70(3):1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein JS, Bjorkman PJ. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog. 2010;6(5):e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467(7315):591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337(6091):183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwong PD, Mascola JR, Nabel GJ. The changing face of HIV vaccine research. J Int AIDS Soc. 2012;15(2):17407. doi: 10.7448/IAS.15.2.17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 14.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449(7158):101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 16.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 17.Trkola A, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11(6):615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 18.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109(47):E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334(6060):1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein F, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492(7427):118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481(7379):81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veselinovic M, Neff CP, Mulder LR, Akkina R. Topical gel formulation of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 confers protection against HIV-1 vaginal challenge in a humanized mouse model. Virology. 2012;432(2):505–510. doi: 10.1016/j.virol.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109(30):E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30(5):423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, et al. Deep sequencing with longitudinal sampling of a VRC01-like-antibody response in a chronically infected individual. Retrovirology. 2012;9(Suppl 2):O36. [Google Scholar]
- 33.Ota T, et al. Anti-HIV B Cell lines as candidate vaccine biosensors. J Immunol. 2012;189(10):4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoot S, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013;9(1):e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson CA, et al. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27(19):2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nabel GJ, Kwong PD, Mascola JR. Progress in the rational design of an AIDS vaccine. Philos Trans R Soc Lond B Biol Sci. 2011;366(1579):2759–2765. doi: 10.1098/rstb.2011.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flajnik MF, Deschacht N, Muyldermans S. A case of convergence: Why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011;9(8):e1001120. doi: 10.1371/journal.pbio.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt AG, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110(1):264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verkoczy L, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci USA. 2010;107(1):181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 42.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26(2):205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;12(2005):12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 45.Klein JS, et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci USA. 2009;106(18):7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.