Significance
Regulated release of neurotransmitters is made possible by activity of transporters that mediate their accumulation into synaptic vesicles. Vesicular monoamine transporter (VMAT), a member of the largest superfamily of transporters, mediates transport of monoamines. One of the structural features of these transporters is the pseudo symmetry between the first and the second half of the protein. We demonstrate the importance of two anchor points between these two domains that provide hinge points about which the two halves of the protein flex and straighten to open and close the translocation pathway, a process that enables alternating exposure of the substrate-binding site.
Keywords: ion coupling, multidrug resistance, membrane proteins, neurotransmitter transporter, homology modeling
Abstract
Vesicular monoamine transporter 2 (VMAT2) catalyzes transport of monoamines into storage vesicles in a process that involves exchange of the charged monoamine with two protons. VMAT2 is a member of the DHA12 family of multidrug transporters that belongs to the major facilitator superfamily (MFS) of secondary transporters. Here we present a homology model of VMAT2, which has the standard MFS fold, that is, with two domains of six transmembrane helices each which are related by twofold pseudosymmetry and whose axis runs normal to the membrane and between the two halves. Demonstration of the essential role of a membrane-embedded glutamate and confirmation of the existence of a hydrogen bond probably involved in proton transport provide experimental evidence that validates some of the predictions inherent to the model. Moreover, we show the essential role of residues at two anchor points between the two bundles. These residues appear to function as molecular hinge points about which the two six transmembrane-helix bundles flex and straighten to open and close the pathways on either side of the membrane as required for transport. Polar residues that create a hydrogen bond cluster form one of the anchor points of VMAT2. The other results from hydrophobic interactions. Residues at the anchor points are strongly conserved in other MFS transporters in one way or another, suggesting that interactions at these locations will be critical in most, if not all, MFS transporters.
Transport and storage of neurotransmitters in synaptic vesicles allow their regulated release from the presynaptic cell into the synaptic cleft. The neurotransmitter molecules are accumulated in synaptic vesicles by vesicular neurotransmitter transporters (1–3). Transport of monoamines (serotonin, dopamine, histamine, adrenaline, and noradrenaline) is carried out by the vesicular monoamine transporter (VMAT) family, which includes two isoforms, VMAT1 and VMAT2, in a process that involves the exchange of two protons for one substrate molecule (1–3). The proton electrochemical gradient necessary for transport is generated by the vesicular H+-ATPase (V-ATPase).
The structural basis for the function of VMAT remains unknown. VMAT2 is a member of the DHA12 family of multidrug transporters that belongs to the major facilitator superfamily (MFS) of secondary transporters. Most MFS transporters contain 12 transmembrane (TM) helices, and crystal structures revealed that the 12 TM helices are arranged in two domains of six TMs each, which are related by a twofold pseudosymmetry with an axis that runs normal to the membrane and between the two halves (4–9). Furthermore, analysis of the lactose permease (LacY) crystal structure revealed the presence of inverted topology repeat units within each of the domains (10). That is, the first three helices of each domain are structurally related to the second three helices of that domain by a twofold pseudosymmetry axis that runs through the center of the six-TM domain and parallel with the membrane plane.
According to the alternating-access mechanism, a single binding site is alternately exposed, by conformational change, to either side of the membrane (11, 12). Different crystal structures of MFS members have been solved in various conformations, representing different steps in the transport cycle (4–9). Together with available biochemical knowledge the structures support and confirm the alternating-state mechanism for MFS transporters (13). It has been suggested that in several families of secondary transporters the basis for the alternating-access mechanism arises from inverted-topology repeats (12). Good examples are the neurotransmitter sodium symporters (14) and excitatory amino acid transporters (15), whose folds differ significantly from the MFS transporter fold. Similarly, the periplasm-facing conformation of LacY can be generated by swapping the conformations of the repeat units in each half of the cytoplasm-facing structure (10).
Based on structural conservation, a homology model of rat VMAT2 (rVMAT2) has been reported previously, with a crystal structure of LacY in the Cin conformation serving as a template (16). However, the large number of MFS transporter sequences from diverse subfamilies now available provides an opportunity to improve the alignment between VMAT2 and its potential templates and thereby increase the accuracy of the model. This opportunity, together with advances in modeling techniques, led to the generation of an improved model, which we present here.
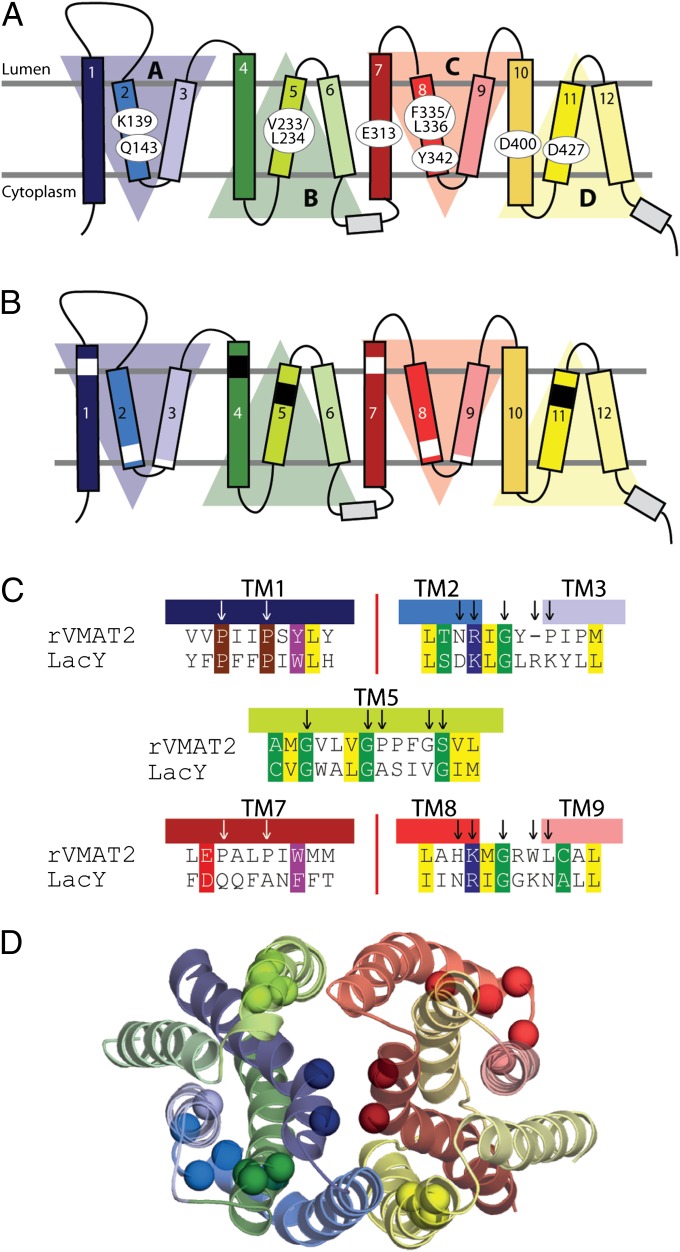
The model presented here indicates key roles for several residues not identified in earlier work (Fig. 1A). Several of those residues are membrane-embedded ionizable residues that are likely to be important for structural or functional reasons. Specifically, the model predicts that six carboxyl residues and a lysine are located in the transmembrane segments. Five of the six carboxyl residues have been studied previously and partially characterized: D33 (TM1), D263 (TM6), D400 (TM10), D427 (TM11), and D461 (TM12). Although D263 and D461 could be replaced with uncharged residues without affecting transport activity, replacements with uncharged residues at the other positions seriously impaired transport activity (17). Analysis of the role of residues corresponding to D400 and D427 in the VMAT1 isoform suggested a possible function in proton translocation or in conformational changes occurring after substrate binding (18). Based on work on the closely related vesicular acetylcholine transporter (VAChT) (19), D400 was suggested to form an ion pair with Y342 (16), although this interaction has not yet been documented for the VMATs. Among the remaining acidic residues, D427 was suggested to form an ion pair with K139 (TM2) (17). However, replacements of the residue corresponding to D427 in VMAT1 retain partial activity (18), and therefore the role of the suggested ion pair should be re-evaluated. Here we propose a different interaction between K139 and D427 that is mediated by Q143 within a network of hydrogen bonds. Also, although our proposed model is not biased or refined to reflect existing biochemical data as the previous model did, D400 and Y342 nevertheless are also located in close proximity and are predicted to form a hydrogen bond. Finally, and unlike predictions based on hydropathy plots, the model predicts that E313 lies in the middle of TM7 and is highly exposed to the cavity. To predict the role of these positions during transport, we also generate a model of rVMAT2 in the vesicle lumen-facing conformation by swapping the conformations of the inverted-topology repeat units within each of the domains.
Fig. 1.
Key residues and motifs in rVMAT2. (A and B) Transmembrane topology of rVMAT2. Shaded triangles indicate the four structural repeat elements in the MFS fold (10). The same coloring of TM helices is used in all subsequent figures. (A) Residues that have been investigated in this study are highlighted. (B) Regions of structural motifs conserved in the entire MFS family (white bars) or only in DHA12 transporters (21) (black bars) are highlighted. (C) Alignment between rVMAT2 and LacY in the motif regions. (D) Model of rVMAT2 in a cytoplasmic-facing state, indicating the locations of the motif residues (spheres), viewed from the vesicular lumen side.
Our results validate the proposed models of VMAT2, confirm the existence of an hydrogen bond, reveal the essential role of the membrane-embedded E313, and support the existence of two clusters of residues functioning as molecular hinge points about which the two helix bundles comprising six TMs each flex and straighten to open and close the pathways on either side of the membrane as required for transport.
Results
Alignment of Key Motifs in the DHA12 Cluster of the MFS Superfamily.
The VMATs share a weak but distinct homology with the potential structural templates used in building our model. Despite their large number and diversity, MFS transporters are characterized by conserved sequence motifs, some of which are found across the entire superfamily and others that are limited to specific MFS families. The presence of such conserved motifs means that the inclusion of members of different families, even those of distant homologs, may help guide a reasonable alignment of MFS transporter sequences, at least in the motif-containing helices. Here, we generated an alignment of rVMAT2 with a structural template (LacY) by combining information from homologs of three different subfamilies for which structural information exists, that is, using a nonredundant set of homologs of LacY, EmrD, GlpT, and VMAT2 (Materials and Methods).
In the resulting alignment (Fig. S1), motifs that are conserved across the MFS in TMs 1 and 7 and in the loops between TMs 2 and 3 and between TMS 8 and 9 are now aligned between the rVMAT2 and LacY sequences (Fig. 1). Compared with an earlier alignment (16) in which those motifs were not aligned, TMs 2, 3, 7, and 8 have each been shifted by one helical turn, i.e., the rVMAT2 sequence is shifted by −4, −3, 4, and 3 residues, respectively. Note that manual adjustments that we made to the present alignment to improve the structural conservation pattern (in TMs 3 and 10) and to remove gaps (in TMs 1, 4, 6, and 11) did not affect the matching of these motifs (Fig. S2).
Within the DHA12 subfamily, there are also conserved motifs in TMs 4, 5, and 11 (20, 21), although these are, by definition, harder to identify in the template. The TM4 motif is absent in the LacY sequence but is present in the sequence of EmrD from Escherichia coli, and even though no structural information was specifically included in the initial alignment, the rVMAT2 motif (RxxxGxG in TM4; Fig. S1) aligns to the LacY sequence in such a way that the positions of the TM4 motif residues in the model correspond to their positions in the crystal structure of EmrD (8), suggesting that the alignment of TM4 is reasonable.
We next consider the DHA12 motifs in TMs 5 and 11 (21). The main feature of these motifs is a sequence of three Gly residues separated from each other by three or four residues (Fig. 1 and Fig. S1). The spacing between the Gly residues is equivalent to one turn of a helix, indicating a possible structural role. Indeed, Ala or Cys mutants of the second Gly in this motif in TM5 decrease transport activity in some members of the DHA12 family (22, 23). Interestingly, in our alignment, motifs in TMs 5 and 11 of rVMAT2 align to Gly-rich segments in the LacY sequence, even though LacY belongs to a different family from the DHA12 proteins. Moreover, the corresponding Gly-rich segment in TM5 of LacY also is important for the transport-associated conformational change; specifically, the mutation of a native Gly (G154) in LacY to Cys resulted in a transporter that is arrested in the cytoplasm-facing conformation (6) and can be rescued by replacing a Gly in a neighboring helix (G24 in TM 1) by Cys (24). The similar functional roles of the aligned positions provide additional support for the alignment in that region of TM5 (Fig. 1 and Fig. S1).
Homology Model of VMAT2.
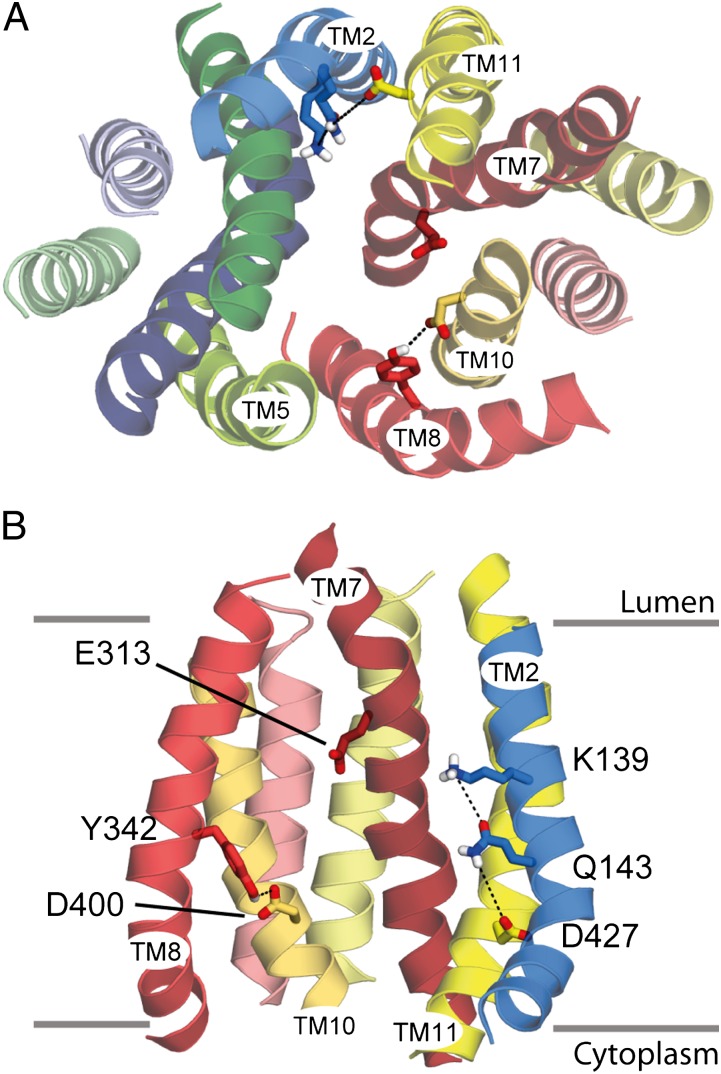
A model of VMAT2 in the cytoplasm-facing conformation was built based on the sequence alignment described above (Fig. 1D and 2). The template used (LacY-CC) was constructed from the crystal structure of LacY from E. coli by replacing the N-terminal domain by a copy of the C-terminal domain to increase the regularity of the helices in the N-terminal domain of the VMAT2 model. Experimental measurements presented here support various aspects of this model, revealing the essential role of E313 and the existence of an important hydrogen bond cluster involving Q143, in addition to K139 and D427.
Fig. 2.
Model of the cytoplasm-facing conformation of rVMAT2. The transporter is shown as helical cartoons, viewed from the cytoplasm (A) or from the plane of the membrane showing only the C-terminal domain and TM2 (B). Side chains of key residues are shown as sticks. Predicted interactions between residues are shown (dashed lines). Approximate boundaries of the membrane are shown as gray lines.
Glu313 Is Irreplaceable.
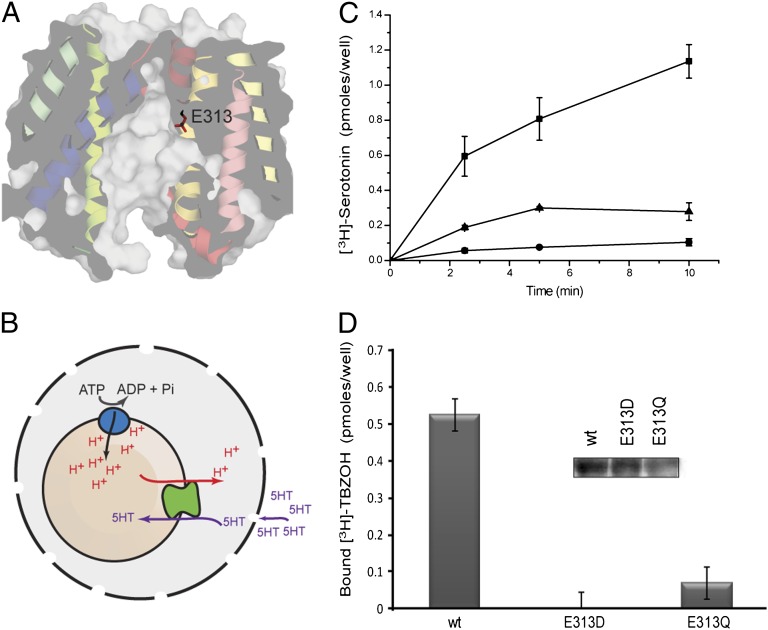
In the model proposed in this work, E313 from TM7 is exposed to the cavity and is located about midway across the membrane (Figs. 2 and 3A), suggesting that it is involved in substrate or proton binding. The positioning of E313 in the pathway in this model is consistent with the alignment of MFS motifs in TM7 of rVMAT2 with those in the structure of LacY (Fig. 1C). According to transmembrane predictions based on hydropathy plots (e.g., www.uniprot.org/uniprot/Q01827), E313 is not considered membrane embedded. In the previously published model of VMAT2, although E313 was located within TM7, it was closer to the vesicular lumen and was not exposed (16). Nevertheless, multiple sequence alignments of a nonredundant set of rVMAT2 homologs identified by PSI-BLAST (Fig. S3A) reveal that E313 is strongly conserved, even if bacterial homologs are included (Fig. S3C), and is fully conserved among higher organisms (Fig. S3B). These observations underscore E313 as a possible key residue and as an interesting position for further study.
Fig. 3.
E313 is crucial for rVMAT2 activity in cells. (A) Predicted location of E313 in the cytoplasm-facing model of rVMAT2, viewed from the plane of the membrane with the cytoplasm at the bottom. Helices are shown as ribbons and using a surface representation, sliced through the center of the protein. The side chain of E313 is shown as sticks. (B) Schematic of the whole-cell transport system. HEK293 cells are permeabilized with digitonin and supplemented with ATP to generate the proton gradient by the V-type ATPase. (C) [3H]-serotonin transport into vesicles in cells transfected with rVMAT2 (■), E313Q (●), and E313D (▲). Digitonin-permeabilized cells were assayed after incubation with [3H]-serotonin. (D) Binding of the noncompetitive inhibitor of VMAT2, [3H]-TBZOH. (Inset) Western blot. Membranes were analyzed as described in Materials and Methods. Results presented are from duplicate experiments; error bars indicate SE. Experiments were repeated at least twice.
To gain insight into the role of this residue, we mutated it to Gln and Asp in rVMAT2. When expressed in HEK293 cells, the wild-type protein binds [3H]-dihydrotetrabenazine ([3H]-TBZOH), an analog of the noncompetitive inhibitor tetrabenazine (TBZ), and accumulates [3H]-serotonin into vesicles in a process that is dependent on the proton gradient generated by a V-type ATPase (Fig. 3 B–D). To assay for both activities, the plasma membrane was permeabilized with digitonin, and the cells were then challenged with the radiolabeled compounds (Fig. 3B). To generate the vesicular pH gradient necessary for the transport reaction, ATP was supplied along with serotonin. The mutated proteins, E313D and E313Q, displayed very low or nil [3H]-TBZOH binding and [3H]-serotonin uptake (Fig. 3 C and D).
Because the [3H]-TBZOH binding and the expression levels of the E313 mutants, as measured by Western blots (Fig. 3D, Inset and Fig. S4), were low, we also collected rVMAT2 from appropriate amounts of cells, and reconstituted it into proteoliposomes after solubilization and partial purification. The amount of membranes used for each proteoliposome preparation was calculated so that similar amounts of the wild-type or mutant proteins were incorporated into the corresponding proteoliposomes, as confirmed by dot blot analysis (Fig. S3E, Inset). To generate a pH gradient, the proteoliposomes were loaded with (NH4)2SO4, diluted into an ammonium-free buffer and assayed for [3H]-serotonin uptake (Fig. S3D, Inset). Replacements of E313 with either Asp or Gln displayed undetectable activity in proteoliposomes (Fig. S3D).
To determine whether E313 also is essential for binding substrate and/or inhibitors, we tested the ability of the mutants to bind [3H]-TBZOH in proteoliposomes. Although the wild-type rVMAT2 binds up to 0.4 pmoles of [3H]-TBZOH, we could not detect any binding to either the mutants, suggesting that E313 also is essential for recognition of TBZ (Fig. S3E) and confirming the results from permeabilized cells (Fig. 3). Taken together with its predicted location in the center of the cavity (Fig. 3A), these results suggest that E313 plays a key role in the transport cycle.
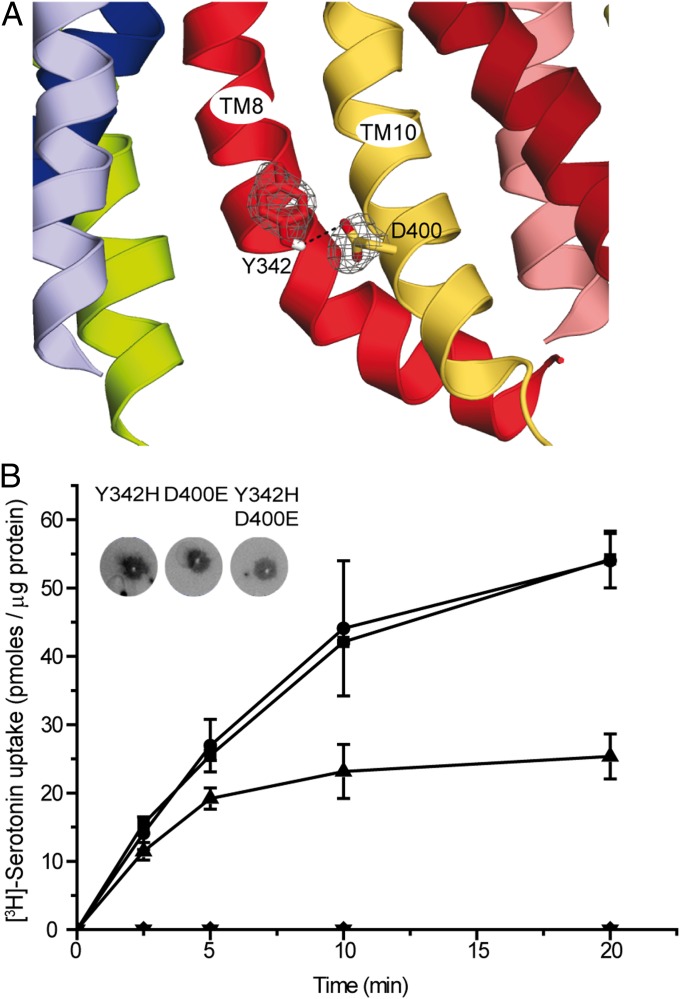
Predicted Hydrogen Bond Between D400 of TM10 and Y342 in TM8.
The existence of an ion pair between D398 and H338 in rVAChT was postulated previously (19). The two corresponding residues in rVMAT2 (D400 in TM10 and Y342 in TM8) are located within a short distance of one another in the model of the cytoplasm-facing state (Figs. 2 and 4A) and also line the central pathway. Here, we evaluated the presence of this predicted hydrogen bond in rVMAT2 experimentally, by constructing a set of mutants of the two positions. D400 was replaced with Glu, Asn, Ser, or Cys, and Y342 was replaced with His, Phe, Gln, or Ser. Similar to findings for rVMAT1 and rVAChT (18, 19), our data suggest that a negatively charged residue at position 400 is essential for proper activity. Specifically, D400S or D400C mutants displayed no transport activity (Fig. 4B and Fig. S5), although the mutant protein D400E still can transport serotonin, albeit at twofold-lower levels than wild-type rVMAT2 (Fig. 4B, Fig. S5, and Table 1). D400N did not express to significant levels. The equivalent replacement in rVMAT1 was shown to be inactive previously (18).
Fig. 4.
Predicted interaction between residues Y342 and D400 in TMs 8 and 10. (A) The model of rVMAT2 in a cytoplasm-facing state is viewed along the plane of the membrane focusing on the cytoplasmic half of the protein, which is oriented with the cytoplasm toward the bottom. Side chains of Y342 and D400 are shown as sticks, and the probability density of their positions in the 100 top models is shown as a gray mesh. (B) [3H]-serotonin uptake. Liposomes (1 µL) reconstituted with rVMAT2 (■) D400E (▲), Y342H (●),Y342H-D400E (▼), and vector with no gene (mock) (◆) were assayed for serotonin transport as described in Materials and Methods. (Inset) Quantitation of protein amounts in proteoliposomes using a dot blot assay. Results presented are from duplicate experiments; error bars indicate SE. Experiments were repeated at least twice.
Table 1.
Kinetic properties of VMAT2 mutated at positions 342 and 400
| Strain | Serotonin transport | TBZ binding (Kd)) (nM) | Displacement of TBZ binding by MPP+
(IC50) (μM) |
|
| Km (nM) | Vmax (pmoles/min per microgram protein) | |||
| Wild type | 178 ± 43 | 9 ± 0.4 | 10 ± 1 | 69 ± 2 |
| D400E | 372 ± 69 | 2 ± 1 | 29 ± 5 | 145 ± 44 |
| D400S | — | — | 18 ± 2 | 273 ± 40 |
| Y342H | 165 ± 97 | 7 ± 1.5 | 5 ± 2 | — |
| Y342H- D400E | — | — | 81 ± 6 | — |
For Kd measurements, 1.5 μL membrane (5–10 μg total protein) produced from HEK293 cells transfected with wild-type or mutated rVMAT2 was incubated in the presence of various concentrations of [3H]-TBZOH for 20 min. For Km measurements, liposomes (1 µL) were incubated for 5 min with various concentrations of [3H]-serotonin. For determination of TBZ binding and displacement by MPP+, membranes prepared from HEK293 cells expressing wild type, D400E, or D400S rVMAT2 were incubated in the presence of various concentrations of MPP+ for 20 min followed by 20-min incubation with [3H]-TBZOH (5 nM) at pH 8.
At position 342, of the various replacements tested, the only one active for transport is Y342H (Fig. 4B and Fig. S5). Replacement with another aromatic residue (Y342F) or with different polar residues (Y342S, Y342Q) abolished the ability to transport [3H]-serotonin (Fig. S5) even though they express to levels similar to those of the wild-type protein (Fig. S4). These results suggest that position 342 of rVMAT2 requires a protonatable residue to interact functionally with D400. Interestingly, both D400E and D400S retained TBZ-binding ability (Table 1). Moreover, both the D400E and D400S mutants can still recognize substrates with a reasonable affinity. This contention is supported by the observation that another substrate of rVMAT2, 1-methyl-4-phenylpyridinium (MPP+), displaces TBZ from D400E at concentrations similar to those that displace TBZ in the wild-type protein (Table 1).
Although each of the replacements alone preserves transport activity, the combination Y342H-D400E eliminates it (Figs. 4B, Fig. S5, and Table 1). Two other replacements at position 342 (Y342Q and Y342S) also were inactive in a D400E background (Fig. S5).
These results suggest that very specific residues are needed at these positions to allow a functional interaction between them that is necessary for transport.
Identification of a Hydrogen Bond Cluster Linking the N- and C-Terminal Domains.
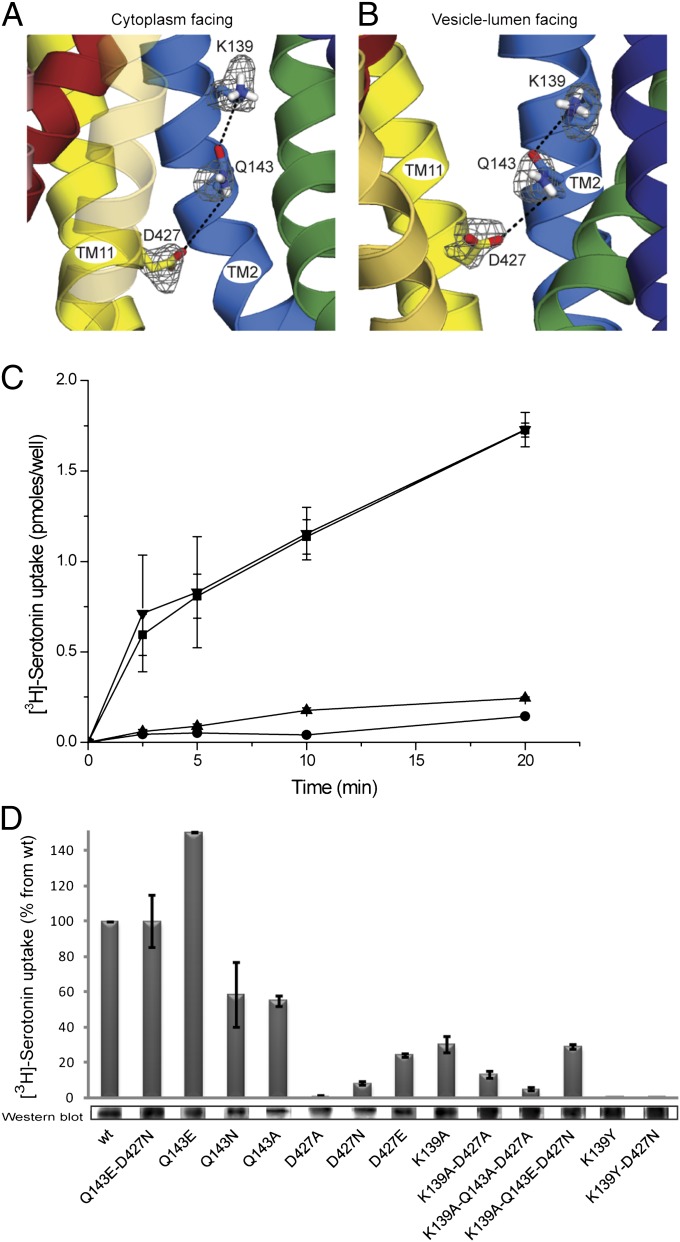
Previous work suggested that D427 from TM11 interacts with K139 from TM2 via an ion pair or salt bridge (17). Position 427 (and equivalents in rVMAT1 and rVAChT) was identified as essential for transport activity, and even the most conservative replacement to glutamate dramatically reduced transport activity (17, 18, 25). Interestingly, however, the rVMAT1 mutant retains the ability to bind the competitive inhibitor reserpine, a process that is accelerated by the proton gradient and involves transport of one of the two H+ ions exchanged with the transported substrates (18, 26). This finding suggested that the lack of transport activity after replacement of D427 was not caused by a deleterious effect on the binding of either the substrate or the first proton out of the two transported by VMAT2. As previously mentioned, D427 and K139 are too far apart to be able to interact directly (Figs. 2 and 5A). However, the model predicts that Q143 may mediate the interaction between them, creating a hydrogen bond network (Figs. 2 and 5A). To test this prediction, we mutated the relevant residues and assayed for activity.
Fig. 5.
Predicted network of interactions involving residues K139, Q143, and D427N in TMs 2 and 11. (A and B) Models of rVMAT2 in a cytoplasm-facing (A) and lumen-facing (B) state viewed along the plane of the membrane oriented with the cytoplasm toward the bottom. The side chains of K139, Q143, and D427 are shown as sticks, and the probability density of their positions in the 100 top models is shown as a gray mesh. (C) [3H]-Serotonin transport into vesicles in cells transfected with rVMAT2 (■) D427N (●), D427E (▲), and Q143E-D427N (▼). (D) [3H]-Serotonin uptake into transfected HEK293 cells and estimation of expression levels by Western blot as described in Materials and Methods. Quantitation of expression levels of all mutants is shown in Fig. S4. Results presented are from duplicate experiments; error bars indicate SE. Experiments were repeated at least twice.
Negative Charge Can Be Moved Within the Cluster.
Position 427 of rVMAT2 was mutated to Ala, Asn, or Glu, and the effect of the different substitutions on vesicular [3H]-serotonin uptake in permeabilized cells was tested. The results (Fig. 5 C and D) confirm previous observations for rVMAT1, rVMAT2, and VAChT (17, 18, 25) showing that position 427 does not tolerate even conservative replacements. In all cases, no significant transport activity into vesicles was measured. Notably, however, the negative charge in the cluster is not required for either inhibitor or substrate binding. All the D427 mutants bound [3H]-TBZOH with similar affinities, in the 30–40 nM range, three- to fourfold lower than the affinity of the wild-type protein (Table 2). Furthermore, the Glu, Gln, and Ala replacements bind substrate, albeit with somewhat lower affinities as judged by the ability of MPP+ to displace TBZ (Table 2).
Table 2.
Tetrabenazine binding to VMAT2 mutated in the cluster K139–Q143–D427
| Strain | Tetrabenazine binding (Kd) (nM) | Displacement of TBZ binding by MPP+ (IC50) (μM) |
| Wild type | 10 ± 1 | 63 ± 2 |
| Q143E | 10 ± 3 | — |
| Q143N | 7 ± 2 | — |
| Q143A | 18 ± 3 | — |
| Q143E-D427N | 20 ± 4 | — |
| D427E | 33 ± 8 | 173 ± 19 |
| D427N | 45 ± 13 | 336 ± 110 |
| D427A | 40 ± 10 | 141 ± 76 |
| K139A | 44 ± 10 | — |
| K139A-Q143E-D427N | 62 ± 14 | — |
| K139A-D427A | 73 ± 6 | — |
| K139A-Q143A-D427A | 159 ± 22 | — |
| K139Y | 51 ± 7 | — |
| K139Y-D427N | 15 ± 3 | — |
Membranes (1.5 μL, 5–10 μg total protein) prepared from HEK293 cells transfected with wild-type or mutated rVMAT2 were incubated in the presence of various concentrations of [3H]-TBZOH for 20 min.
In an attempt to invert the polarity of the putative pair K139/D427 in VMAT1, Merickel et al. (17) previously reported no activity of the D139/K427 mutant. Because we now suggest that Q143 may be part of the hydrogen bond cluster, we tested how permissive the transporter is to the location of the negative charge. We constructed a double mutant, Q143E-D427N, which combines a mutation that abolishes transport (D427N; Fig. 5C) with one that introduces the negative charge in the predicted neighboring residue, Q143. Strikingly, combination of the lethal mutation D427N with Q143E restores wild-type levels of [3H]-serotonin transport activity to rVMAT2 (Fig. 5C). This finding provides strong support for the existence of an interaction between the two residues, consistent with the close proximity of these positions in TM2 and TM11 and supporting the predicted hydrogen bond network involving K139, Q143, and D427 (Fig. 5A).
K139–Q143–D427 Cluster Has a Role in the Transport Cycle.
The hydrophilic interaction between TM2 and TM11 is highly conserved at this position in many proteins of the MFS superfamily. In a large group of bacterial homologs, the equivalent to K139 is Tyr, and the equivalent to D427 is usually Ser or Asn (Fig. S6). Although Q143 is not fully conserved in TM2, other polar residues close to the Asn in TM11 also may form part of the cluster (e.g., Gln at position 430; Fig. S6). These residues could provide a polar environment and create an equivalent hydrogen bond network between TM2 and TM11 in the bacterial transporters.
To gain further knowledge about the significance of the polar environment and the interaction between these two helices, we generated single and multiple Ala replacements. Although the single replacements in D427 resulted in complete loss of activity, single replacements in the TM2 residues have less dramatic effects. In the case of the Q143 replacements, the mutant with Glu transports [3H]-serotonin like the wild-type protein, whereas the Asn and Ala substitutions significantly impair the transport reaction (Fig. 5C). However, all three mutants, Q143E, Q143A, and Q143N, bind [3H]-TBZOH at levels similar to the wild-type rVMAT2 (Table 2). These results indicate the importance of a polar residue at this position for wild-type levels of transport activity but also suggest that this residue is not required for binding TBZ.
Although the single mutations K139A and Q143A (Fig. 5D) resulted in about 35% and 50% of wild-type transport activity, respectively, the double mutation K139A-D427A resulted in only 20% of wild-type levels, and the triple mutation K139A-Q143A-D427A completely abolished the ability of rVMAT2 to transport [3H]-serotonin (Fig. 5D). The single K139A replacement displayed similar transport activity whether it was in a wild-type background or in a Q143E/D427N mutant (Fig. 5D).
High-affinity binding of [3H]-TBZOH was also affected significantly by the Ala replacements. Specifically, the Kd values increased by two- to fourfold for the single replacements and by seven- and 15-fold for the double and triple replacements, respectively, compared with the wild-type protein (Table 2). Together, these results indicate that the polar environment supplied by the residues in the cluster is necessary for high-affinity recognition of substrates and inhibitors.
To test further whether other polar interactions are sufficient for inhibitor binding, we generated a mutant with residues identified in bacterial homologs (Fig. S6). Notably, K139Y-D427N retains the ability to bind [3H]-TBZOH with relatively high affinity (Table 2); however, K139Y-D427N does not show significant transport activity (Fig. 5D). It is interesting that removing a single charge reduces Kd by fivefold, whereas removing the other charge as well brings the affinity back to about wild-type levels (Table 2).
Taken as a whole, these results support the contention that a negative charge in the hydrogen bond cluster is not essential for binding. Complete abolition of the cluster by a triple-Ala replacement inhibits transport and dramatically impairs the ability of rVMAT2 to bind [3H]-TBZOH. We suggest that the influence on binding may be caused by a more general structural effect in which the two symmetric halves of the transporter are not properly connected, because the double mutant K139Y-D427N retains the polar environment and can bind [3H]-TBZOH with an affinity comparable to that of the wild type.
Hydrophobic Interactions Between the N and C Domain Are Important for Transport Activity.
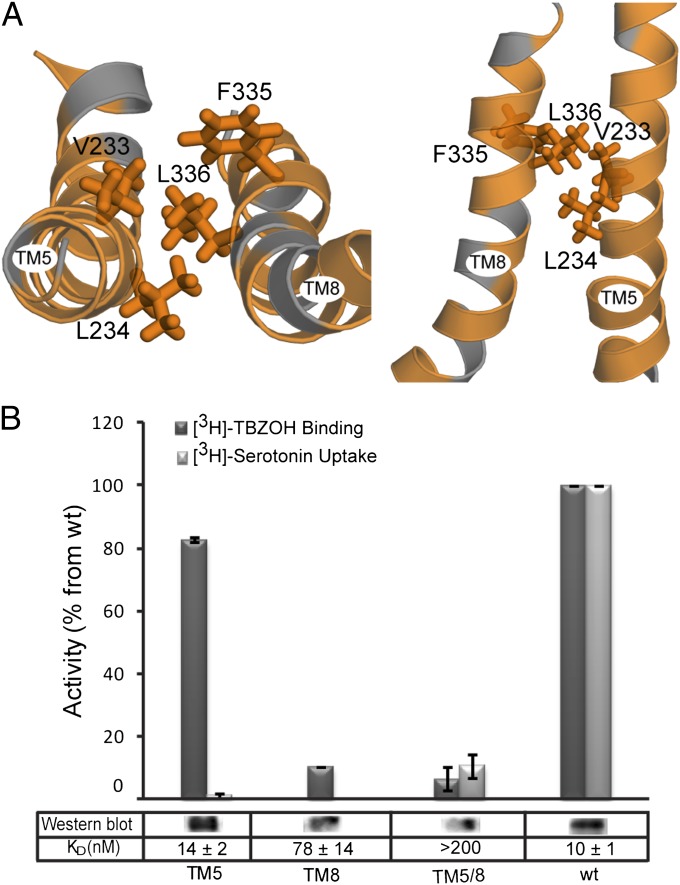
Because each of the domains consists of pseudosymmetry-related elements, we investigated whether a second set of interactions exists between TMs 8 and 5 that connect the domains on the opposite side of the central pathway from TMs 2 and 11. The model of VMAT2 predicts a set of hydrophobic interactions between the two helices that may connect them (Fig. 6A). Replacement of two hydrophobic residues (V233 and L234) in TM5 with Ala resulted in a dramatic reduction of serotonin uptake and only a small decrease in the level of TBZ binding (Fig. 6B). Replacement of F335 and L336 in TM8 abolished both serotonin uptake and TBZ binding (Fig. 6B).
Fig. 6.
Predicted hydrophobic interactions between TM5 and TM8. (A) Close-up of TMs 5 and 8 in the cytoplasm-facing model of rVMAT2 viewed as in Fig. 2A (Left) and along the plane of the membrane (Right) with the cytoplasm toward the bottom. Hydrophobic residues (Ala, Gly, Val, Ile, Leu, Phe, Met, and Trp) are colored orange. Side chains of L336, F335, L234, and V233 are shown as sticks. (B) [3H]-TBZOH binding to HEK239 permeabilized cells, [3H]-serotonin transport for V233A/L234A (TM5), F335A/L336 (TM8), and the quadruple mutant V233A/L234A/F335A/L336 (TM5/8), and Western blot relative to wild-type rVMAT2. Results presented are from duplicate experiments; error bars indicate SE. Experiments were repeated at least twice.
Interactions Between the N and C Domains Are Conserved Within Families.
Given the structural homology across the MFS, we hypothesized that similar hinge points are required for function in other transporters in this superfamily. Therefore, the available crystal structures of other members of the MFS were analyzed. Although not conserved across families (Table S1 and Fig. S7), residues forming the interfaces between TM2 and TM11 and between TM5 and TM8 are among the most conserved positions within each family, with median values of 8 and 9, respectively, for the normalized conservation scores from ConSurf (Table S1). Residues in LacY at this region (Fig. S7) have been shown to be important for function (27, 28). The fact that highly conserved interactions at the midpoints of these helices are found connecting the two domains of the transporter in all these families lends further support to their significance.
Discussion
Homology modeling was used previously to construct a model of VMAT2, based on hydropathy plot-guided, manually adjusted sequence alignments (16). The availability of a greater number of more diverse sequences can help generate more accurate alignments, the most crucial factor determining the accuracy of a homology model for a given template. This availability, together with improvements in techniques for evaluating models, enabled the generation of a refined model of rVMAT2 unbiased a priori by biochemical data, which is presented here. This model subsequently was validated by analysis of sequence motif conservation and by biochemical studies. The inclusion of a variety of different sequence homologs allowed the evolutionary relationships to guide the alignment of the MFS and DHA12 motifs better. As a result, the model we present here differs from the previous one in several aspects, especially in the alignment of TMs 2 and 7, allowing us to identify new roles for several residues and interactions that are critical for transport.
In contrast to predictions based on hydropathy plots, this model predicts that E313 is midway along TM7 and is highly exposed to the cavity (Fig. 3A). We also show that E313 is highly conserved even among bacterial homologs (Fig. S3) and is fully conserved among eukaryotic homologs, revealing its likely importance for neurotransmitter transport. Indeed, even conservative replacements such as E313D and E313Q dramatically affect all transporter activities tested (inhibitor binding and substrate transport), pointing at the significance of this residue (Figs. 3 and 4).
Further work is required to elucidate whether E313 is involved in proton coupling, in substrate recognition and binding, or in both. A dual role for carboxyl residues was demonstrated in EmrE, where a conserved Glu (E14) can alternately bind either the substrate or proton and thus couple the two processes (29, 30). Similar mechanisms also were proposed in the transport cycle of other MFS transporters such as LmrP (31) and MdfA (32). Interestingly, when the position equivalent to E313 in the close homolog VAChT was replaced by Asp or Gln, the mutant protein was still able to transport acetylcholine (33). Only replacement by Ala leads to inactivity, indicating that in VAChT the polarity of this residue, and not necessarily the charge and the precise size of the residue, is important for activity. Thus, we suggest that in VAChT this residue participates in substrate recognition and not necessarily in proton translocation. Nevertheless, it is possible that the coupling mechanism of VAChT differs from that of VMAT2, and a role for E313 in the coupling mechanism cannot be ruled out.
We next turn to the predicted hydrogen bond between Y342 (TM8) and D400 (TM10) (Figs. 2 and 4A). D400 has been the subject of intensive research in all members of the vesicular amine transporter (SLC18) family (17, 18, 25). In all cases, a carboxyl group at this position was found to be crucial for proper transport activity. Experiments with reserpine in rVMAT1 identified this residue as a possible site for proton binding (18). The involvement of hydrogen bonds in proton coupling has been postulated in several ion-coupled transporters (34–36). It has been suggested that hydrogen bonds (and ion pairs) can serve as the core of the coupling mechanism (37). The dynamics of these bonds (e.g., because of deprotonation) can lead to conformational changes. Therefore it was very tempting to examine the predicted hydrogen bond in rVMAT2. According to our findings it is clear that proper transport activity requires the presence of a carboxyl group at position 400 and a protonatable residue at position 342, as would be consistent with a functional interaction between the two groups. This conclusion is supported by the finding that, although the single replacements Y342H and D400E maintain transport ability, their combination abolishes it, likely because any residual interaction between the two mutated residues in the single mutants is lost in the double mutant. Our findings regarding the function of these hydrogen bonds in the transport cycle fit well with other paradigms suggested for MFS transporters, based on available crystal structures (4, 6, 35). In particular, in LacY it was suggested that a proton is shared between Glu269 from TM8 and His322 from TM10, and biochemical evidence supports these interactions (38). These interactions are not observed in the cytoplasm-facing crystal structure but are observed in a published model of the periplasm-facing conformation (10). Here, we supply evidence for a similar mechanism and suggest that D400 is involved in proton transport. Although speculative, we suggest that protonation or deprotonation of D400 breaks the hydrogen bond between D400 and Y342, mediating a conformational change required for the next step in the transport cycle and thus coupling the proton movement to the substrate translocation. This suggestion would be consistent with evidence indicating that the equivalent residue in VMAT1 plays a role in the transport cycle rather than in substrate recognition (18).
The hydrogen bond cluster connecting the two bundles each comprising six TMs is essential and appears to play a more complex role. The model presented here predicts that Q143 can bridge the interactions between K139 and D427, and our experimental data support this prediction. Specifically, even though all the single replacements at position 427 (D427E/N/A) abolish transport ability, the effect of the D427N mutation is suppressed when in combination with Q143E, indicating that the two residues are likely to be close in space. Notably, none of the mutations of this cluster had any effect on the transporter’s ability to bind TBZ, which provides an indication of correct folding. Furthermore, although D427E/A/N no longer can transport substrate, MPP+ is still able to inhibit the binding of TBZ. Taken as whole, these data suggest a network of hydrogen bonds between K139, Q143, and D427 in which the negative charge plays an essential role in the transport cycle. No one of the residues in TM2 is fully essential for suboptimal activity, but both are needed for a fully active transporter.
The finding that the hydrophilic environment is conserved among vesicular neurotransmitter transporters (Fig. S6) encouraged us to probe further the possible role of the cluster. We showed here that the interactions between TM2 and TM11, established by the polar cluster, are highly significant for the transporter's structure and function. Partial or complete abolition of the cluster (the double and the triple Ala replacement, respectively) decreases both TBZ binding levels and transport levels. Replacement by polar residues (K139Y-D427N) maintains proper binding activity, albeit not serotonin uptake. Therefore the significant decrease in TBZ binding ability exhibited by the double and triple Ala replacements but not by K139Y-D427N supports the suggestion that this cluster of polar groups is required to create and stabilize the structure of the protein.
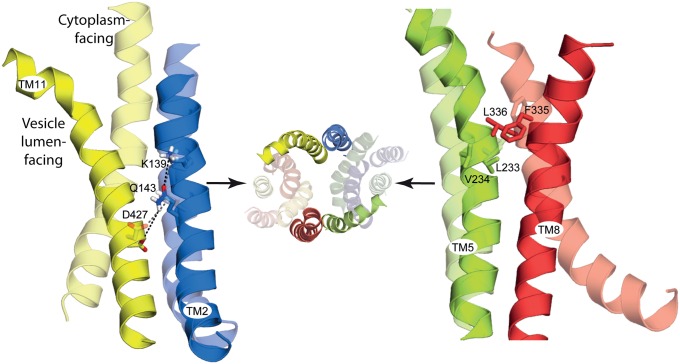
Given the common architecture of the MFS transporters [for example, as seen in the structures ofGlpT (4) and LacY (6),] an interaction between TM2 and TM11 is expected to be conserved, independent of the local sequence. In OxlT, for example, cross-linking studies indicated that TM2 and TM11 are in close proximity (39, 40). A possible ion pair connecting TM2 and TM11 was also found in the melibiose carrier (MelB) (41). To understand better the nature and the possible mechanistic role of this cluster of residues in VMAT2, the vesicle lumen-facing conformation of the transporter was modeled by swapping the conformations of the two internal repeats in the cytoplasm-facing model (Fig. S8). Comparison of the two models indicates that TM2 and TM11 undergo dramatic conformational changes relative to one another during the transport cycle, with the ends of the helices separating on the side that is opening and approaching each other on the side that is closing (Fig. 7). Despite these large changes, the cluster connecting the two helices is predicted to maintain the same relative position in both conformations (Fig. 7).
Fig. 7.
The relative position of the proposed molecular hinge residues is essentially unchanged between the cytoplasm- and vesicle lumen-facing models. The models of the cytoplasm-facing (transparent) and vesicle lumen-facing conformations (opaque) of rVMAT2 are superposed using the N-terminal domain (TMs 1–6), showing the predicted change in orientation of the C-terminal domains (TMs 7–12) around the proposed molecular hinge-point residues (sticks). Predicted polar interactions are indicated (dashed lines). Helices are shown as cartoons and viewed along the plane of the membrane with the cytoplasm to the bottom, for TMs 2 and 11 (Left) and TMs 5 and 8 Right). For reference, the vesicle lumen view of all 12 TMs is shown also (Center), with key helices opaque.
One of the structural features of the MFS family is the pseudosymmetry between the first and the second sets of six-TM helices (42). Following this symmetry relationship, the helices “equivalent” to 2 and 11 are 8 and 5, respectively. Key interactions between TM5 and TM8, either direct or mediated by the substrate, have been identified in LacY and GlpT (4, 6), and therefore we also examined those helices in rVMAT2. Analysis of the predicted structure of TMs 5 and 8 in VMAT2 revealed a set of hydrophobic interactions connecting the two helices that are essential for activity and appear to be the symmetry equivalents of the polar interactions between TM2 and TM11.
We speculate that these interactions provide important anchor points between the two domains, functioning as hinge points about which the two bundles flex and straighten to open and close the two pathways. These hinge points may mediate conformational changes while retaining the integrity of the transporter structure. In the GlpT transport cycle, it was hypothesized that the interface between TM2/11 and TM5/8 would enable a “rocker-switch” type movement in which the cytoplasmic ends of these helices come closer as the periplasmic ends move further apart (43). More recent structures [e.g., PepT (35)] and biophysical analyses (44, 45) indicate the additional involvement of occluded states of MFS transporters in which both sides are closed simultaneously. Here, we extend the proposals for GlpT, ascribing a role for specific interactions in TMs 2/11 and 5/8 in the cycle of outward-open, occluded, and inward-open states of VMAT2, and suggest that interactions at this location (Table S1) will be critical in most, if not all, MFS transporters.
Materials and Methods
Computational Methods.
Constructing the alignment for modeling the cytoplasm-facing state of rVMAT2.
The sequences of E. coli LacY (GI 16128328), E. coli GlpT (GI 16130175), E. coli EmrD (GI 110807632), and rat VMAT2 (GI 34978379) were selected as query sequences. For each query sequence, a PSI-BLAST (46) search was performed, as provided in the National Center for Biotechnology Information BLAST package (version 2.2.17) (47). Five iterations were used to search the nonredundant protein database (as of December 2, 2009). We selected 250 hits for each of the query sequences. To exclude incomplete sequences (fragments) and sequences possessing atypically long terminal domains or insertions, the PSI-BLAST hits were filtered so that their length was in the range σ ± δ, where σ is the mean length of all of the hits and δ is the SD thereof. From the hits for each query sequence, the most informative sequences were selected according to a sequence identity cutoff of 60% using CD-HIT (48). This selection resulted in 39, 14, 36, and 32 sequence homologs for LacY, GlpT, EmrD, and rVMAT2, respectively. The four subsets were combined, and then all the sequences were aligned using MUSCLE (49) (Fig. S1). The preliminary pairwise sequence alignment of LacY and rVMAT2 was extracted from this multiple sequence alignment.
The Rate4site-algorithm (50) implemented in the ConSurf web server (51) was used to quantify the evolutionary conservation of each amino acid in the alignment; mapping these values onto the resultant model guided the placement of residues in variable and conserved positions so that they face the lipid environment and protein interior, respectively. Specifically, we adjusted the preliminary alignment in TMs 3 and 10 according to the ConSurf results, as well as in TMs 1, 4, 6, and 11 to remove gaps from the preliminary alignment.
In the final alignment of LacY and rVMAT2 (Fig. 1 and Fig. S2), 12% of the residues are identical, calculated over the whole sequence but excluding the loops between TMs 1 and 2 and between 6 and 7. Typically, homology models built with this sequence identity range can be expected to have an error of ∼3 Å in the positions of the Cα atoms (52).
Modeling the cytoplasm-facing state of rVMAT2.
Models of rVMAT2 were constructed based on the structure of LacY [Protein Data Bank (PDB) ID 1PV7] using Modeler version 9.7 (53). All of the three potential cytoplasm-facing templates considered (LacY, GlpT, and EmrD) share ∼10% identical residues with rVMAT2. Of these, LacY was selected because of the higher resolution of its structure relative to EmrD and because reliable alignments also are available for construction of a repeat-swapped model (10). Even without further adjustments to the alignment shown in Fig. S1, the model based on LacY is entirely consistent with a cytoplasm-facing state of rVMAT2. Nevertheless, we noticed that TMs 3 and 4 were distorted in this model; we assumed these distortions are not general features of MFS transporters but instead are specific to the LacY template structure, because such distortions are not observed in structures of GlpT, PepT, or FucP. Therefore, an alternative artificial template for homology modeling (referred to as “LacY-CC”) was constructed from the cytoplasm-facing crystal structure of E. coli LacY by replacing the N-terminal domain of LacY with a copy of its C-terminal domain. Although the alignment of TMs 3 and 4 of VMAT2 to LacY was relatively good, replacing the N-terminal domain with the C-terminal domain led to more reasonable models in terms of the ConQuass score (see below).
The alignment of the N-terminal domains of the rat rVMAT2 and LacY-CC sequences was guided by the structural analogy between the N- and C-terminal domains of LacY, so that the vertical position and lipid exposure of residues from TMs 1–6 in the resulting model was similar to that in the model built on the crystal structure of LacY from E. coli. The alignment between rVMAT2 and LacY-CC in the C-terminal domain region was the same as that used for the LacY template (Fig. S2). The loops between TMs 1 and 2 and between TMs 6 and 7 were not modeled because their lengths are beyond the range accessible to current loop prediction methodologies.
The final cytoplasm-facing homology model of rVMAT2 based on LacY-CC has a ConQuass (54) score of 0.10 ± 0.01 (mean ± SD over 1,000 Modeler models for residues 18–51 and 126–472). These models therefore are more reasonable than those built using LacY as the template, whose ConQuass scores are 0.08 ± 0.01, and compare well with the LacY crystal structure, which has a ConQuass score of 0.18. The structure with the lowest Modeler discrete optimized protein energy (DOPE) score of 1,000 optimization cycles was selected to be the representative and is available on the Protein Model Databank (http://mi.caspur.it/PMDB/), identifier PM0078823. This model has excellent quality according to PROCHECK (55), with only one residue occupying the generously allowed regions of the Ramachandran plot. In addition to this representative model, the 100 models with the lowest DOPE scores were selected for further refinement by energy minimization (see below).
Constructing the alignment for modeling the vesicle lumen-facing state of VMAT2.
Constructing a model of the vesicle lumen-facing state requires exchanging the conformations of the internal structural repeats (10). For rVMAT2, an initial pairwise sequence alignment was constructed first by extracting the pairs of residues found to be close in space after superposing the structural repeats from the model of rVMAT2 in the cytoplasm-facing state onto one another. Specifically, we superposed repeat unit A containing TMs 1–3 (residues 18–51 and 126–182) onto repeat unit B containing TMs 4–6 (residues 183–270), and we superposed repeat unit C containing TMs 7–9 (residues 292–378) onto repeat unit D containing TMs 10–12 (residues 379–472) using the structure alignment program DaliLite (56), resulting in two pairs of alignments. The rmsd in the Cα positions between segments A and B was 3.0 Å, whereas the rmsd between segments C and D was 2.8 Å. The fragments of sequence alignment then were concatenated to obtain a preliminary pairwise alignment between the model sequence (VMAT2) and the corresponding template sequence, which is the rVMAT2 sequence whose repeat units are rearranged in the order BADC. As with the cytoplasm-facing state model, the sequence identity of VMAT2 to its template in the final alignment (Fig. S9) is 12%.
Modeling the vesicle lumen-facing state of VMAT2.
The repeat-swapped model of rVMAT2 was constructed using the corresponding segments of the cytoplasm-facing rVMAT2 model as templates, according to the alignment in Fig. S9. The ConQuass (54) score of the resultant 1,000 models was 0.11 ± 0.01 (mean ± SD of 1,000 Modeler models for residues 20–51 and 126–472). The structure with the lowest DOPE score of 1,000 optimization cycles was selected to be the representative. This model has reasonable quality according to PROCHECK (55), with four and one residues occupying generously allowed and disallowed regions, respectively, of the Ramachandran plot. The model is available on the Protein Model Databank (http://mi.caspur.it/PMDB/), identifier PM0078824. We also selected the 100 models with the lowest DOPE scores for further refinement by energy minimization (see below).
Energy minimization of the models.
All models were energy minimized before analysis using CHARMM (version 3.4) (57). Before minimization, the positions of all hydrogen atoms and Asn, Gln, or His side-chain rotamers were predicted using REDUCE (version 2.21) (58). Subsequent energy minimization was performed using the CHARMM22 force field (59) and was carried out in three stages, each with 250 steps of steepest decent minimization, followed by 250 steps of conjugated gradient minimization. First, all nonhydrogen atoms were kept fixed, and only the hydrogen atoms were allowed to move. Second, the nonhydrogen atoms of the backbone were kept fixed, and only the hydrogen and side-chain atoms were allowed to move. In the final stage all atoms were allowed to move. The minimization was performed using the CHARMM22 force field (59). To account for the large central aqueous cavity, a dielectric constant of 80 was used for the calculation of electrostatic interactions.
Laboratory Methods.
Plasmids.
The rat VMAT2 gene (SLC18A2), with an HA tag in the second loop between positions 96 and 105 as well as 10 His residues at the C terminus (60), was cloned into pcDNA3.1 plasmid (Invitrogen). Cloning was carried out using PCR, with HindIII and NotI restriction sites. Site-directed mutagenesis was done with the Quickchange II Site-directed Mutagenesis Kit (Stratagene). The sequences of all constructs were verified by DNA sequencing.
Cell culture and transfections.
HEK293 cells were grown at 37 °C under a 5% CO2 atmosphere in DMEM with 10% (wt/vol) FBS (Biological Industries) and 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 0.25 µg/mL amphotericin B.
For rVMAT2 expression, cells grown in 10-cm plates to 40–50% confluence were transfected with 7 µg of plasmid DNA coding for wild-type or mutant rVMAT2, using the PEI transfection reagent (Sigma). After 40–48 h, cells were treated with trypsin and collected, centrifuged for 6 min at 800 × g at 4 °C, and washed twice with PBS. The cell pellet was frozen in liquid air and stored at −70 °C. For whole-cell assays, cells were grown in 12-well plates to 40–50% confluence and were transfected with 5.5 µL of DNA as described above. The transfected cells were assayed after 40–48 h.
Membrane preparation, solubilization, purification, and reconstitution.
For membrane preparation, frozen cells were thawed, resuspended in lysis buffer [0.3 M sucrose, 15 mM Hepes (pH 7.4), 5 mM MgCl2, 15 µg/mL DNase I (Sigma) and protease inhibitor mixture (Sigma)], and disrupted using a homogenizer. The membrane fraction was collected by ultracentrifugation for 1 h at 213,500 × g at 4 °C and was resuspended in the buffer above, divided into aliquots, frozen in liquid air, and stored at −70 °C. Membranes containing ∼5–10 µg/µL total membrane protein were solubilized, purified, and reconstituted essentially as described in ref. 61.
Western blot analysis.
Samples were analyzed essentially as previously described (61), except that for detection the blot was incubated with DyLight-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories) diluted 1:4,000 and was imaged using MF-chemibis 32 (DNR Bio-Imaging Systems). Protein amounts were quantified using Gel-Quant software (DNR Bio-Imaging Systems). For dot blot analysis, 5 µL of liposomes were spotted on PVDF membranes and then were treated as described above.
Binding of [3H]-TBZOH.
Membranes prepared as described above (1.5 µL) were added to 200 µL of buffer containing 0.3 M sucrose, 10 mM K-Hepes (pH 7.4), and increasing concentrations of [3H]-TBZOH (77 Ci/mmol) (Vitrax Radiochemicals) at room temperature. The reaction was stopped after 20 min by dilution in ice-cold buffer with 125 µM TBZ and filtered through grade GF/F glass filters (Whatman) presoaked with 125 µM TBZ. Nonspecific binding measured in the presence of 125 µM TBZ was subtracted from the total binding levels.
To assess inhibition by MPP+, membranes were incubated in the presence of various concentrations of MPP+ for 20 min before incubation with [3H]-TBZOH (5 nM). Data analysis for all the experiments was performed using Origin 8.1 software (OriginLab). All experiments were done in duplicate and were repeated at least twice (the lowest R-squared value is 0.95).
Uptake of [3H]-Serotonin.
Whole cells.
Transfected cells were washed once with uptake buffer containing 110 mM Na-K-tartarate, 5 mM glucose, 20 mM K-Hepes (pH 7.4), 0.2% (wt/vol) BSA, and 5 mM MgCl2. Cells were permeabilized for 10 min at 37 °C in uptake buffer containing 10 mM digitonin. The medium was removed and replaced with fresh buffer without digitonin containing 5 mM MgATP, 1 mM ascorbic acid, and the indicated concentration of the radiolabeled serotonin, usually 100 nM [3H]-serotonin (28 Ci/mmol) (Perkin-Elmer). Nonspecific accumulation of [3H]-serotonin was measured in the presence of 125 µM TBZ and was subtracted from the total transport. The reaction was stopped at given time periods by aspirating the reaction buffer, and the cells were washed twice with ice-cold uptake buffer containing 2 mM MgSO4 instead of MgCl2. The cells then were collected with 0.1% SDS, and radioactivity assessed by liquid scintillation.
Proteoliposomes.
Liposomes were thawed and sonicated to clarity in a bath-type sonicator. The uptake assay was performed in reaction buffer containing 140 mM K2-tartrate, 10 mM tricine, 10 mM Tris, and 5 mM MgCl2, at pH 8.5. Liposomes (1 µL) were diluted into 200 µL of reaction buffer with 50 nM valinomycin and the indicated concentrations of the radiolabeled serotonin, usually 100 nM [3H]-serotonin. Nonspecific accumulation of [3H]-serotonin was measured as above. The reaction was stopped at the indicated time points by dilution of the mixture in 2 mL ice-cold buffer and filtering on 0.22-µm GSWP filters (Millipore). Radioactivity in the liposomes was measured using liquid scintillation.
Supplementary Material
Acknowledgments
S.S. is the Mathilda Marks-Kennedy Professor of Biochemistry at the Hebrew University of Jerusalem. This work was supported by National Institutes of Health Grant NS16708 (to S.S.), the German Research Foundation Collaborative Research Center 807 “Transport and Communication across Biological Membranes,” and the Behrens-Weise Foundation (L.R.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220497110/-/DCSupplemental.
References
- 1.Eiden LE, Schäfer MK, Weihe E, Schütz B. The vesicular amine transporter family (SLC18): Amine/proton antiporters required for vesicular accumulation and regulated exocytotic secretion of monoamines and acetylcholine. Pflugers Arch. 2004;447(5):636–640. doi: 10.1007/s00424-003-1100-5. [DOI] [PubMed] [Google Scholar]
- 2.Schuldiner S, Shirvan A, Linial M. Vesicular neurotransmitter transporters: From bacteria to humans. Physiol Rev. 1995;75(2):369–392. doi: 10.1152/physrev.1995.75.2.369. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhry FA, Edwards RH, Fonnum F. Vesicular neurotransmitter transporters as targets for endogenous and exogenous toxic substances. Annu Rev Pharmacol Toxicol. 2008;48:277–301. doi: 10.1146/annurev.pharmtox.46.120604.141146. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301(5633):616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 5.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30(2):417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 7.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467(7316):734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 8.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312(5774):741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490(7420):361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 10.Radestock S, Forrest LR. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J Mol Biol. 2011;407(5):698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 12.Forrest LR, Rudnick G. The rocking bundle: A mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50(45):9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrest LR, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA. 2008;105(30):10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crisman TJ, Qu S, Kanner BI, Forrest LR. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc Natl Acad Sci USA. 2009;106(49):20752–20757. doi: 10.1073/pnas.0908570106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vardy E, Arkin IT, Gottschalk KE, Kaback HR, Schuldiner S. Structural conservation in the major facilitator superfamily as revealed by comparative modeling. Protein Sci. 2004;13(7):1832–1840. doi: 10.1110/ps.04657704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merickel A, Kaback HR, Edwards RH. Charged residues in transmembrane domains II and XI of a vesicular monoamine transporter form a charge pair that promotes high affinity substrate recognition. J Biol Chem. 1997;272(9):5403–5408. doi: 10.1074/jbc.272.9.5403. [DOI] [PubMed] [Google Scholar]
- 18.Steiner-Mordoch S, Shirvan A, Schuldiner S. Modification of the pH profile and tetrabenazine sensitivity of rat VMAT1 by replacement of aspartate 404 with glutamate. J Biol Chem. 1996;271(22):13048–13054. doi: 10.1074/jbc.271.22.13048. [DOI] [PubMed] [Google Scholar]
- 19.Kim MH, Lu M, Kelly M, Hersh LB. Mutational analysis of basic residues in the rat vesicular acetylcholine transporter. Identification of a transmembrane ion pair and evidence that histidine is not involved in proton translocation. J Biol Chem. 2000;275(9):6175–6180. doi: 10.1074/jbc.275.9.6175. [DOI] [PubMed] [Google Scholar]
- 20.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60(4):575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vardy E, Steiner-Mordoch S, Schuldiner S. Characterization of bacterial drug antiporters homologous to mammalian neurotransmitter transporters. J Bacteriol. 2005;187(21):7518–7525. doi: 10.1128/JB.187.21.7518-7525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwaki S, Tamura N, Kimura-Someya T, Nada S, Yamaguchi A. Cysteine-scanning mutagenesis of transmembrane segments 4 and 5 of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a permeability barrier in the middle of a transmembrane water-filled channel. J Biol Chem. 2000;275(30):22704–22712. doi: 10.1074/jbc.m910354199. [DOI] [PubMed] [Google Scholar]
- 23.Varela MF, Sansom CE, Griffith JK. Mutational analysis and molecular modelling of an amino acid sequence motif conserved in antiporters but not symporters in a transporter superfamily. Mol Membr Biol. 1995;12(4):313–319. doi: 10.3109/09687689509072433. [DOI] [PubMed] [Google Scholar]
- 24.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Interhelical packing modulates conformational flexibility in the lactose permease of Escherichia coli. Biochemistry. 2005;44(21):7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 25.Kim MH, Lu M, Lim EJ, Chai YG, Hersh LB. Mutational analysis of aspartate residues in the transmembrane regions and cytoplasmic loops of rat vesicular acetylcholine transporter. J Biol Chem. 1999;274(2):673–680. doi: 10.1074/jbc.274.2.673. [DOI] [PubMed] [Google Scholar]
- 26.Rudnick G, Steiner-Mordoch SS, Fishkes H, Stern-Bach Y, Schuldiner S. Energetics of reserpine binding and occlusion by the chromaffin granule biogenic amine transporter. Biochemistry. 1990;29(3):603–608. doi: 10.1021/bi00455a002. [DOI] [PubMed] [Google Scholar]
- 27.Bailey J, Manoil C. Missense mutations that inactivate Escherichia coli lac permease. J Mol Biol. 1998;277(2):199–213. doi: 10.1006/jmbi.1998.1627. [DOI] [PubMed] [Google Scholar]
- 28.Kaback HR. Structure and mechanism of the lactose permease. C R Biol. 2005;328(6):557–567. doi: 10.1016/j.crvi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Adam Y, Tayer N, Rotem D, Schreiber G, Schuldiner S. The fast release of sticky protons: Kinetics of substrate binding and proton release in a multidrug transporter. Proc Natl Acad Sci USA. 2007;104(46):17989–17994. doi: 10.1073/pnas.0704425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794(5):748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Schaedler TA, van Veen HW. A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J. 2010;24(10):3653–3661. doi: 10.1096/fj.10-156927. [DOI] [PubMed] [Google Scholar]
- 32.Tirosh O, et al. Manipulating the drug/proton antiport stoichiometry of the secondary multidrug transporter MdfA. Proc Natl Acad Sci USA. 2012;109(31):12473–12478. doi: 10.1073/pnas.1203632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khare P, Ojeda AM, Chandrasekaran A, Parsons SM. Possible important pair of acidic residues in vesicular acetylcholine transporter. Biochemistry. 2010;49(14):3049–3059. doi: 10.1021/bi901953j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruivo R, et al. Mechanism of proton/substrate coupling in the heptahelical lysosomal transporter cystinosin. Proc Natl Acad Sci USA. 2012;109(5):E210–E217. doi: 10.1073/pnas.1115581109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newstead S. Towards a structural understanding of drug and peptide transport within the proton-dependent oligopeptide transporter (POT) family. Biochem Soc Trans. 2011;39(5):1353–1358. doi: 10.1042/BST0391353. [DOI] [PubMed] [Google Scholar]
- 36.Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc Natl Acad Sci USA. 2008;105(26):8896–8901. doi: 10.1073/pnas.0803577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudker O, Verdon G. Structural perspectives on secondary active transporters. Trends Pharmacol Sci. 2010;31(9):418–426. doi: 10.1016/j.tips.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He MM, Kaback HR. Interaction between residues Glu269 (helix VIII) and His322 (helix X) of the lactose permease of Escherichia coli is essential for substrate binding. Biochemistry. 1997;36(44):13688–13692. doi: 10.1021/bi9715324. [DOI] [PubMed] [Google Scholar]
- 39.Ye L, Maloney PC. Structure/function relationships in OxlT, the oxalate/formate antiporter of Oxalobacter formigenes: Assignment of transmembrane helix 2 to the translocation pathway. J Biol Chem. 2002;277(23):20372–20378. doi: 10.1074/jbc.M111140200. [DOI] [PubMed] [Google Scholar]
- 40.Kim YM, Ye L, Maloney PC. Helix proximity in OxlT, the oalate:formate antiporter of oxalobacter formigenes. Cross-linking between TM2 and TM11. J Biol Chem. 2001;276(39):36681–36686. doi: 10.1074/jbc.M106079200. [DOI] [PubMed] [Google Scholar]
- 41.Franco PJ, Jena AB, Wilson TH. Physiological evidence for an interaction between helices II and XI in the melibiose carrier of Escherichia coli. Biochim Biophys Acta. 2001;1510(1-2):231–242. doi: 10.1016/s0005-2736(00)00353-9. [DOI] [PubMed] [Google Scholar]
- 42.Hirai T, Heymann JAW, Maloney PC, Subramaniam S. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J Bacteriol. 2003;185(5):1712–1718. doi: 10.1128/JB.185.5.1712-1718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemieux MJ, Huang Y, Wang N. Crystal structure and mechanism of GlpT, the glycerol-3-phosphate transporter from E. coli. J Electron Microsc (Tokyo) 2005;54(Suppl 1):i43–i46. doi: 10.1093/jmicro/54.suppl_1.i43. [DOI] [PubMed] [Google Scholar]
- 44.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104(42):16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madej MG, Soro SN, Kaback HR. Apo-intermediate in the transport cycle of lactose permease (LacY) Proc Natl Acad Sci USA. 2012;109(44):E2970–E2978. doi: 10.1073/pnas.1211183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. FASEB J. 1998;12:A1326–A1326. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Li WZ, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 49.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: An algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl 1):S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 51.Glaser F, et al. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 52.Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91(2):508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 54.Kalman M, Ben-Tal N. Quality assessment of protein model-structures using evolutionary conservation. Bioinformatics. 2010;26(10):1299–1307. doi: 10.1093/bioinformatics/btq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 56.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 57.Brooks BR, et al. Charmm - A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 58.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285(4):1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 59.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 60.Elbaz Y, Danieli T, Kanner BI, Schuldiner S. Expression of neurotransmitter transporters for structural and biochemical studies. Protein Expr Purif. 2010;73(2):152–160. doi: 10.1016/j.pep.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adam Y, Edwards RH, Schuldiner S. Expression and function of the rat vesicular monoamine transporter 2. Am J Physiol Cell Physiol. 2008;294(4):C1004–C1011. doi: 10.1152/ajpcell.00348.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.