Significance
Lactococcal siphophages infect Lactococcus lactis, a Gram-positive bacterium used in commercial dairy fermentations. The phage TP901-1 baseplate (BP) recognizes and binds specifically to polysaccharides covering the host cell. We raised llama nanobodies against the BP as tools to dissect the molecular determinants of phage infection. Using complementary techniques, we identified BP binders and determined their affinity and epitopes for their targets and their impact on phage infectivity. X-ray structures revealed that two nanobodies block the BP saccharide binding site, and viral infection assays showed that they neutralize infection, a possible way to circumvent phages detrimental effect on dairy fermentation.
Abstract
Lactococcal phages belong to a large family of Siphoviridae and infect Lactococcus lactis, a Gram-positive bacterium used in commercial dairy fermentations. These phages are believed to recognize and bind specifically to pellicle polysaccharides covering the entire bacterium. The phage TP901-1 baseplate, located at the tip of the tail, harbors 18 trimeric receptor binding proteins (RBPs) promoting adhesion to a specific lactococcal strain. Phage TP901-1 adhesion does not require major conformational changes or Ca2+, which contrasts other lactococcal phages. Here, we produced and characterized llama nanobodies raised against the purified baseplate and the Tal protein of phage TP901-1 as tools to dissect the molecular determinants of phage TP901-1 infection. Using a set of complementary techniques, surface plasmon resonance, EM, and X-ray crystallography in a hybrid approach, we identified binders to the three components of the baseplate, analyzed their affinity for their targets, and determined their epitopes as well as their functional impact on TP901-1 phage infectivity. We determined the X-ray structures of three nanobodies in complex with the RBP. Two of them bind to the saccharide binding site of the RBP and are able to fully neutralize TP901-1 phage infectivity, even after 15 passages,. These results provide clear evidence for a practical use of nanobodies in circumventing lactococcal phages viral infection in dairy fermentation.
Tailed bacteriophages (Caudovirales order) typically possess a tail distal machinery used to recognize the host with high specificity as well as ensure genome delivery. The Caudovirales phylum encompasses three families: phages belonging to the Myoviridae family possess a contractile tail (e.g., T4) (1), and Podoviridae phages have a short tail (e.g., P22) (2, 3), whereas members of the Siphoviridae bear a long noncontractile tail such as HK97 (4), SPP1 (5), or TP901-1 (6–8).
The siphophages distal machinery can be described as a straight tail tip, a morphology identified in phages binding to a membrane protein as receptor, such as Escherichia coli phage-λ (9, 10), Bacillus subtilis phage SPP1 (5, 11, 12), or Lactococcus lactis phage C2 (13). Phages recognizing and binding to saccharidic receptors possess a massive organelle, the baseplate, carrying a large number (12 or more) of antireceptor proteins, also called receptor binding proteins (RBPs) (6, 14, 15). This setting probably provides avidity of multiple receptor binding events to compensate for the moderate affinity of a single saccharidic receptor for an individual RBP.
In recent years, we have reported on the structures of the RBPs and the baseplate of two phages, p2 and TP901-1, which infect the Gram+ bacterium L. lactis, and we have identified their different strategies used to ensure infection (6, 8, 14–19). Although Ca2+ ions trigger large conformational changes of phage p2 baseplate to orientate the RBPs to the host (15), the RBPs of phage TP901-1 baseplate point to the appropriate direction without the need for conformational change or Ca2+ requirement (6). These structural data were confirmed and extended to members of the 936 (p2) and P335 (TP901-1) lactococcal phages groups using in vivo infection experiments (6). We hypothesized that, as in the case of phage SPP1 (5, 20), host adhesion would generate a signal, which after propagation along the tail, would promote opening of the portal complex and ejection of the dsDNA genome out of the capsid. The origin of such a signal should reside in the baseplate, because it is in direct contact with the host cell envelope. Although the large conformational change of p2 baseplate is an obvious candidate for initiating such a signal, the case of TP901-1 is less straightforward. We have proposed that subtle conformational changes occur on binding of a TP901-1 tripod [a complex of three RBP trimers linked to an upper baseplate protein (BppU) holder] (6) to the host and thus, may generate the initial signal. Here, we designed an approach, based on the use of camelid antibodies fragments, nanobodies, or variable domain of heavy-chain antibody (vHH) (21, 22), to explore this hypothesis. These nanobodies have been found to be able to neutralize phages and viruses (23–25) by blocking their RBDs (14, 15, 26).
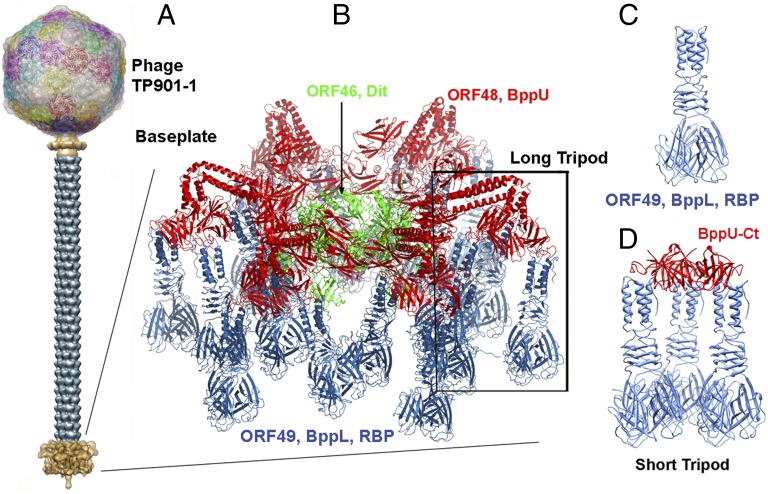
The TP901-1 baseplate is composed of four proteins: the two proteins aligned along the phage axis, Dit (distal tail) and Tal (tail-associated lysozyme), and the two peripheral proteins, BppU and lower Bpp (BppL) (the RBP) (8). The TP901-1 complete baseplate comprises 6 Dit, 3 Tal, 18 BppU, and 54 BppL. We have expressed a baseplate of TP901-1 (BPTP901-1) containing all of the baseplate components except Tal (i.e., a complex of 78 proteins and 1.8 MDa) (6, 17) (Fig. 1). We have also expressed and purified subcomponents of the baseplate: Dit alone as a monomer, the trimer of BppL (the RBP) (Fig. 1) (16, 27), a complex of three BppU and nine BppL/RBP (the long tripod) (6, 17), and the trimer of a BppU C-terminal fragment (encompassing residues 195–299 of the full-length BppU) in complex with nine RBPs/BppL (the short tripod) (Fig. 1) (6). We also cloned and purified Tal N terminus (1–386) as a monomer.
Fig. 1.
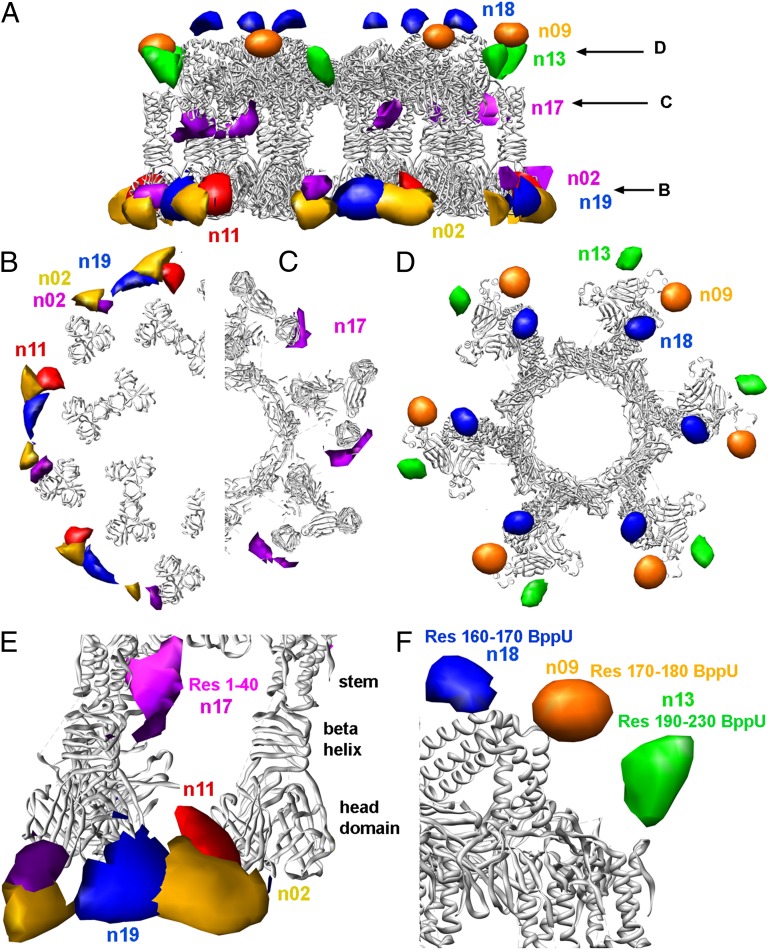
Phage TP901-1, its baseplate, and the baseplate components used in this study. (A) The complete phage TP901-1 (8) and the baseplate (brown) at the tip of the tail. (B) The baseplate and the long tripod. (C) The BppL/RBP trimer. (D) The short tripod (three BppU-Ct + nine BppL/RBP).
In this report, we generated camelid nanobodies by immunizing a llama with BPTP901-1, produced these nanobodies to determine their affinity for the baseplate and localize their epitopes, and analyzed their in vivo effect on L. lactis infection by TP901-1. Four distinct nanobodies were shown to bind to the RBP (BppL), of which three bind to the C-terminal head domain carrying the receptor binding site and one binds to the N-terminal stem domain. Two head-binding nanobodies neutralize infection, even after 15 passages, whereas the remaining nanobody has no effect on the infection efficiency. None of the nanobodies binding to BppU, connecting the RBPs to the core of BPTP901-1, had any effect on infection, whereas those nanobodies binding to the N-terminal domain of Tal, the tail’s most distal protein, modulate infection and provide up to 25% neutralization.
Results
Nanobodies Selection and Production.
Lymphocytes were isolated from blood samples of an immunized llama, and a nanobody phage display library was generated (28). Specific nanobodies were selected by panning (29). Nine BPTP901-1–specific binders were selected and sequenced (Fig. S1) before being subcloned into the pHEN6 expression vector (28). For the Tal N terminus, we obtained four different nanobodies, of which two could be expressed and thus, were further characterized (Fig. S1).
Affinity of the Different Nanobodies for the Baseplate Components.
Using surface plasmon resonance (SPR), we analyzed the affinity of nine nanobodies for the baseplate and three nanobodies for Tal, all selected among nanobodies characterized by ELISA (Materials and Methods) on the basis of their apparent affinities. We devised a strategy to determine the specificity of the expressed nanobodies by coupling the different nanobodies to a CMD500M sensor chip followed by successive injection of the flow cell with four molecular entities: the RBP, the short tripod, the long tripod, and BPTP901-1 (Fig. S1). In the case of the anti-Tal nanobodies, we injected Tal monomer. Distinction of nanobody specificity (other than nanobodies that were specific for Tal) was performed based on the following rationale. A nanobody binding to four injected targets (i.e., RBP, short tripod, long tripod, and BPTP901-1) should be BppL-specific; a nanobody not binding to BppL but binding to the three other targets should be specific of the BppU C terminus. A nanobody binding only to the long tripod and the baseplate should be specific of BppU but not its C terminus (residues 195–299), and finally, a nanobody binding only to the baseplate should be specific of Dit. Indeed, it cannot be excluded that nanobodies identified as binding to a given ORF may be located at the interface between that ORF and a neighboring one.
We observed that nanobodies 2, 11, 17, and 19 (Table 1) bind to the four tested molecular species and hence, should be ascribed as RBP/BppL binders. Their KD values to the RBP/BppL were measured at ∼15 ± 5 nM, except for nanobody 17, which exhibits a lower affinity (KD of 35 nM). Nanobodies 13 and 18 do not bind to RBP/BppL but bind to the three other molecular species; therefore, they are assigned as specific binders of the C terminus of BppU (Table 1). Their KD values are lower than those values of the BppL binders (in the ∼60–90 nM range for the long tripod and the 20–50 nM range for the baseplate) (Table 1). Noteworthy, nanobody 18 epitope probably encompasses the junction between BppU N and C termini, because it exhibits a KD value for the short tripod approximately fourfold larger than for the long one (Table 1). Nanobody 13, in contrast, binds only to the long tripod and the baseplate, indicating that its epitope does not overlap with the C terminus of BppU. Nanobody 9 exhibits a very low affinity for the short tripod and therefore, seems to bind essentially to the N terminus of BppU, with a stoichiometry of six nanobodies per baseplate. Finally, nanobodies 3 and 7 bind only to the baseplate, and hence, they are counted as Dit binders. Their affinities for the baseplate are much lower than the affinities measured for the other nanobodies (around 300 ± 130 nM), and they were not further characterized. The affinity measurements of the two nanobodies, T11 and T41, for the N-terminal domain of Tal yielded KD values of 32 and 30 nM, respectively (Table 1).
Table 1.
Analysis by SPR of the interactions between nanobodies and phage TP901-1–expressed components
| Baseplate | BppL | Tripod short | Tripod long | Epitope | |
| KD values (nM) BPTP901-1 nanobodies | |||||
| Nanobody 2 | 10 ± 2 | 15 ± 3 | 13 ± 3 | 10 ± 4 | BppL |
| Nanobody 11 | 19 ± 3 | 13 ± 1 | 20 ± 2 | 19 ± 3 | BppL |
| Nanobody 17 | 300 ± 10 | 35 ± 3 | Nd | 20.6 ± 1 | BppL |
| Nanobody 19 | 12 ± 2 | 12 ± 2 | 15 ± 2 | 7 ± 2 | BppL |
| Nanobody 9 | 29 ± 3 | — | 1,000 ± 500 | 120 ± 40 | BppU Nt (+Ct) |
| Nanobody 13 | 48 ± 8 | — | 64 ± 7 | 92 ± 11 | BppU Ct |
| Nanobody 18 | 22 ± 2 | — | 280 ± 40 | 65 ± 9 | BppU Nt (+Ct) |
| Nanobody 3 | 420 ± 50 | — | — | — | Dit |
| Nanobody 7 | 170 ± 15 | — | — | — | Dit |
| KD values (nM) Tal-Nt nanobodies | Isolated Tal-Nt 1–386 | ||||
| Nanobody T11 | 32 ± 2 | ||||
| Nanobody T41 | 30 ± 2 |
The measurements have been performed in triplicate. Nd, not determined.
Affinity of the Different Nanobodies for the TP901-1 Virion.
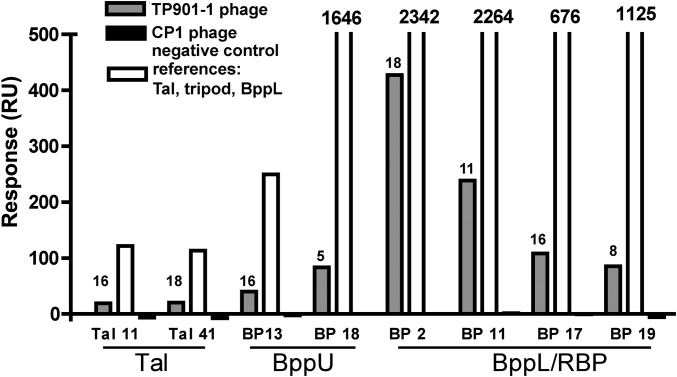
Using the same coupled sensor chips as above, we injected intact, purified TP901-1 virion in the flow cell (8) and measured the response unit in triplicate (Fig. 2). As a negative control, we injected another unrelated phage (phage Cp-1) (30) in the flow cell. We observed that anti-BppL nanobodies 2, 11, 17, and 19 bind efficiently to the TP901-1 virion, although with a lower signal than for the molecular reference, whereas as expected, no binding was observed for the unrelated phage (Fig. 2). Similarly, anti-BppU nanobodies 13 and 18 also bind efficiently to TP901-1 but failed to do so to the negative control. Interestingly, anti-Tal nanobodies T11 and T41, which could not be assayed in the context of a complete baseplate, generated a positive response with TP901-1 virion (Fig. 2).
Fig. 2.
SPR analysis of the interactions between nanobodies and TP901-1 virion or phage components. Binding (expressed in response units) of phage TP901-1 virion to the immobilized nanobodies (gray). The positive controls (white) are performed with Tal-Nt, the long tripod, and Bpp for the nanobodies against Tal-Nt, BppU, or BppL as indicated. The negative controls (black) are performed with phage Cp-1.
Epitope Mapping of the Different Nanobodies on the Baseplate Components.
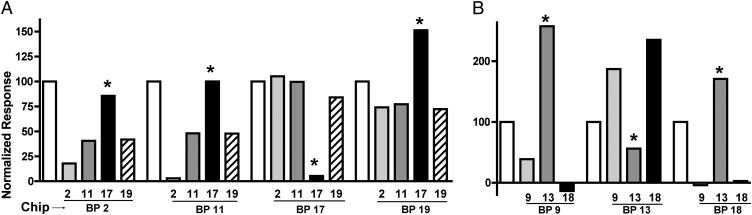
We subsequently wished to determine more precisely whether the epitopes of the different nanobodies on their molecular target were overlapping. To this end, we injected BppL alone over an anti-BppL nanobody (the reference) followed successively by the injection of complexes of BppL with its four specific nanobodies (Fig. 3A). We expected to obtain a decrease of the SPR signal with the overlapping nanobodies compared with the reference and a conservation or increase of the signal if the epitopes were distinct. In this manner, we observed that nanobodies 2, 11, and 19 recognize the same or an overlapping epitope, because the signal decreased; however, nanobody 17 recognizes an epitope distinct from the three other nanobodies, because the produced signal increased (Fig. 3A). Conversely and consistent with the above observation, when nanobody 17 was bound to the chip, the signal of the other nanobodies was not different from the signal obtained for the positive control (Fig. 3A).
Fig. 3.
Epitope mapping of the anti-BPTP901-1 nanobodies performed by SPR. (A) Anti-BppL nanobodies. (B) Anti-BppU nanobodies. (A) BppL and (B) BppU were injected over the immobilized nanobodies (Chip) as positive controls (white) and have been normalized to 100. Then, (A) BppL and (B) BppU were injected in presence of the different anti-BppL and anti-BppU nanobodies, and the response signal was normalized to the positive control. *Nanobodies presenting a response pattern dissimilar to the other patterns.
We then used the same strategy of epitope mapping with the BppU binders using the same rationale (Fig. 3B) and observed that the epitope of nanobody 13 does not overlap with nanobody 9 (increased signal); this finding is consistent with the finding presented above that nanobody 13 binds exclusively to the C terminus of BppU, whereas nanobody 9 binds essentially to the BppU N terminus. Noteworthy, the epitope of nanobody 13 does not overlap also with the epitope of nanobody 18 (increased signal), although the latter seems to recognize (part of) the BppU C terminus (preceding paragraph and Table 1). In contrast, the epitopes of nanobodies 9 and 18 overlap (decreased signal), indicating that nanobody 9 binds rather close to nanobody 18 and confirming that both bind close to the BppU N terminus/C terminus (Nt/Ct) junction area. Therefore, the epitopes of the three anti-BppU nanobodies seem to cluster at—or near—the BppU C terminus domain. Finally, epitope mapping of the Tal nanobodies T11 and T41 revealed that both nanobodies bind to the same epitope of the monomeric molecule.
Specificity of the Nanobodies for TP901-1 Compared with Tuc2009 Baseplate.
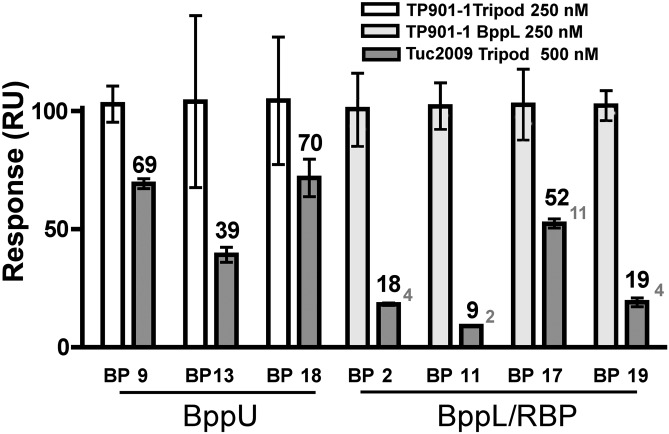
We then examined the specificity of our nanobodies for the phage TP901-1 and its components compared with the closely related phage Tuc2009 (31–33). Both phages share overall ∼96% sequence identity but not distributed evenly. BppU N terminus is similar between the two phages (98% identity), whereas similarity drops sharply at the C terminus. In contrast, the N termini of the RBPs share significant similarity (residues 1–60), whereas the C termini are entirely different.
We used the same nanobodies coupled to a sensor chip and injected successively the baseplate components of TP901-1 and Tuc2009. The response units (RUs) measured for nanobodies 2, 11, and 19 with Tuc2009 RBP are only 2–5% of RUs measured with TP901-1 (Fig. 4), thus indicating a high specificity of these nanobodies for TP901-1 components. Noteworthy, nanobody 17 generates a slightly higher RU, indicating some significant binding to the Tuc2009 RBP/BppL. We, therefore, determined the KD of nanobody 17 for the Tuc2009 long tripod and found a KD value of 3 μM, ∼100 times higher than for TP901-1. These data suggest that nanobody 17 binds to the more conserved BppL C terminus, whereas the other nanobodies may bind the C-terminal head domain (the receptor binding domain) (6, 16). In contrast, affinity of the anti-BppU nanobodies 9 and 18 for the Tuc2009 long tripod is comparable with affinity for TP901-1 counterparts (Fig. 4), which is in agreement with the conserved sequences of the binding area. Notably, the affinity of nanobody 13 for Tuc2009 tripod is slightly lower than the affinity obtained for TP901-1 tripod, consistent with its recognition of the less-conserved BppU C-terminal domain (Fig. 4).
Fig. 4.
Specificity of nanobodies to expressed phage components of TP901-1 vs. Tuc2009 analyzed by SPR. TP901-1 tripod and BppL were injected as a control (white) over the immobilized anti-BppU (Left) and anti-BppL (Right) nanobodies, and the response signal was normalized to 100. Tuc2009 tripod was injected over the immobilized nanobodies (gray), and the response was normalized to the positive control (black letters). For clarity, the response of Tuc2009 tripod vs. anti-BppL nanobodies is rationalized to the molecular weight (gray letters).
EM Reconstructions of the Nanobodies–BPTP901-1 Complexes.
We investigated the baseplate–nanobody complexes by negative staining EM to confirm our epitope mapping analysis and obtain the exact positions and stoichiometries of these complexes. To this end, we generated complexes of the TP901-1–purified baseplate, with one or even two nanobodies concomitantly. We first analyzed the baseplate alone and observed that it appears as a monomeric species in the EM reconstruction. Then, we generated complexes of the purified TP901-1 baseplate with one or two nanobodies simultaneously.
Adding nanobodies to the baseplate led to its dimerization, except for in nanobody 9 (Fig. S2). Dimerization occurs with the BppL head domains facing each other, except for nanobodies 7 + 13 (for which baseplate Dit rings are stacked against each other) (Fig. S2). We observed that three anti-RBP/BppL nanobodies (nanobodies 2, 11, and 19) are bound to the head domain. Nanobodies 2 and 11 were shown to exhibit a stoichiometry of 12 monomers per baseplate, whereas nanobody 19 is present as 6 monomers per baseplate (Fig. 5 A, B, and E). Careful inspection of the bound nanobody location seems to indicate that nanobodies 2 and 11 are almost totally overlapping, whereas nanobody 19 is bound at a slightly different position. In contrast and as suggested by the SPR experiments, nanobody 17 binds to the RBP/BppL stem domain (Fig. 5 A, C, and E). Its stoichiometry was shown to be six nanobodies per baseplate. As expected, nanobody 13 binds to the most external BppU C-terminal domain (residues 190–230), with six nanobodies bound per baseplate (Fig. 5 A, D, and F). Probably, the two other BppU C-terminal binding sites are hidden, because they are covered by the BppU helices joining the tripod to the baseplate central core. In contrast, nanobody 18 binds to the upper BppU helix (residues 160–170), joining the tripod to the baseplate central core not far from the C terminus, which was suggested by the SPR experiments (Fig. 5 A, D, and F). Here again, there are clearly six nanobodies bound per baseplate, because this exposed epitope is only represented six times (Fig. 5 A and D). Nanobody 9 binds the junction hinge of BppU (residues 170–180) not far from nanobody 18 (Fig. 5 A, D, and F), which is in agreement with the epitope mapping experiments (Fig. 3).
Fig. 5.
Negative staining single-particle EM analysis of the binding of the various nanobodies to the BPTP901-1. (A) The ribbon view of the BPTP901-1 X-ray structure (6) and the difference maps calculated for nanobodies 2, 11, 17, and 19 (anti-BppL) and nanobodies 9, 13, and 18 (anti-BppU). (B) Section observed at the level of the BppU/BppL junction. (C) Section observed at the level of the BppL head domain. (D) Section observed at the level of the BppL stem domain. (E) Close-up view of the BppU/BppL junction. (F) Close-up view of the BppL area.
Crystal Structure Determination of the RBP in Complex with Nanobodies.
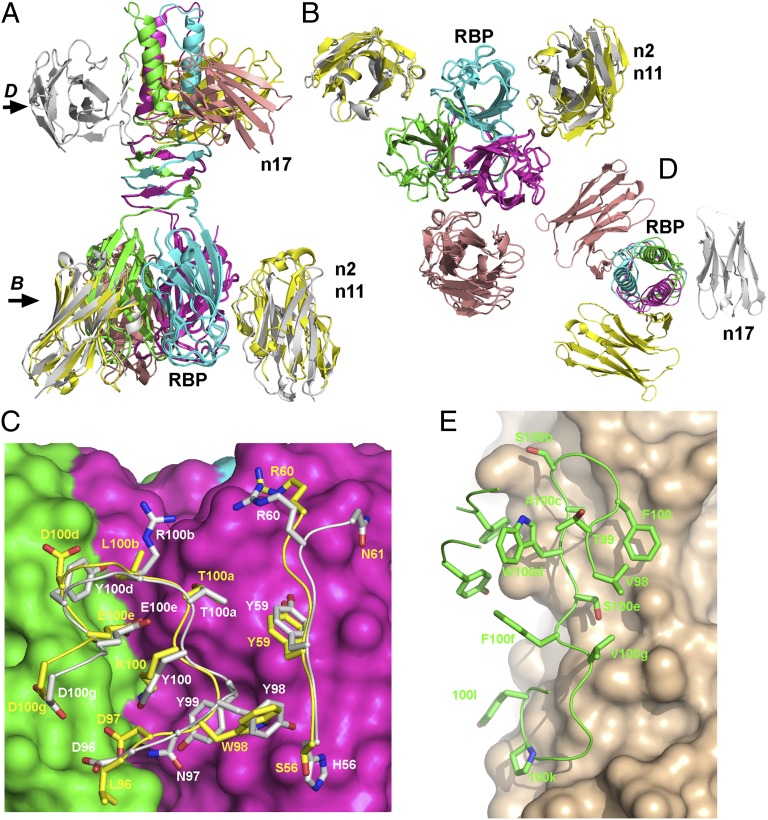
With a view to zoom in the structure of the nanobodies with their binding partner, we successively crystallized nanobodies 2, 11, and 17 with the RBP/BppL, and we solved the complex structures by molecular replacement (Table 2). Each complex counts three nanobodies bound to an RBP trimer. As previously seen (14, 15, 26, 27), the RBP has been cleaved serendipitously by proteases during crystallization, leading to missing segments before residue ∼30 for the nanobody 2/BppL complex and before residue 63 for the nanobody 11/BppL complex (Fig. S3 A–C). The nanobodies were observed to be in contact with the head domain, obstructing the putative saccharidic receptor binding site (14, 16, 26, 27). The BppL was found to be intact in the complex with nanobody 17, which binds to the stem domain of the RBP and probably masks the cleavage zone to proteases.
Table 2.
Data collection and refinement statistics (highest resolution value is in bold)
| ORF 49-n02 | ORF 49-n17 | ORF 49-n11 | |
| Data collection | |||
| Protein Data Bank ID code | 4HEM | 4HEP | 4IOS |
| Source | Soleil Proxima 1 | Soleil Proxima 1 | Soleil Proxima 1 |
| Space group; cell dimensions | P212121; 71.0, 104.2, 122.2 | H32; 74.4, 74.4, 375.9 | H3; 148.3, 148.3, 104.6 |
| Resolution limits* (Å) | 50.0–1.65 (1.69–1.65) | 50.0–1.75 (1.80–1.75) | 50.0–2.40 (2.40–2.54) |
| Rmerge* (%) | 5.4 (47.0) | 2.9 (43.6) | 8.6 (53.4) |
| No. of observations* | 1,401,086 (104,352) | 315,367 (23,537) | 226,472 (36,310) |
| No. of unique reflections* | 108,049 (7,798) | 41,184 (2,999) | 33,444 (5,342) |
| Mean (SD)* | 25.9 (5.7) | 33.5 (4.4) | 15.5 (2.25) |
| Completeness* (%) | 98.7 (97.5) | 99.9 (100.0) | 99.8 (99.7) |
| Multiplicity* | 13.0 (13.4) | 7.7 (7.8) | 6.3 (6.15) |
| Refinement | |||
| Resolution* (Å) | 24.2–1.65 (1.69–1.65) | 50.0–1.75 (1.80–1.75) | 22.1–2.40 (2.47–2.40) |
| No. of reflections* | 108,009 (7,782) | 41,168 (2,873) | 33,442 (2,852) |
| No. of protein/water | 5828/937 | 2,169/325 | 6,299/253 |
| No. of test set reflections | 3,234 | 1659 | 1,672 |
| Rwork/Rfree* (%) | 16.8/17.7 (17.9/21.6) | 19.8/20.4 (22.60–22.1) | 20.1/22.7 (23.1/28.7) |
| rmsd bonds (Å)/angles (°) | 0.01/1.09 | 0.009/1.13 | 0.009/1.21 |
| B Wilson/B average | 25.5/31.6 | 36.5/42 | 56.9/54.3 |
| Ramachandran preferred/allowed/outliers (%) | 97.8/2.2/0 | 95.8/3.2/1 | 94.6/4.2/1.2 |
Numbers in parentheses refer to the highest resolution bin.
Nanobody 2 covers a surface of 740–850 Å2 of the RBP head domain at the interface of two monomers (Fig. 6 A and B), with a surface area of 500–520 Å2 for one monomer and 240–340 Å2 for the other monomer. The CDR2 (residues 57–62) covers 203 Å2 of the RBP first monomer at the periphery of the sugar binding site, whereas the complementarity determining region (CDR) 3 is projected inside the crevice and covers 300 Å2 of the first monomer and 252 Å2 of the second monomer. Noteworthy, the same residues of the CDR3 interact with both monomers, such as the four Tyr residues 98, 99, 100, and 100d, which account for one-half of the total buried surface area (342 Å2) (Fig. 6C). Nanobody 2 contacts the residues involved directly in saccharide binding, which are found in the X-ray structure of the TP901-1 RBP in complex with glycerol (27). It establishes strong hydrogen bonds with Arg-155 (Table S1) and van der Waals contacts with His-133, Asp-135, and Phe-145.
Fig. 6.
X-ray structures of the various complexes formed by BppL and nanobodies 2, 11, and 17. (A) Ribbon view of the superposition of the three structures of the complexes; (B) 90° view (compared with A) of the BppL complexes with nanobodies 2 and 11 sliced at the level of the head domains. (C) Detailed view of the interaction area between BppL head domains (pink and green surfaces) and CDR2 and -3 of nanobodies 2 (white sticks) and 11 (yellow sticks); (D) 90° view (compared with A) of the BppL complexes with nanobody 17 sliced at the level of the stem domains. (E) Detailed view of the interaction area between BppL stem and β-helix domains (beige surface) and CDR3 of nanobody 17 (green sticks). In C and E, numbering is according to the work by Kabat et al. (47)
Nanobody 11 exhibits a position very close to the position of nanobody 2 (Fig. 5 A and B), with an rmsd of 0.36 Å on the Cα atoms and a buried surface area of 845 Å2 (505 Å2 for one monomer and 340 Å2 for the second monomer). CDR2 and -3 are also involved in the interaction, covering a surface area of 205 and 640 Å2, respectively. It is worth mentioning that the three CDRs display the same residue length but have quite different sequences, with 25% and 35% sequence identity for CDR2 and -3 of nanobodies 2 and 11, respectively. The most striking example of such divergence is the replacement of the key CDR3 residues Tyr-98, -99, -100, and -100d by Trp, Gly, Lys, and Asp, respectively. However, the conformations of CDR2 and -3 between the two nanobodies are amazingly conserved (Fig. 6C). Nanobody 11 also establishes strong hydrogen bonds with Arg-155 (Table S1), an RBP key residue involved in saccharide bonding (27).
The X-ray structure of the complex nanobody 17/RBP confirms that, in contrast to nanobodies 2 and 11, it does not bind the head domain but does bind to the helical stem and β-helix neck regions, covering RBP residues 2–6, 19, 20, 23, 24, 27, 28, 30–34, and 38–45 (Fig. 6 A, C, and E). The total buried surface area of the RBP is 888 Å2 (676 Å2 for one monomer and 212 Å2 for the second monomer). Most of this surface (694 Å2, 572/122) and most of the contacts originate from the very long CDR3, which counts 22 vs. 14 residues for nanobodies 2 and 11 and forms an extended β-hairpin (residues 96–100h) parallel to the RBP stem helices (Fig. 6 D and E). A loop (residues 100i–100n) further completes the interaction by contacting the two first turns of the β-helix of the neck (Fig. 6 A and E).
Infection Inhibition Potential of the Isolated Nanobodies.
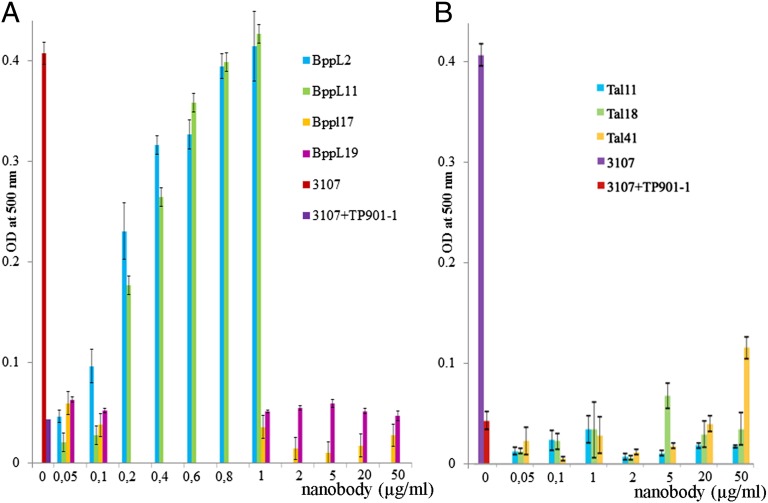
We previously assayed the neutralization potency of a nanobody raised against p2 phage (26, 27) and a designed ankyrin repeat protein (DARPIN) selected against TP901-1 RBP/BppL (16). Similar to this approach, we assayed all of the nanobodies studied here to check their potential neutralizing or modulating effect on infection by TP901-1 of its L. lactis host strain 3107 (Fig. 7A). Two of three nanobodies binding to the head domain, nanobodies 2 and 11, were shown to fully neutralize TP901-1 infection of L. lactis at concentrations of 1 μg/mL (70 nM) (Fig. 7). This full neutralization was maintained after 15 consecutive passages of phage exposure, indicating an extraordinary resistance to mutation escape compared with phage p2 (26). Furthermore, knowing the number of phages in solution, it was possible to evaluate in vivo KD for the virion’s BppL of nanobodies 2 and 11 at values of 20 and 46 nM, respectively, which are comparable with the values when measured in vitro. Surprisingly, nanobody 19 did not exert any neutralization effect, even at concentrations as high as 10 μg/mL (0.7 μM). Furthermore and as expected, nanobody 17, which binds the stem domain of RBP, was inefficient in neutralization. Also, we observed that the five other nanobodies, anti-Dit, and anti-BppU did not exhibit any neutralization of phage infection. We then examined the effect of the anti-Tal nanobodies on TP901-1 infectivity (Fig. 7B). We observed that only nanobody T41 exerts ∼25% neutralization at 50 µg/mL (3.0 µM), indicating some negative effect on phage infection fitness. Finally, neutralization assays of phage Tuc2009 by all nanobodies proved to be negative, including those assays involving nanobodies 2, 11, and 19.
Fig. 7.
Neutralization study of L. lactis strain 3107 infection by phage TP901-1 in the presence of nanobodies. (A) OD at 500 nm of the culture in the presence of 105 pfu TP9010-1 and BppL VHH2, -11, -17, and -19 across the concentration range of 0.05–50 µg/mL after 7 h coincubation at 30 °C. (B) The same conditions were applied with Tal VHH11, -18, and -41. Controls of uninfected and infected culture in the absence of nanobodies were included, and all results are the average of at least triplicate assays.
Discussion
Immunization by a large molecular assembly, such as the TP901-1 baseplate, proved to be an efficient technique to obtain nanobodies against the exposed surface of the various molecular components of the complex to evaluate the effect of binding on infectivity. Although the three components of the baseplate were targeted, BppU and BppL dominated the immunological response, with some epitopes being clearly favored. The head domain of BppL and its saccharide binding groove were targeted by two TP901-1–neutralizing—strongly binding—nanobodies. This groove was also found to be the epitope of a nanobody-neutralizing (VHH5) phage p2 (14, 15, 24, 26). This groove-directed mode of binding is a characteristic of nanobodies, because this mode has also been observed on several occasions for catalytic crevices of enzymes (22, 34, 35). Noteworthy, nanobodies are distinctly different from DARPINs, because this latter family of artificial binders binds preferentially to flat or convex surfaces. We observed this kind of interaction in a TP901-1 RBP/DARPIN complex, in which the DARPIN was observed as binding on top of the trimeric head domain (16). EM maps indicate that nanobody 19 binds the head domain close to but distinct from the binding positions of nanobodies 2 and 11. Because its X-ray structure is not available, we suggest that it binds to the more convex surface of a head domain monomer without reaching the saccharide binding crevice. This binding is observed at low resolution in the EM maps of its complex with the baseplate. A second favored epitope of TP901-1 baseplate is the hinge junction between the two BppU α-helices (residues 160–180). Nanobodies 9 and 18 bind in this very convex and exposed area, an observation that is in marked contrast with the most general behavior of nanobodies. In contrast, the C-terminal domain of BppU (residues 190–230), to which nanobody 13 binds, seems to be more concave. Finally, nanobody 17 exhibits a very interesting mode of binding on a rather flat area, deeply buried within the bush of RBPs/BppLs of the BPTP901-1. To exert this interaction, nanobody 17 possesses an extralong hairpin-like CDR3 positioned parallel to the RBP stem helices.
Strikingly, nanobodies 2 and 11 provide complete protection against TP901-1 infection, and this neutralization is maintained after 15 consecutive passages of exposing the lactococcal host to TP901-1 without being disrupted by so-called escape mutants. This finding is in sharp contrast with what was observed with phage p2, which could escape through the acquisition of mutations generated after three to five passages (26). The mechanism by which neutralization occurs is not as straightforward as it seems at first glance. The binding of only 12 nanobodies to the peripheral BppL head sites of the baseplate is not surprising because of steric hindrance. Indeed, the protrusion of the nanobodies CDR3 in these binding clefts does prevent the receptor (which is presumed to be the pellicle polysaccharide) from binding, because the affinity for the RBPs is in favor of the nanobodies compared with the saccharides. Surprising, however, is the observation that nanobodies binding to only 12 external RBP clefts (of a total of 54) are sufficient to neutralize infection, although 42 sites are still available for binding to pellicle polysaccharides. It is reasonable to postulate, however, that the peripheral sites of the baseplate are the most important for an initial interaction with the host, whereas binding of the most internal sites might occur only when the virion has been immobilized by this initial interaction. This mechanism might be similar to the two-step binding procedure often described for other tailed phages (36).
We and others have proposed that the signal triggering the portal complex (portal–connector–stopper) opening and the DNA release and injection might occur initially within the baseplate and might be transmitted by the tail tube components major tail protein (MTP) or tape measure protein (TMP) (5, 6). We suggested that the origin of the baseplate signal might be a large conformational change of the baseplate, which was observed in phage p2 (15) or myophage T4 (36), or might be the result of small structural perturbations resulting from baseplate binding stress (6). The present work did not identify a nanobody preventing these small oscillations, if it exists; however, nanobody 17 is a perfect candidate to block the tripod oscillations, and nanobodies 9 and 18 (not 13) are perfectly positioned to rigidify the hinge formed by BppU α-helices. In contrast, we observed that the anti-Tal nanobody T41 is able to reduce infectivity significantly but not totally. It has been shown that the Siphoviridae Tal N-terminal domain is in a closed position at rest and able to open to allow DNA passage (5, 12, 15). Indeed, binding of a nanobody to the confined space around Tal-Nt in the context of the baseplate (8, 18) should prevent its opening. However, the strong impact of DNA on ejection might detach Tal from the baseplate, a result suggested in the case of phage SPP1 (5, 12), and hence, counteract the effect of T41 nanobody binding. Noteworthy, it was found that treating Tuc2009 with anti-Tal polyclonal antibodies also caused reduction of infection efficacy (37).
We showed here that llama immunization by TP901-1 baseplate gave rise to many potent nanobody binders, targeting in a privileged manner the RBP binding cleft, although nanobodies recognizing other RBP regions or other baseplate gene products have been isolated. Among the characterized nanobodies, several could neutralize phage infection without allowing the generation of phage escape mutants. These results provide, therefore, evident opportunities for exploring the practical use of nanobodies to circumvent viral infection in dairy fermentations.
Materials and Methods
Llama Immunization and Nanobody Library Construction.
A llama (Llama glama) was immunized with the purified BPTP901-1 and Tal N-terminal domain (encompassing residues 1–386) (Capralogics INC). Lymphocytes were isolated from blood samples, and a nanobody phage display library was generated using standard procedures (28). The nanobody library contained about 3 × 108 independent transformants, of which 95% of the clones harbored an insert of expected nanobody gene size as determined by PCR on individual colonies. Selection of specific binders from the library was done by phage display according to standard procedures (29). A clear enrichment of antigen-specific clones was observed after three consecutive rounds of selection on solid phase-coated antigen; 48 for the baseplate and 96 for Tal-Nt randomly chosen colonies—after the third round—were grown for expression of their specific nanobody as soluble protein. Of the crude periplasmic extracts tested in an ELISA, 44 were shown to be specific to the TP901-1 baseplate, and 14 were shown to be specific to Tal N terminus. The sequence analysis of positive clones showed nine and four different clones, respectively, being subcloned into pHEN6 expression vector. Nanobodies expression and purification were performed as described (28).
SPR.
The SPR binding studies were performed using a BIAcore T-200 Instrument (GE Healthcare). The carboxymethyldextran hydrogel sensor chip was purchased from XanTech Bioanalytics GmbH. The buffer (10 mM Hepes, pH 7.4, 150 m M NaCl, 0.005% vol/vol Surfactant P20) and the amine coupling kit were purchased from GE Healthcare. The sensor chip was first equilibrated in running buffer and then activated with a 1:1 mixture of 0.1 M N-ethyl-N′-(dimethylamino-propyl) carbodiimide and 0.1 M N-hydroxysuccinimide. The different nanobodies were injected at a concentration of 100 μg/mL in running buffer. Remaining N-hydroxysuccinimide esters were deactivated with a pulse of 1 M ethanolamine, pH 8.5. On average, 500 RUs of the nanobodies were immobilized. The assayed proteins or complexes were diluted at five concentrations in the running buffer and analyzed by passing them across the immobilized nanobodies. Virions were injected as a concentration of 2–3.108 pfu/mL. For epitope mapping experiments, proteins and nanobodies were injected at concentrations of 250 nM and 2 μM, respectively. The ligand surface was regenerated with 10 mM HCl. Measurements were performed in triplicate. The sensorgrams were processed using the double referencing method (38). First, the binding response from the reference surface was subtracted from the binding response of the surface containing the immobilized nanobody to correct for bulk refractive index changes. Second, the response from an average of the blank injections was subtracted to remove any systematic artifacts observed between the reaction and the reference flow cell. The KD, Bmax, and R2 values were obtained using GraphPad Prism version 4.00 for Macintosh (GraphPad Software).
EM.
The complexes were prepared by mixing an excess of nanobody with BPTP901-1. The samples were filtered to remove nanobody excess and incubated on glow-discharged carbon grids at a concentration of 0.05 mg/mL. After blotting away excess, buffer grids were stained with 2% (wt/wt) uranyl acetate. Approximately 100 and 50 images were collected for samples of the baseplate incubated with the different nanobodies and the baseplate alone as a control, respectively, on a Tecnai Spirit Microscope operated at 120 kV at 45,000× magnification, resulting in a pixel size of 4.9 Å/pixel. On average, ∼3,000 particles were extracted from the collected images for each sample using the program boxer from the EMAN2 package (39). Particles were pretreated using the SPIDER package (40) and submitted to maximum likelihood classification and alignment (41) using the Xmipp Package (42). For every dataset, an initial model was built from a side-view average, imposing D6 or C6 symmetry in the case of nanobody 9 and the control baseplate. The initial models were refined by maximum likelihood refinement first and spider later across the entire dataset, with a sampling rate of 5°. Difference maps were calculated using Chimera against control models built by fitting two single control baseplates into the dimeric structures obtained for the baseplate and the nanobodies.
Crystallography.
The BppL/RBPs were expressed and purified as described previously (16). Crystals were grown by hanging-drop vapor diffusion; datasets were collected at the synchrotron Soleil (PROXIMA-1), and data were treated by XDS and XSCALE (43). The structure was initially solved using molecular replacement with the BppL and nanobodies structures (16, 35) using Molrep (44). Refinement was performed with AutoBUSTER (45) and model (re)construction by COOT (46).
Phage Infectivity.
bromcresol purple (BCP) broth (3 mL) was supplemented with 10 mM calcium chloride, and to this mixture, 105 pfu TP901-1 were added. Initially, all nanobodies were assessed at the levels of 0.05, 0.1, and 1.0 µg/mL for neutralizing effects. The neutralization curve was prepared at nanobody concentrations in the range of 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, and 1.0 µg/mL. Where weak or no neutralization was observed under standard conditions, 2, 5, 20, and 50 µg/mL relevant nanobodies were also tested. Controls were included to validate the assay, whereby the host alone (L. lactis 3107) or the host and its infecting phage (TP901-1) were applied to show growth and phage infection, respectively. The mixtures were incubated at 30 °C for 1 h, after which time 45 µL L. lactis 3107 were added; incubation at 30 °C was continued for another 7 h. Color change from purple to yellow indicated acidification and therefore, growth of the host bacteria, whereas a purple color indicated phage infection. OD changes at 500 nm were also monitored to allow the detection of more subtle changes where a weak neutralizing effect was observed. For the two nanobodies that fully neutralized TP901-1 infection, we attempted to generate escape mutant of TP901-1. To this end, the neutralization assay was repeated 15 times using the phage in the clarified supernatant of the previous assay in the subsequent assay. Between each round of the assay, the phage titer was determined to ensure that consistent levels of phage (105 pfu) were applied in each successive round of the neutralization assay.
Supplementary Material
Acknowledgments
Molecular graphics images were produced using the University of California at San Francisco Chimera Package from the Resource for Biocomputing, Visualization, and Informatics at the University of California at San Francisco (supported by National Institutes of Health Grant P41 RR-01081). This work was supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05-01, and, in part, by Agence Nationale de la Recherche Grant ANR-11-BSV8-004-01 (“Lactophages”). D.v.S. is a recipient of Science Foundation Ireland (SFI) Principal Investigatorship Award Reference Number 08/IN.1/B1909 through the Irish Government National Development Plan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HEM, 4IOS, and 4HEP).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301336110/-/DCSupplemental.
References
- 1.Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10(9):688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 2.Lander GC, et al. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17(6):789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 4.Veesler D, Johnson JE. Virus maturation. Annu Rev Biophys. 2012;41(2012):473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plisson C, et al. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007;26(15):3720–3728. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veesler D, et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci USA. 2012;109(23):8954–8958. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75(3):423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bebeacua C, et al. Visualizing a complete Siphoviridae member by single-particle electron microscopy: The structure of lactococcal phage TP901-1. J Virol. 2013;87(2):1061–1068. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Hofnung M, Charbit A. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J Bacteriol. 2000;182(2):508–512. doi: 10.1128/jb.182.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigle J. Assembly of phage lambda in vitro. Proc Natl Acad Sci USA. 1966;55(6):1462–1466. doi: 10.1073/pnas.55.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veesler D, et al. Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1): A baseplate hub paradigm in gram-positive infecting phages. J Biol Chem. 2010;285(47):36666–36673. doi: 10.1074/jbc.M110.157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulet A, et al. The opening of the SPP1 bacteriophage tail, a prevalent mechanism in Gram-positive-infecting siphophages. J Biol Chem. 2011;286(28):25397–25405. doi: 10.1074/jbc.M111.243360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valyasevi R, Sandine WE, Geller BL. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J Bacteriol. 1991;173(19):6095–6100. doi: 10.1128/jb.173.19.6095-6100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spinelli S, et al. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol. 2006;13(1):85–89. doi: 10.1038/nsmb1029. [DOI] [PubMed] [Google Scholar]
- 15.Sciara G, et al. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci USA. 2010;107(15):6852–6857. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veesler D, et al. Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: Comparison of DARPin and camelid VHH binding mode. J Biol Chem. 2009;284(44):30718–30726. doi: 10.1074/jbc.M109.037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campanacci V, et al. Solution and electron microscopy characterization of lactococcal phage baseplates expressed in Escherichia coli. J Struct Biol. 2010;172(1):75–84. doi: 10.1016/j.jsb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Bebeacua C, et al. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J Biol Chem. 2010;285(50):39079–39086. doi: 10.1074/jbc.M110.175646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd DA, Veesler D, Lichiere J, Ashcroft AE, Cambillau C. Unraveling lactococcal phage baseplate assembly by mass spectrometry. Mol Cell Proteomics. 2011;10(9):M111.009787. doi: 10.1074/mcp.M111.009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lhuillier S, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106(21):8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 22.Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: The superfluous luxury of paired domains. Trends Biochem Sci. 2001;26(4):230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- 23.Ledeboer AM, et al. Preventing phage lysis of Lactococcus lactis in cheese production using a neutralizing heavy-chain antibody fragment from llama. J Dairy Sci. 2002;85(6):1376–1382. doi: 10.3168/jds.s0022-0302(02)74204-5. [DOI] [PubMed] [Google Scholar]
- 24.De Haard HJ, et al. Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J Bacteriol. 2005;187(13):4531–4541. doi: 10.1128/JB.187.13.4531-4541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hultberg A, et al. Lactobacillli expressing llama VHH fragments neutralise Lactococcus phages. BMC Biotechnol. 2007;7:58. doi: 10.1186/1472-6750-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tremblay DM, et al. Receptor-binding protein of Lactococcus lactis phages: Identification and characterization of the saccharide receptor-binding site. J Bacteriol. 2006;188(7):2400–2410. doi: 10.1128/JB.188.7.2400-2410.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinelli S, et al. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J Biol Chem. 2006;281(20):14256–14262. doi: 10.1074/jbc.M600666200. [DOI] [PubMed] [Google Scholar]
- 28.Conrath K, et al. Camelid nanobodies raised against an integral membrane enzyme, nitric oxide reductase. Protein Sci. 2009;18(3):619–628. doi: 10.1002/pro.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoogenboom HR, et al. Antibody phage display technology and its applications. Immunotechnology. 1998;4(1):1–20. doi: 10.1016/s1380-2933(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 30.Häuser R, Sabri M, Moineau S, Uetz P. The proteome and interactome of Streptococcus pneumoniae phage Cp-1. J Bacteriol. 2011;193(12):3135–3138. doi: 10.1128/JB.01481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vegge CS, et al. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J Bacteriol. 2006;188(1):55–63. doi: 10.1128/JB.188.1.55-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciara G, et al. A topological model of the baseplate of lactococcal phage Tuc2009. J Biol Chem. 2008;283(5):2716–2723. doi: 10.1074/jbc.M707533200. [DOI] [PubMed] [Google Scholar]
- 33.Mc Grath S, et al. Anatomy of a lactococcal phage tail. J Bacteriol. 2006;188(11):3972–3982. doi: 10.1128/JB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desmyter A, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3(9):803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 35.Desmyter A, et al. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J Biol Chem. 2002;277(26):23645–23650. doi: 10.1074/jbc.M202327200. [DOI] [PubMed] [Google Scholar]
- 36.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118(4):419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 37.Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol. 2004;186(11):3480–3491. doi: 10.1128/JB.186.11.3480-3491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 2000;323:325–340. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 39.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3(12):1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SH. Classification of structural heterogeneity by maximum-likelihood methods. Methods Enzymol. 2010;482:295–320. doi: 10.1016/S0076-6879(10)82012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH, Núñez-Ramírez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3(6):977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 45.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. 1991. Sequences of Proteins of Immunological Interest, ed Services USDoHaH (Public Health Service, National Institutes of Health, Bethesda, MD), 5th Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.