Abstract
The centromere is the part of the chromosome that organizes the kinetochore, which mediates chromosome movement during mitosis and meiosis. A small fragment from chromosome 3, named Duplication 3a (Dp3a), was described from UV-irradiated materials by Stadler and Roman in the 1940s [Stadler LJ, Roman H (1948) Genetics 33(3):273–303]. The genetic behavior of Dp3a is reminiscent of a ring chromosome, but fluoresecent in situ hybridization detected telomeres at both ends, suggesting a linear structure. This small chromosome has no detectable canonical centromeric sequences, but contains a site with protein features of functional centromeres such as CENH3, the centromere specific H3 histone variant, and CENP-C, a foundational kinetochore protein, suggesting the de novo formation of a centromere on the chromatin fragment. To examine the sequences associated with CENH3, chromatin immunoprecipitation was carried out with anti-CENH3 antibodies using material from young seedlings with and without the Dp3a chromosome. A novel peak was detected from the ChIP-Sequencing reads of the Dp3a sample. The peak spanned 350 kb within the long arm of chromosome 3 covering 22 genes. Collectively, these results define the behavior and molecular features of de novo centromere formation in the Dp3a chromosome, which may shed light on the initiation of new centromere sites during evolution.
Keywords: epigenetics, neocentromere
The centromere is a unique chromosomal domain that ensures accurate segregation of chromosomes during mitosis and meiosis in eukaryotes. The centromere was first described in 1882 as the primary constriction in the chromosome (1). Centromeres are essential for chromosome orientation and separation and usually contain highly repetitive DNA sequences, which associate with kinetochore proteins. The centromere is paradoxical in that its basic function is highly conserved but the DNA sequences are divergent (2). The centromere-specific H3 variant, CENH3, associates with the centromeric region, which provides the basal foundation for recruiting the kinetochore proteins (3, 4).
The positions of centromeres on chromosomes are generally conserved. However, neocentromeres have been identified in human chromosomes at ectopic sites of unique DNA where functional kinetochore assembly occurs. These neocentromeres rescue chromosome fragments for subsequent transmission and segregation (5). Recently, studies have shown that CENH3 chromatin directs and induces the formation of new centromeres (6–8).
Maize centromeres contain two basic types of DNA elements. One is the centromeric satellite referred to as CentC, a 156-bp unit present at all primary constrictions of the 10 chromosomes (9). Another element is a retrotransposon family named Centromeric Retrotransposon of Maize (CRM) (10, 11). The term “neocentromere” in maize originally was used to refer to a specific behavior of heterochromatic knobs, which exhibit centromere activity only in meiosis and only in the presence of abnormal chromosome 10 (12) and do not have the usual fundamental centromeric proteins (13). In other plants, the term “neocentromere” applies to plants with centromere activities throughout the life cycle, which have been described for a barley chromosome (14) and a maize chromosome fragment in oat (15). Here we provide evidence that a small chromosome fragment in maize, which was recovered from UV irradiation decades ago, lacks CentC and CRM sequences but nevertheless has a de novo site with centromere function throughout the life cycle. This small chromosome fragment has a 350-kb CENH3-binding region, which is involved in new centromere formation and originates from chromosome arm 3L.
Results and Discussion
Cytological Examination of Somatic Metaphase Chromosomes in Duplication 3a.
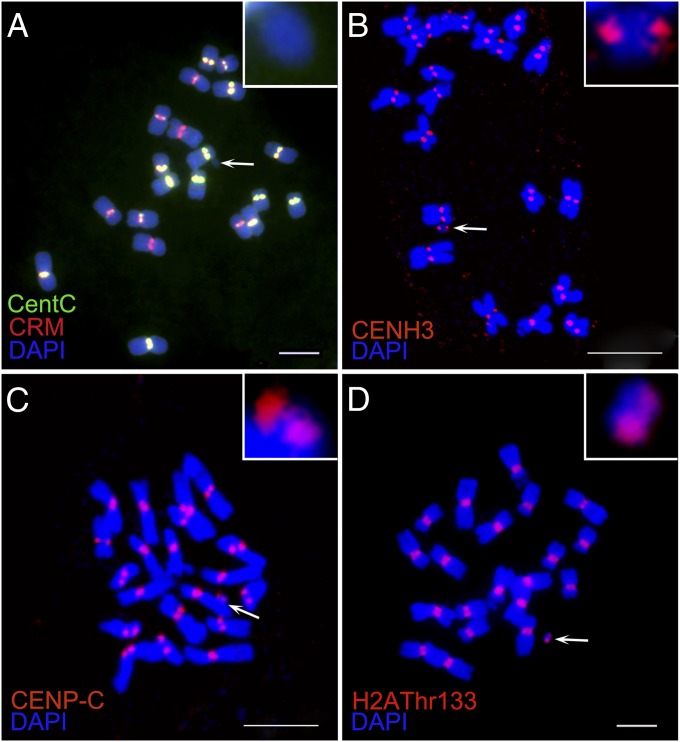
Duplication 3a (Dp3a) is a chromosome fragment derived from the long arm of maize chromosome 3 and recovered by Stadler and Roman (16). It contains the dominant alleles of the anthocyaninless1 (A1) and shrunken2 (Sh2) genes and typically exhibits a mosaic phenotype due to somatic instability (17) (Fig. S1). We performed fluorescent in situ hybridization (FISH) on somatic chromosome spreads from root tips containing Dp3a, using CentC and CRM as probes. All normal chromosomes exhibited CentC and CRM signals at the centromere regions, but no signals were found on the Dp3a chromosome even after overexposure (Fig. 1A), indicating the absence of a normal centromere or indeed any detectable canonical centromere repeats.
Fig. 1.
FISH patterns and immunolocalization analysis of Dp3a. (A) FISH with CentC (green) and CRM (red) as probes. There are no detectable CentC and CRM signals on the Dp3a chromosome (arrow) even with overexposure. (B–D) Immunostaining of the centromere-specific proteins CENH3, CENPC, and phosphorylated H2AThr133, respectively. The red CENH3, CENPC, and H2A antibody signals on Dp3a confirm the presence of a functional centromere. Arrows indicate the Dp3a chromosome. Insets display an enlarged image of Dp3a in each panel. (Scale bars, 5 μm.)
It was originally not clear whether the Dp3a chromosome was linear or circular (16). The regular somatic loss and apparent rearrangement of the chromosome was reminiscent of a ring chromosome. However, using a maize telomere sequence probe, we detected signals at the ends of the Dp3a chromosome by fluorescent in situ hybridization (FISH), suggesting a linear structure in its present form (Fig. 2). The signal strength was routinely stronger at one of the ends. Also, as noted by Stadler and Roman (16) and our own observations, the structure of Dp3a at the pachytene stage of meiosis appears linear (Fig. S2) and has never been observed in an open circular form but with the caveat that nonhomologous sequences could pair to form a collapsed ring. Chromosome breaks in maize will heal in the sporophytic generation (17) by the addition of telomere repeats (18), which could have occurred during Dp3a formation. It is not known whether the detected telomere sequences originate from a normal chromosome end or from addition to a break.
Fig. 2.
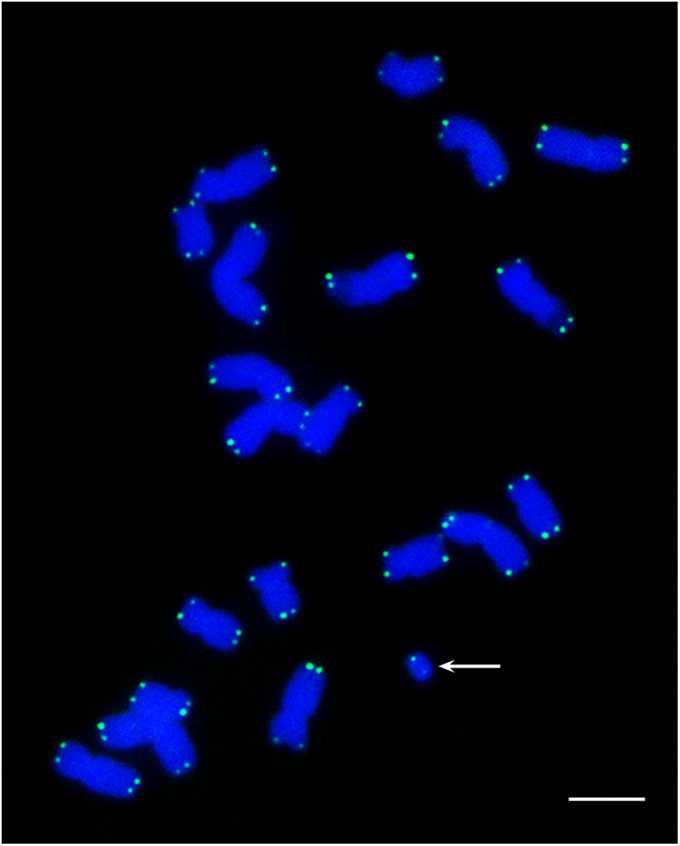
Telomeres are present on Dp3a. FISH analysis using a maize telomeric DNA probe (green) to detect the structure of the Dp3a chromosome (arrow) in root-tip metaphase cells. Telomere signals are detected at both ends of the chromosome. DAPI-stained chromosomes are blue.
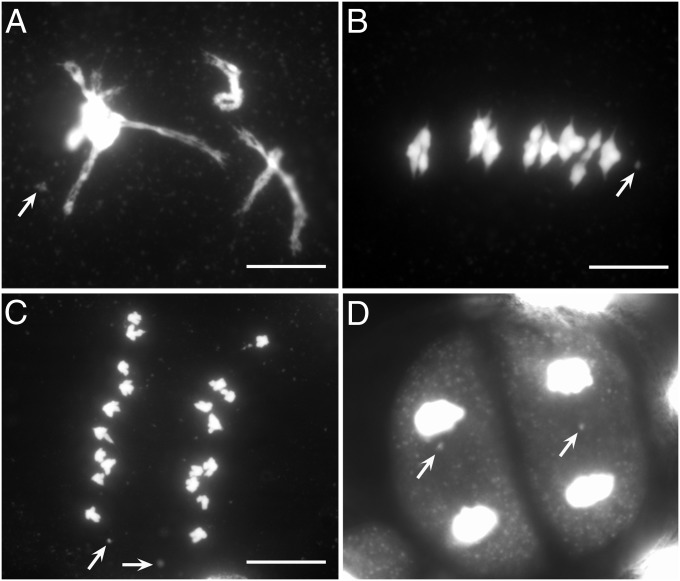
During meiosis, the Dp3a chromosome did not pair with normal chromosome 3 (Fig. 3 A and B and Fig. S2). As with other small chromosomes in maize, sister chromatids of the Dp3a chromosome separated at anaphase I (Fig. 3C) and then moved to the tetrads in meiosis II (Fig. 3D).
Fig. 3.
Meiotic behavior of the Dp3a chromosome. In diplonema (A) and metaphase I (B), the Dp3a chromosome did not pair with chromosome 3. (C) In anaphase I, sister chromatids of Dp3a chromosome separated to opposite poles. (D) In meiosis II, the Dp3a chromosomes were observed in tetrads. (Scale bar, 10 μm.)
Centromeric Chromatin Assembly on the Dp3a Chromosome.
Given the lack of canonical centromeric DNA on Dp3a, we tested for the presence of centromeric chromatin. Immunostaining revealed the coincident presence of CENH3 and CENPC in root-tip cells (Fig. 1 B and C) and in pollen mother cells (Fig. S3). Another feature of centromeric histones is phosphorylation, which is necessary for centromere function and maintenance (19, 20). We used three histone phosphorylation antibodies (against H2A-Thr133, H3-Ser10, and H3-Thr3) to study the Dp3a histone modifications. All three types of phosphorylation were detected (Fig. 1D and Figs. S4 and S5).
Three-Hundred-Fifty-Kilobase Sequence Is involved in Dp3a Centromere Formation.
To define the DNA sequences associated with the centromere-specific histone CENH3 in Dp3a, we used plants grown from kernels from the same ear with and without the Dp3a chromosome to perform chromatin immunoprecipitation (ChIP) using maize CENH3 antibodies. Before ChIP-sequencing, we used FISH to test the relative enrichment in the immunoprecipitated DNA sequences from centromeric regions. The recovered DNA was labeled as FISH probes and used to test whether signals were localized to the centromere regions. The centromeric regions of the chromosomes were strongly labeled as anticipated. Although potential CENH3-associated sequences from Dp3a would be expected to constitute a small fraction of the probe, a detectable signal was found on Dp3a (Fig. S6), and further data connecting the CENH3-associated region and Dp3a are presented below.
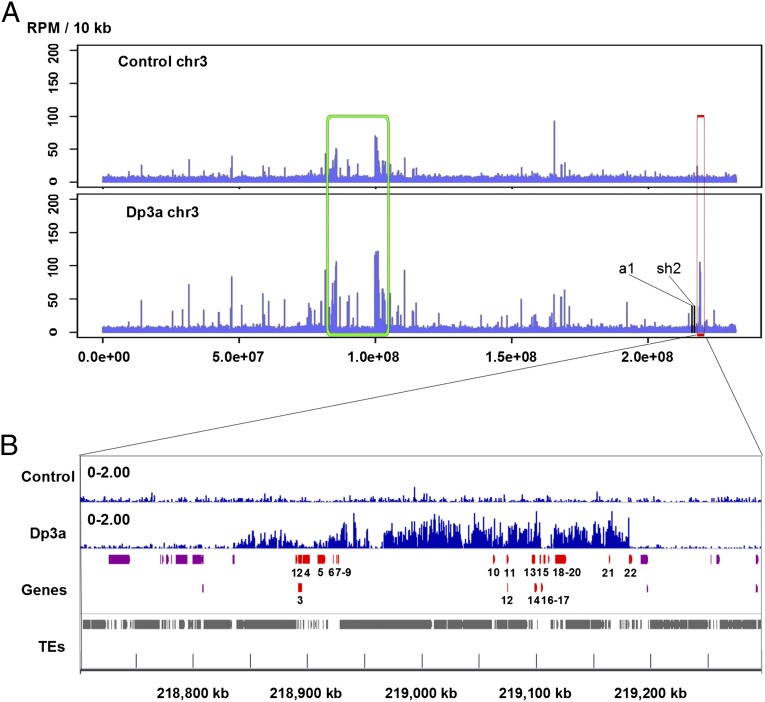
Having validated cytologically the composition of the recovered DNA, we then subjected these samples to Illumina sequencing. The reads were mapped to the maize genome using the BWA software (21). About 76% of the total reads had perfect genomic matches to the maize B73 genome (22) (Table S1). We used ChIP-Seq reads per million (RPM) in 10-kb genomic regions to compare the CENH3-binding intensity between plants with and without the Dp3a chromosome. Generalized CentC and CRM sites within the genome sequence matched the sequences enriched by ChIP. In addition, we identified a distinct peak of CENH3 association in the long arm of chromosome 3 in the Dp3a sample, which was absent in the control sample without the extra chromosome (Fig. 4). This region served as a candidate for CENH3 association on Dp3a. The peak region included 22 protein-encoding genes, but only 11 genes had CENH3-binding signals above the control levels (Fig. 4 and Table S2). We used probes (Table S2) from one of these genes (GRMZM2G045275), which was chosen for its size and clear position within the CENH3-associated region, to perform FISH and detected hybridization signals on both the Dp3a chromosome and the long arm of chromosome 3, confirming that the 350 kb was from chromosome 3 (Fig. 5). From the above results, we hypothesize that chromosome 3 was fragmented during the UV irradiation process to produce Dp3a and that CENH3 was seeded within the 350-kb region, at least to some degree, to organize a kinetochore. The initial deposition of CENH3-containing nucleosomes might not have spanned the 350-kb region but must have been sufficient for recovery of the chromosome. This fragment was transmitted to daughter cells with a normal chromosome 3 and has been subsequently inherited over generations.
Fig. 4.
Genomic distribution of ChIP-seq reads on chromosome 3 (control and Dp3a with CENH3-associated DNA). (A) Read density represented by the number of unique mapping reads (RefGen ZmB73 Release 5a) derived from ChIP with CENH3 antibodies in 10-kb windows. The x axis shows the position on chromosome 3. The generalized region of endogenous centromere 3 is outlined with green. A 350-kb CENH3-binding domain, outlined in red, was found in the Dp3a sample compared with the control. The positions of the a1 and sh2 genes, known genetically to be present on Dp3a, are noted. (B) Enlarged view of the CENH3-binding region and immediately surrounding sequences showing the distribution of genes and transposons (TEs). A total of 22 protein-encoding genes are contained in this region and are numbered. A description of the 22 genes in the region is given in Table S2.
Fig. 5.
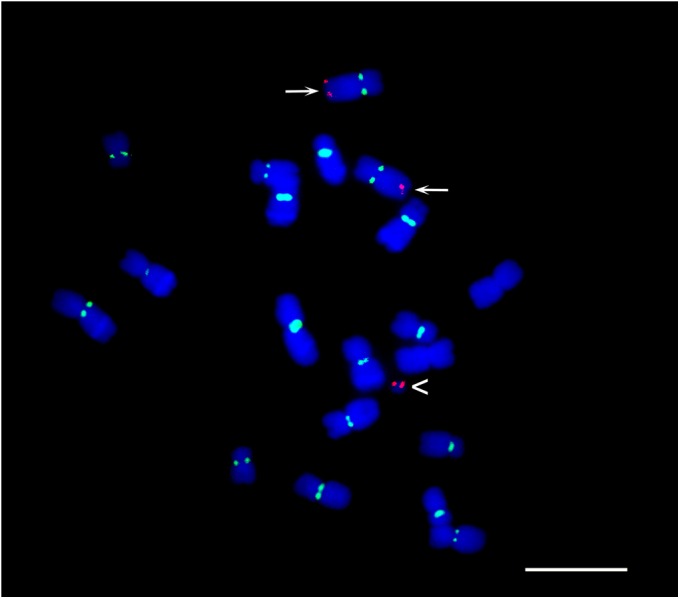
The 350-kb region on Dp3a confirmed by FISH. FISH analysis of metaphase cells using 5.8 kb of DNA (GRMZM2G045275 is gene 4 in Fig. 4) from the 350-kb region as a probe (red). Arrowhead indicates the Dp3a chromosome. Both chromosomes 3 (arrows) and Dp3a have red FISH signals. CentC centromere signals are in green. (Scale bar, 10 μm.)
In summary, we demonstrated that the small chromosome Dp3a originated from the long arm of chromosome 3 and has features that suggest it is a linear chromosome. The evidence for de novo centromere formation is as follows: (i) Dp3a contained no centromere array typical of a normal chromosome or even any detectable CentC or CRM sequences; (ii) all of the biochemical features of centromere function such as the presence of CENH3, CENP-C, and phosphorylation characteristics are present; (iii) ChIP of CENH3 identifies a 350-kb region unique to the sample carrying Dp3a that is present in 3L near the location of genetic markers (A1 Sh2) known to be present on the chromosome; and (iv) FISH using a probe consisting of a gene within the region of CENH3 association localizes to Dp3a. Together, the data suggest that a 350-kb region of 3L spanning unique genes became associated with CENH3 and confers kinetochore formation.
It seems likely that de novo centromere formation occurred immediately after UV irradiation. However, the chromosome is highly unstable (Fig. S1), suggesting that the centromere function is subnormal. The size of the CENH3-binding domain is considerably smaller than those of the normal sequenced centromeres 2 and 5, which are ∼1.8 and 4.2 Mb, respectively (11). Experimental reduction of the size of the B chromosome centromere to the same size range of the CENH3-associated sequences in Dp3a will result in impaired function and instability (23–25). If centromeres in maize do not function optimally below this potential size threshold, the initiation of new sites for centromeres during karyotype evolution would need to span a greater length or spread from an initial focus. Centromere inactivation has been documented in several cases in maize, including in irradiated material (26, 27). Thus, both inactivation and de novo formation have been found in rearranged genomes. However, because de novo formation is potentially restricted in size below the limits of normal function, these considerations are consistent with the fact that, over evolutionary time, centromere death is more common than centromere birth (28). These results provide further evidence of the epigenetic basis of centromere function in maize, in the case described here, by the initiation of a centromere over unique sequences. The results illustrate the features of an incipient centromere and will provide a model system for understanding how new centromeres are formed.
Materials and Methods
Immunolocalization and FISH.
Dp3a seeds were kindly supplied by Pat Schnable (Iowa State University, Ames, Iowa) and the Maize Genetics Stock Center. FISH and immunolocalization in mitosis and meiosis was performed as described (27, 29). The images were captured as a confocal Z-stack (Zeiss LSM 710 NLO), and a flat projection of the 3D image was created with the ZEN 2009 Light Edition (Zeiss), processed with Photoshop CS 3.0 (Adobe). Fluorochromes used were Texas Red and Alexa488, which are referred to as red and green, respectively.
Chromatin Immunoprecipitation.
ChIP was performed according to previously described protocols (30) with only minor modifications. Antibody used for ChIP was rabbit polyclonal anti-CENH3, which was affinity-purified, and the concentration was 0.83 mg/mL. Approximately 10 g of fresh leaf tissue was collected. Extracted nuclei were digested with micrococcal nuclease (Sigma; N3755). The digested chromatin was used for ChIP experiments using maize CENH3 antibody (5 µL antibodies per 25 µg chromatin). The CENH3 antibody is a rabbit polyclonal against the peptide RPGTVALREIRKYQKSSTSATPERAAGTGGR. The antibody was produced and supplied by GL Biochem.
ChIP-Seq and Data Analysis.
Plant materials with and without Dp3a were subjected to the ChIP protocol as described above. We used 30 ng of DNA from each ChIP sample for library preparation following the instructions of the Illumina ChIP-seq sample-prep kit. Fragments (280–500 bp) were selected by gel extraction. The Illumina HiSeq2000 platform was used for sequencing, generating paired-end 100-nt reads. Alignment of the ChIP-seq data to the B73 reference genome (Release 5a) was carried out using BWA software (21), allowing at most three mismatches. Reads that were mapped to a unique genomic location were chosen, and only a random one was used for copy duplicates (reads mapped to the same location). To compare different samples, RPM values were calculated with 10-kb windows along the genome. Figures were produced using R scripts. The CENH3 ChIP data have been deposited in the Gene Expression Ominibus database under accession nos. GSM1057274 and GSM1057275.
Supplementary Material
Acknowledgments
We thank Gernot Presting for comments on the manuscript. This work was supported by National Science Foundation Grants DBI 0922703 and DBI 0701297; the National Basic Research Program of China (973 Program, 2011CB944601); the National Natural Science Foundation of China (31130033 and 31071083); and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCXZ-YW-N-074).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSM1057274 and GSM1057275).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303944110/-/DCSupplemental.
References
- 1.Flemming W. Zellsubstanz, Kern und Zelltheilung [Cell substance, nucleus and cell division] Leipzig, Germany: FCW Vogel; 1882. German. [Google Scholar]
- 2.Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293(5532):1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 3.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet. 2008;9(12):923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144(4):471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82(2):261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9(1):33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 7.Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334(6056):686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 8.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477(7364):354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ananiev EV, Phillips RL, Rines HW. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci USA. 1998;95(22):13073–13078. doi: 10.1073/pnas.95.22.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang J, et al. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA. 1996;93(24):14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfgruber TK, et al. Maize centromere structure and evolution: Sequence analysis of centromeres 2 and 5 reveals a major role of retrotransposons. PLoS Genet. 2009;5:e1000723. doi: 10.1371/journal.pgen.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhoades MM, Vilkomerson H. On the anaphase movement of chromosomes. Proc Natl Acad Sci USA. 1942;28(10):433–436. doi: 10.1073/pnas.28.10.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawe RK, Hiatt EN. Plant neocentromeres: Fast, focused, and driven. Chromosome Res. 2004;12(6):655–669. doi: 10.1023/B:CHRO.0000036607.74671.db. [DOI] [PubMed] [Google Scholar]
- 14.Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA. 2005;102(28):9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topp CN, et al. Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res. 2009;124(3–4):228–238. doi: 10.1159/000218128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler LJ, Roman H. The effect of X-rays upon mutation of the gene A in maize. Genetics. 1948;33(3):273–303. doi: 10.1093/genetics/33.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26(2):234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao S, Gardiner JM, Melia-Hancock S, Coe EH., Jr Physical and genetic mapping of chromosome 9S in maize using mutations with terminal deficiencies. Genetics. 1996;143(4):1785–1794. doi: 10.1093/genetics/143.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birchler JA, Han F. Maize centromeres: Structure, function, epigenetics. Annu Rev Genet. 2009;43:287–303. doi: 10.1146/annurev-genet-102108-134834. [DOI] [PubMed] [Google Scholar]
- 20.Dong Q, Han F. Phosphorylation of histone H2A is associated with centromere function and maintenance in meiosis. Plant J. 2012;71(5):800–809. doi: 10.1111/j.1365-313X.2012.05029.x. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 23.Kaszás E, Birchler JA. Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics. 1998;150(4):1683–1692. doi: 10.1093/genetics/150.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phelps-Durr TL, Birchler JA. An asymptotic determination of minimum centromere size for the maize B chromosome. Cytogenet Genome Res. 2004;106(2–4):309–313. doi: 10.1159/000079304. [DOI] [PubMed] [Google Scholar]
- 25.Jin W, et al. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell. 2005;17(5):1412–1423. doi: 10.1105/tpc.104.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han F, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA. 2006;103(9):3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Z, Fu S, Dong Q, Han F, Birchler JA. Inactivation of a centromere during the formation of a translocation in maize. Chromosome Res. 2011;19(6):755–761. doi: 10.1007/s10577-011-9240-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Bennetzen JL. Centromere retention and loss during the descent of maize from a tetraploid ancestor. Proc Natl Acad Sci USA. 2012;109(51):21004–21009. doi: 10.1073/pnas.1218668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han FP, Gao Z, Birchler JA. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell. 2009;21(7):1929–1939. doi: 10.1105/tpc.109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaki K, et al. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163(3):1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.