Abstract
Jawless vertebrates are pivotal representatives for studies of the evolution of adaptive immunity due to their unique position in chordate phylogeny. Lamprey and hagfish, the extant jawless vertebrates, have an alternative lymphocyte-based adaptive immune system that is based on somatically diversifying leucine-rich repeat (LRR)-based antigen receptors, termed variable lymphocyte receptors (VLRs). Lamprey T-like and B-like lymphocyte lineages have been shown to express VLRA and VLRB types of anticipatory receptors, respectively. An additional VLR type, termed VLRC, has recently been identified in arctic lamprey (Lethenteron camtschaticum), and our analysis indicates that VLRC sequences are well conserved in sea lamprey (Petromyzon marinus), L. camtschaticum, and European brook lamprey (Lampetra planeri). Genome sequences of P. marinus were analyzed to determine the organization of the VLRC-encoding locus. In addition to the incomplete germ-line VLRC gene, we have identified 182 flanking donor genomic sequences that could be used to complete the assembly of mature VLRC genes. Donor LRR cassettes were classifiable into five basic structural groups, the composition of which determines their order of use during VLRC assembly by virtue of sequence similarities to the incomplete germ-line gene and to one another. Bidirectional VLRC assembly was predicted by comparisons of mature VLRC genes with the sequences of donor LRR cassettes and verified by analysis of partially assembled intermediates. Biased and repetitive use of certain donor LRR cassettes was demonstrable in mature VLRCs. Our analysis provides insight into the unique molecular strategies used for VLRC gene assembly and repertoire diversification.
The ability to achieve specific immune responses to a virtually unlimited variety of antigens is unique to vertebrates. Both jawed and jawless vertebrates have humoral and cellular arms of adaptive immunity (reviewed in refs. 1 and 2). Antigen recognition is achieved in the jawed vertebrates through Ig-based B-cell receptors (BCRs) and T-cell receptors (TCRs), the latter of which typically recognize peptide fragments of antigens presented by major histocompatibility complex (MHC) class I and II proteins. BCRs and TCRs are generated by somatic rearrangement of variable (V), diversity (D), and joining (J) gene segments during the development of B and T lymphocytes, respectively (3). In contrast, adaptive immunity in jawless vertebrates is mediated by leucine-rich repeat (LRR)-based receptors for antigens, which are known as variable lymphocyte receptors (VLRs) (2, 4, 5).
BCR, TCR, and MHC genes have not been found in the jawless vertebrates, and VLR genes have not been found in jawed vertebrates. This suggests that the two types of anticipatory receptors evolved as convergent solutions for specific antigen recognition in the ancestors of jawless and jawed vertebrates.
Three VLR genes (VLRA, VLRB, and VLRC) have been found in lampreys, and only two VLR genes (VLRA and VLRB) have been identified so far in hagfish (4, 6–8). Lamprey VLRA and VLRB genes are expressed in a monogenic and monoallelic fashion by discrete populations of lymphocytes (9); thus, each VLRA+ or VLRB+ lymphocyte expresses a unique VLRA or VLRB gene. VLRA is expressed on lymphocytes with T cell-like characteristics, whereas VLRB is expressed on lymphocytes that are B cell like and that can be activated by antigenic stimulation to differentiate into plasma cells secreting multivalent VLRB antibodies (9–12). Current evidence suggests that VLRA+ lymphocytes are generated in a thymus-equivalent structure (termed the thymoid) located at the tips of the gill folds and neighboring secondary gill filaments, whereas VLRB+ lymphocytes appear to be generated in hematopoietic tissues (13). The germ-line VLR genes are incomplete in that they have sequences encoding the invariant N- and C- terminal VLR portions that are separated by noncoding intervening sequences, but lack coding sequences for the internal LRR sequence elements (4, 6, 10, 14) used for antigen binding (15–17).
During lymphocyte development in jawless vertebrates, the intervening sequences of incomplete germ-line VLR genes are modified by the insertion of donor cassettes that encode additional LRR units. The stepwise incorporation of donor cassette sequences into incomplete germ-line VLRA and VLRB genes can be initiated from the 5′ or 3′ ends (10, 14). Short stretches of nucleotide homology (∼10–30 bp long) between donor and acceptor sequences appear to be sufficient for each step in VLR gene assembly. Thus, short stretches of sequences situated at the ends of germ-line elements or previously incorporated LRR-encoding donor cassettes likely determine the choice of flanking donor cassettes for successive steps (6, 10, 17). Unlike the recombination-activating gene- mediated recombination events in the Ig/TCR locus in jawed vertebrates, the assembly of VLRA and VLRB genes in jawless vertebrates is thought to involve a gene conversion-like mechanism mediated by two cytosine deaminases, CDA1 and CDA2, which are homologs of activation-induced cytosine deaminase (AID) (6).
Recent efforts have led to the assembly of the draft genome sequence of the sea lamprey (Petromyzon marinus; Ensembl assembly: Petromyzon_marinus_7.0), providing the opportunity to examine the structure of VLR loci in more detail. In the present study, we analyzed the genomic composition of the recently discovered VLRC locus and examined the mechanism of VLRC assembly by comparing genomic sequences with the sequences of partially and completely assembled VLRC genes.
Results
Characterization of Mature VLRC Gene Assemblies.
To characterize the structure of completely assembled mature VLRC genes and VLRC-encoding genomic donor cassettes in P. marinus, we obtained 60 mature VLRC cDNA sequences (30 sequences from a single P. marinus larva and 30 sequences from different larvae; GenBank accession nos. KC244050–KC244109). A conceptual translation of mature VLRC cDNA sequences indicates that VLRC receptors have a multidomain structure and consist of a signal peptide of 19 amino acid residues, a 47-aa N-terminal LRR (LRRNT) domain, an 18-aa LRR1, between three and five LRRV modules (each module generally comprising 24 amino acids), a 13-aa connecting peptide (CP), a 50-aa (if encoded by the incomplete germ-line gene) or 52-aa (if encoded by a donor cassette) C-terminal LRR (LRRCT) element, and a 71-aa C-terminal domain (Fig. 1). Sequence comparisons indicate that VLRC sequences are well conserved among P. marinus, arctic lamprey (Lethenteron camtschaticum), and European brook lamprey (Lampetra planeri) (Fig. S1), even though these species belong to different genera and live in different ecosystems.
Fig. 1.
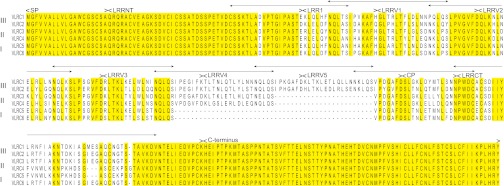
Sequence alignment of mature VLRCs of P. marinus. VLRCs can be classified into three groups (groups I, II, and III) based on the number of LRRV modules. Two representative conceptually translated sequences from each group are shown. The signal peptide (SP), N terminus, LRRNT, LRR1, LRRV, connecting peptide (CP), LRRCT, and C-terminal regions are indicated. The arrowheads above the alignment correspond to the predicted coding regions of the respective genomic donor cassettes; the precise junctions may vary by 3–15 bp on both the 5′ and 3′ ends. Note that individual LRR segments in complete assemblies are encoded by more than one donor cassette; this phenomenon is discussed in the text.
We assigned the mature VLRCs of P. marinus to three groups (I, II, and III), distinguished by the number of constituent LRRV modules: three LRRV modules in group I sequences, four in group II sequences, and five in group III sequences. Alignments of representative protein sequences in groups I, II, and III are shown in Fig. 1. We also analyzed 102 previously deposited mature VLRC sequences from L. camtschaticum (7) together with the 60 sequences from P. marinus. Of these 162 mature VLRC sequences, ∼72% belonged to group II, ∼11% to group I, and ∼17% to group III. Only 1 sequence of the 102 mature VLRC sequences from L. camtschaticum (GenBank accession no. AB507313) contained two LRRV modules.
Genomic Organization of VLRC Locus.
The incomplete germ-line VLRC gene of ∼11 kb in P. marinus consists of two exons and one large intron (∼9.65 kb). Exon 1 includes part of the 5′-UTR, whereas exon 2 includes the remaining 5′-UTR, the N-terminal coding region (signal peptide, C-terminal region of LRRNT), a short intervening sequence, the C-terminal coding region (N-terminal region of CP, LRRCT, and C terminus), and the 3′-UTR (Fig. 2). Using an interative similarity search strategy (Fig. S2), we determined the minimal number of genomic donor cassettes flanking the incomplete germ-line VLRC gene and their physical map from the draft genome sequences of P. marinus (Fig. 2 and Tables S1 and S2). We identified the incomplete germ-line VLRC gene and 182 different donor genomic cassettes in the 24 genomic scaffolds indicated in Fig. 2 and Table S2. Although these numbers are minimal estimates, owing to incomplete currently available lamprey genome sequences, the 5× whole-genome sequence coverage of the lamprey genome used for this analysis lends credence to our contention that the estimated number of genomic donor cassettes is close to the actual number.
Fig. 2.
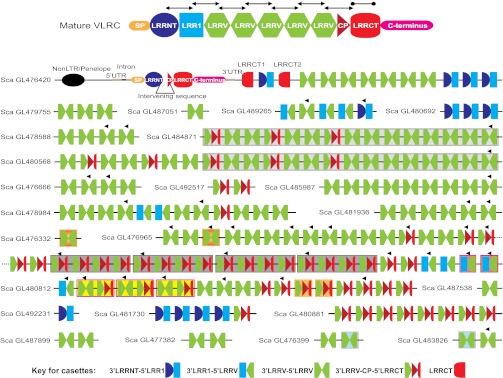
Genomic organization of the P. marinus VLRC components. (Upper) Schematic diagram of a mature group III-type VLRC assembly. (Lower) Schematic diagram of the predicted structures of VLRC elements. The order of the individual scaffolds is arbitrary, given that the fragmented genome assembly precludes determination of their relative order and orientation. Arrowheads above individual genomic donor cassettes indicate their reverse orientation relative to other donor cassettes in a particular scaffold. The LRRCT region can be encoded either by the germ-line sequence of the incomplete VLRC gene or by one of the two donor cassettes located downstream. Presumptive duplication events in individual scaffolds are represented by different background colors for the genomic cassettes. The illustrated components are not drawn to scale.
The donor genomic cassettes can be classified into five groups based on the LRR modules to which they may contribute in the mature assembled VLRCs (Fig. 2 and Table S1): (i) 3′ LRRNT-5′ LRR1 cassettes, which encode C-terminal parts of LRRNT modules and N-terminal parts of LRR1 modules; (ii) 3′ LRR1-5′ LRRV cassettes, which encode C-terminal parts of LRR1 modules and N-terminal parts of LRRV modules; (iii) 3′ LRRV-5′ LRRV cassettes, which encode C-terminal parts of preceding LRRV modules and N-terminal parts of subsequent LRRV modules; (iv) 3′ LRRV-CP-5′ LRRCT cassettes, which encode C-terminal parts of LRRV modules, a 13-aa CP region, and N-terminal parts of LRRCT modules; (v) and LRRCT cassettes, which encode the major portion of the LRRCT module.
We found 13 3′ LRRNT-5′ LRR1, 10 3′ LRR1-5′ LRRV, 103 3′ LRRV-5′ LRRV, 54 3′ LRRV-CP-5′ LRRCT, and 2 LRRCT cassettes in the sea lamprey genome (Table S1). The germ-line VLRC gene is located on scaffold GL476420 (Fig. 2), which also contains 13 different donor cassettes. Two LRRCT-encoding cassettes, LRRCT1 and LRRCT2, located downstream of the incomplete VLRC gene could encode ∼80% of the LRRCT module. Thus, in a mature VLRC, this region of the LRRCT module can be encoded either by the incomplete germ-line gene or by replacement with one of the two downstream cassettes. Whereas ∼97% nucleotide identity exists between LRRCT1 and LRRCT2 donor cassettes, these cassettes exhibit only 66% and 69% nucleotide identity, respectively, with the corresponding LRRCT-encoding region of the incomplete VLRC gene. The coding capacities of these LRRCT genomic cassettes are two amino acids greater than the corresponding region encoded by the incomplete germ-line gene. Among the 60 mature VLRC sequences, 39 used the LRRCT-encoding region of the incomplete VLRC gene, 16 used LRRCT1 donor cassettes, and 5 used LRRCT2 donor cassettes. Of note, the different types of donor cassettes are interspersed in the VLRC locus, and the physical distances between consecutive genomic donor cassettes vary, except in the regions sharing large block duplications, described in the next section. Furthermore, whereas donor cassettes encoding two or more LRR modules have been identified in the VLRA (6), and VLRB loci (4, 14), fused forms of multiple cassettes were not found in the VLRC locus.
We found that the different members of the 182 genomic cassettes were not used equally as donor sequences for mature VLRC assembly. In fact, we did not find 94 of the genomic donor cassettes in assembly of the 60 mature VLRC sequences in the P. marinus dataset. The trend of donor genomic cassette use in this dataset instead indicates preferential use of certain donor cassettes during development of the VLRC repertoire (Table S2). We found that donor genomic cassettes can be incorporated into mature VLRC sequences as either full-length or partial sequences, apparently based on short nucleotide sequence similarities between donor and acceptor sequences, as noted previously for VLRA and VLRB assemblies (6, 10, 14). The regions of short sequence similarities between donor and acceptor sequences are generally well conserved (Fig. S3), although some of the donor cassettes either feature internal stop codons or exhibit highly diverse sequences at the 5′ or 3′ ends. Nevertheless, our analysis suggests that LRR cassettes with such sequences can be partially incorporated into mature sequences, using sequence similarities in the middle or at one end of the cassettes (Fig. S3) to facilitate their contribution during VLRC assembly.
Evolutionary Relationships of VLRC-Encoding Genomic Donor Cassettes.
Given that the genomic donor cassettes necessarily contain elements of the LRR-encoding motif, we used neighbor-joining (NJ) and maximum likelihood (ML) methods to conduct phylogenetic analyses to examine the evolutionary relationship between donor cassettes and search for potential duplicates (18, 19). The tree topologies and the phylogenetic interpretations of the NJ and ML analyses were similar (Figs. S4 and S5). Phylogenetic analysis of nucleotide sequences of the VLRC-encoding genomic donor cassettes indicated clustering of the 3′ LRRNT-5′ LRR1 cassettes with 99% bootstrap support (Fig. S4), owing to their unique sequence signature (Fig. S3). In a phylogenetic tree condensed at the 50% bootstrap support level, the 3′ LRRV-CP-5′ LRRCT cassettes formed a single cluster with 85% bootstrap support, because of the specific sequence signature of the CP-5′ LRRCT part (Fig. S3). Similarly, all 3′ LRR1-5′ LRRV cassettes clustered together (96% bootstrap support), owing to a distinct sequence signature in the 3′ LRR1-encoding region of the cassettes (Fig. S3). No clear-cut classification could be assigned for 3′ LRRV-5′ LRRV donor cassettes; however, a cluster with relatively low bootstrap support (76%) included donor cassettes contributing to the C-terminal region of the first LRRV module and the N-terminal region of the subsequent LRRV module.
The sequences of scaffolds containing multiple VLRC-donor genomic cassettes provide evidence of duplication events, some involving large blocks with different types of donor cassettes (Fig. 2). Presumptive duplication events of cassette sequences were identified by phylogenetic analysis, genomic orientation, and positional clustering of the cassettes; their designation was also supported by a high degree of sequence similarity (≥95%) that extended into noncoding flanking sequences. Another distinguishing feature of the regions sharing block duplications was the invariant spacing of the constituent cassettes. A large block duplication comprising 15 donor cassettes was identifiable in scaffolds GL484871 and GL480568 (Fig. 2). Tandem block duplication events involving four 3′ LRRV-CP-5′ LRRCT cassettes were found in the scaffold GL476965, and additional short tandem duplication events were detected as well. Whether or not these putative duplication events occurred recently in the P. marinus lineage could not be determined, owing to the lack of a suitable outgroup for phylogenetic comparisons.
VLRC Assembly Mechanism.
We sought to gain insight into the mechanism of VLRC assembly and repertoire development using the currently available P. marinus sequence and comparative analysis of the collection of mature VLRC sequences. The degree of coverage and diversity exhibited by the group of 30 sequences from a single lamprey larva as determined by comparison with the genome database was equivalent to that of 30 sequences from different individuals, suggesting that the analysis was not biased by the presence of interindividual sequence polymorphisms. Thus, we combined 30 sequences from a single individual and 30 sequences from different individuals into a query dataset of 60 mature VLRCs to identify donor genomic cassettes and characterize their use in the assembly of mature VLRC sequences. As shown previously for VLRA (6) and VLRB (10, 14), donor VLRC cassettes appear to be incorporated in a stepwise manner (Fig. 3). The 3′ portion of the germ-line LRRNT segment and the 5′ portion of the LRRCT element could provide sufficient sequence similarities to the anchor 3′ LRRNT-5′ LRR1 and 3′ LRRV-CP-5′ LRRCT cassettes, respectively, during the assembly process. If the N-terminal part of LRRCT is encoded by one of the two flanking donor cassettes, it could also serve to anchor the 3′ LRRV-CP-5′ LRRCT cassettes. Thus, it appears possible that the stepwise assembly of VLRC can begin from either end of the incomplete germ-line VLRC gene (Fig. 3A).
Fig. 3.
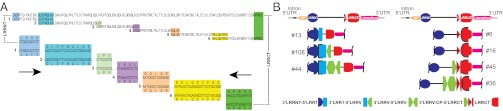
Stepwise assembly of VLRC-encoding donor cassettes. (A) Composite structure of the diversity region of a representative mature VLRC sequence of P. marinus. The conceptual translation of the mature sequence (GenBank accession no. KC244056) is shown at the top; the conceptually translated individual genomic donor cassettes are shown underneath. The overlapping junctional regions are indicated by different colors. The extent of nucleotide similarity at the corresponding junctional regions (i.e., both ends of donor cassettes) is shown at the bottom; the middle portions of donor cassettes are not shown and are represented by dots. The LRRNT and LRRCT regions are indicated by wave lines. The strong sequence similarities between genomic acceptor and donor sequences may allow assembly in either direction (arrows). (B) Structure of partially assembled VLRC genes of L. planeri. Examples of 5′ assemblies and 3′ assemblies are shown beneath a schematic of the germ-line VLRC locus. The numbers refer to clones.
To obtain more compelling evidence for the mode of gene assembly, we analyzed the sequences of partial VLRC genes amplified from genomic DNA extracted from thymoid tissue of L. planeri (13). As expected from a tissue distinguished by ongoing VLR assembly, we could readily obtain VLRC sequences representing intermediate steps of the assembly process (Fig. 3B and Table S3). In VLRC genes in which assembly began at the 3′ end, the structures of the intermediates are indicative of stepwise assembly; for instance, we found sequences exhibiting insertion of either of the two LRRCT donor cassette sequences. In other cases, we observed additional insertion of a CP-encoding cassette.
Interestingly, in two different partial assemblies using the same CP-encoding module, the sequences at the 5′ end of the VLRC gene were identical (Fig. 3B). In yet another instance of 3′-directed assembly, an additional LRRV element was incorporated, producing a partial assembly reflecting three successive insertion events (Fig. 3B). In partial assemblies beginning from the 5′ end, sequences resulting either from single insertions of a LRR1 module or from two successive insertions of LRR1 and LRRV1 modules were observed; remarkably, downstream double-strand breaks in the VLRC gene were often located in the LRRCT region, suggesting that 5′ assemblies require insertion of extragenic LRRCT modules to generate a functional mature VLRC gene. These results, in conjunction with sequence comparisons of the genomic cassettes, unambiguously indicate that the assembly process of the VLRC gene can begin at either end of the locus (Fig. 3 and Table S3).
Generation of VLRC Diversity.
During the VLRC assembly process, any of the five different types of flanking donor cassettes may be incorporated in an ordered and nonrepetitive fashion based on a short region of sequence homology (∼6–30 nt). Although most of the donor cassettes were used only once in a particular VLRC assembly, evidence of repetitive use of a 3′ LRRV-5′ LRRV donor cassette was observed as well (Fig. S6). For example, one genomic cassette (scaffold GL478984: nucleotides 25,026–25,112) apparently was used twice in a mature sequence (GenBank accession no. KC244071), given that no duplicate sequence of that cassette was seen in the P. marinus genome sequence.
Our analysis of mature VLRC sequences indicates that the combinatorial use of donor cassettes with different sequences is a major contributor to VLRC diversity. The diversification process also involves the use of variable numbers of the 3′ LRRV-5′ LRRV cassettes (Fig. 1). Depending on the position of the DNA double-strand breaks at the junctional region during stepwise assembly using donor cassettes, sequence diversity can be introduced in the assembled mature sequence (Fig. 4A). For instance, when two different mature VLRC sequences use the same cassette for a specific region, that specific region could differ for two mature VLRCs, owing to differences in the position of the double-strand break in the junctional region (Fig. 4A). However, we found no evidence of junctional diversity as a result of template-independent processes, a typical feature of the VDJ recombination process of Ig and TCR genes in jawed vertebrates. In principle, sequence diversity of the mature sequence could be introduced at the sites of double-strand breaks that presumably occur when the donor cassette sequences are being copied into partially assembled intermediates. Of note, our analysis of junctional regions in mature VLRCs suggests the absence of nucleotide insertions and deletions (Fig. 4A).
Fig. 4.

Origin of junctional and positional diversity in mature VLRC assemblies. (A) The assembly process generates junctional diversity owing to the variable position of the double-strand break (indicated by X) associated with assembly. A representative example of this mechanism in the LRRNT and LRR1 regions is shown. The sequences of two representative mature VLRCs indicate the use of the same 3′ LRRNT-5′ LRR1 donor cassette to encode the corresponding regions of LRRNT and LRR1. Nonetheless, the sequences of these two mature VLRCs are different because of differences in the position of the potential double-strand break in the overlapping junctional region between the 3′ end of the LRRNT encoding region of the incomplete VLRC gene and the 5′ end of the 3′ LRRNT-5′ LRR1 donor cassette. (B) Positional diversity arising during the assembly VLRC-encoding donor cassettes. A representative example for this mechanism in the LRRNT and LRR1 regions is shown. During the assembly of two mature VLRC genes, one gene incorporated the full length of a 3′ LRRNT-5′ LRR1 donor cassette (GL480692; 26,146–26,323 nt), presumably by means of matched nucleotide sequences at both ends, whereas another gene incorporated two donor cassettes (one common for both VLRCs) to encode the same region, presumably by means of matched internal nucleotide sequences. The presumed positions of double-strand breaks during assembly are indicated by X, as are conceptual translations of mature VLRC sequences.
In the assembly of mature VLRC sequences, genomic donor cassette sequences are used in either full-length or partial form by using sequence homologies either at the ends or in the middle of the cassettes, respectively. One of the two representative mature VLRC sequences shown in Fig. 4B incorporated the full-length sequence of a 3′ LRRNT-5′ LRR1 donor cassette using matching nucleotides at both ends, whereas the other used two donor cassettes (one of which is used in common for two mature VLRC sequences) to encode the same region by means of a nucleotide match in the middle of the cassettes. As a result of this patchwork assembly mode, these mature VLRCs differ from one another in this region. A fraction of the genomic cassettes exhibited internal stop codons or highly divergent sequence composition at the 5′ or the 3′ ends compared with the sequences of frequently used genomic cassettes (Table S1 and Fig. S3). Although these highly divergent cassettes may not be used as full-length templates, they nevertheless could be incorporated into mature VLRC sequences using stretches of sequence identity in the homologous parts of the cassettes.
Discussion
The genomic organization of the VLRC locus of P. marinus reveals that the incomplete germ-line VLRC gene is flanked by five different types of genomic donor cassettes, the sequences of which are used in the assembly of mature VLRC genes. Although the structure of LRR-based antigen receptors in jawless vertebrates is quite distinct from that of Ig domain-based antigen receptors in jawed vertebrates, the overall genomic organization and the evolutionary dynamics of the VLRC locus have similarities to the genomic architecture and mode of evolution of Ig and TCR multigene families. From a genomic standpoint, the incomplete germ-line VLRC gene serves as an equivalent of the constant gene segment, whereas the clusters of different types of donor genomic cassettes represent the functional correlates of the clusters of V, D, and J gene segments of Ig or TCR loci in jawed vertebrates (20–22). Interestingly, the donor LRR-encoding cassettes are not used equally for VLR assembly, in analogy with the preferential use of certain V, D, and J segments in Ig/TCR recombinations in jawed vertebrates (23–26).
The genomic architecture of the VLRC locus and the structure of mature VLRC sequences indicate that the intervening sequence of the germ-line VLRC gene is replaced by different types of donor cassettes in a stepwise manner similar to that previously shown for the VLRA and VLRB loci (4, 6, 10, 14). Some important differences exist with respect to VLRC locus architecture, however. In particular, none of the LRR1- and LRRV-encoding donor cassette sequences encodes an entire module, thereby dictating that each LRR module in the mature VLRC sequences is a chimeric unit encoded by two or more donor cassettes. Using short stretches of nucleotide homology between donor sequences and acceptor sequences, the assembly process proceeds from either the 5′ end or the 3′ end. As in VLRA and VLRB (6), the mature VLRC assemblies have variable numbers of internal LRRV modules, although four internal LRRV elements were found in most instances. This finding suggests that four LRRV modules are optimal for VLRC assembly and may have structural and functional advantages.
Previous structural analyses of antigen-binding VLRB and VLRA proteins indicated that LRR1, the LRRVs, the CP, and the N-terminal portion of LRRCT can contribute to antigen binding during an immune response (15–17, 27). In agreement with those findings, we found that a large number of different 3′ LRRV-5′ LRRV and 3′ LRRV-CP-5′ LRRCT donor cassettes contribute to VLRC repertoire diversity, whereas the LRRCT donor sequences make only a minor contribution to the VLRC diversity. Unlike in the VLRA and VLRB loci (15–17), there are only three LRRCT options in the VLRC locus, and the only two donor LRRCT-encoding cassettes are very similar. Moreover, none of the 5′ LRRCT region sequences of the VLRCs encodes an extended loop (7) like those that play prominent roles in antigen binding by the VLRA and VLRB proteins (4, 6, 10, 14).
Despite their distinct sequence signatures, the different types of donor cassettes are interspersed in the VLRC-related scaffolds, a situation reminiscent of the interspersed arrangements of individual members of V gene families of Ig and TCR loci (20–22). We found evidence of multiple duplication events for the different types of donor cassettes in the VLRC locus; however, in the absence of a suitable outgroup, the number and the evolutionary timing of the apparent duplication events in the VLRC locus cannot be determined precisely. Nevertheless, these multiple duplication events, the sequence divergence in each type of donor cassettes, and the interspersion of different cassette types in the clusters indicate that the evolutionary dynamics of the VLRC locus, like Ig and TCR loci, are subject mainly to the birth-and-death model of evolution (28, 29) rather than to concerted evolution (30) through a continuous change in the copy number of donor cassettes.
In conclusion, our analysis of the genomic VLRC organization and the composition of mature VLRC sequences has revealed previously unappreciated details of the stepwise assembly process and suggests a similar degree of VLRC diversity as that of the lamprey VLRA and VLRB anticipatory receptors.
Materials and Methods
VLRC cDNA Sequences.
To obtain mature VLRC sequences, total RNA was extracted from P. marinus leukocytes. After reverse RNA transcription, PCR was performed using a set of primers designed to amplify the region spanning 5′-UTR to 3′-UTR (Table S4) under conditions described previously (9), cloned to the pCR4-TOPO vector (Invitrogen) and then sequenced. Sixty mature VLRC sequences (30 sequences from a single animal and 30 sequences from multiple animals) were used in this study.
VLRC Genomic Sequences.
To enrich for partially assembled VLRC sequences, genomic DNA was procured from the thymoid region of gill filaments of L. planeri larvae by laser capture microdissection, essentially as described previously (13). VLRC genes were amplified using the primers listed in Table S4, cloned into the pGEM-T vector, and sequenced. The sequences of the incomplete VLRC gene and representative partial assemblies were deposited in the GenBank database (accession nos. KC247673–KC247679).
Identification of VLRC-Encoding Donor Cassettes.
Donor VLRC-encoding genomic cassettes were identified through two rounds of BLASTn searches performed with an E-value ≤10−5 against the current version (Petromyzon_marinus_7.0) of the P. marinus genome sequence publicly available from the Ensembl database (www.ensembl.org/Petromyzon_marinus/Info/Index). In the first round, nucleotide sequences of 60 mature VLRCs (GenBank accession nos. KC244050–KC244109) were used as queries. The putative genomic donor cassette sequences, including approximately 300 nucleotides upstream and downstream, were conceptually translated into six frames, and the likely boundaries (varying up to 15 bp upstream and/or downstream) of each donor cassette were determined using information on protein domains present in the SMART database (31). In a subsequent round of homology searches, the procedures were repeated using the donor cassettes identified in the first round to identify additional donor sequences. In a final verification step, scaffolds were designated as VLRC-related when the sequence of at least one donor cassette was used in the mature VLRC sequences derived independently. A flowchart of the procedure is shown in Fig. S2, and the genomic locations of all identified donor cassettes are summarized in Table S2.
Phylogenetic Analysis.
Sequences were aligned using CLUSTALW (32), and the alignments were manually inspected and corrected where necessary. Graphical representations of the sequence similarity were generated using WebLogo (33). Phylogenetic analysis was performed using the NJ (18) and ML (19) methods in MEGA version 5 with the pairwise deletion option (34). The evolutionary distances for the NJ tree were computed using the p-distance method (35). The Tamura–Nei substitution model (36) was used to construct the ML tree. Support for each node of the phylogenetic tree was tested with 1,000 bootstrap replicates.
Sequence Analysis.
The polyA site was predicted using HCpolya (37). The signal peptide was determined using PrediSi (38) and verified with SignalP (39) and SOSUIsignal (40). Repetitive elements located in the VLRC locus were identified using the CENSOR tool (41).
Supplementary Material
Acknowledgments
We thank Brantley R. Herrin, Jianxu Li, Qifeng Han, Michael Schorpp, and Stephen Holland for their assistance and comments. S.D., M.H. and M.D.C. were supported by National Institutes of Health Grants R01 AI072435 and R01 GM100151 and the Georgia Research Alliance. N.A., B.B., and T.B. were supported by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC244050–KC244109 and KC247673–KC247679).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302500110/-/DCSupplemental.
See Commentary on page 5746.
References
- 1.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 2.Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–157. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 3.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: Similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 4.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430(6996):174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 5.Boehm T, et al. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–220. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8(6):647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 7.Kasamatsu J, et al. Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci USA. 2010;107(32):14304–14308. doi: 10.1073/pnas.1001910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102(26):9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459(7248):796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310(5756):1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 11.Alder MN, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol. 2008;9(3):319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 12.Herrin BR, et al. Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA. 2008;105(6):2040–2045. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bajoghli B, et al. A thymus candidate in lampreys. Nature. 2011;470(7332):90–94. doi: 10.1038/nature09655. [DOI] [PubMed] [Google Scholar]
- 14.Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8(2):206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- 15.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321(5897):1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velikovsky CA, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat Struct Mol Biol. 2009;16(7):725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng L, et al. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci USA. 2010;107(30):13408–13413. doi: 10.1073/pnas.1005475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 20.Das S, Nozawa M, Klein J, Nei M. Evolutionary dynamics of the immunoglobulin heavy chain variable region genes in vertebrates. Immunogenetics. 2008;60(1):47–55. doi: 10.1007/s00251-007-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S. Evolutionary origin and genomic organization of micro-RNA genes in immunoglobulin lambda variable region gene family. Mol Biol Evol. 2009;26(5):1179–1189. doi: 10.1093/molbev/msp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra ZE, et al. Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genomics. 2008;9:111. doi: 10.1186/1471-2164-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malynn BA, Yancopoulos GD, Barth JE, Bona CA, Alt FW. Biased expression of JH-proximal VH genes occurs in the newly generated repertoire of neonatal and adult mice. J Exp Med. 1990;171(3):843–859. doi: 10.1084/jem.171.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraj P, et al. The human heavy chain Ig V region gene repertoire is biased at all stages of B cell ontogeny, including early pre-B cells. J Immunol. 1997;158(12):5824–5832. [PubMed] [Google Scholar]
- 25.Day EB, et al. Structural basis for enabling T-cell receptor diversity within biased virus-specific CD8+ T-cell responses. Proc Natl Acad Sci USA. 2011;108(23):9536–9541. doi: 10.1073/pnas.1106851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler JE, Wertz N, Sun J, Sacco RE. Comparison of the expressed porcine Vbeta and Jbeta repertoire of thymocytes and peripheral T cells. Immunology. 2005;114(2):184–193. doi: 10.1111/j.1365-2567.2004.02072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchdoerfer RN, et al. Variable lymphocyte receptor recognition of the immunodominant glycoprotein of Bacillus anthracis spores. Structure. 2012;20(3):479–486. doi: 10.1016/j.str.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M, Rogozin IB, Piontkivska H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc Natl Acad Sci USA. 2000;97(20):10866–10871. doi: 10.1073/pnas.97.20.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Nikolaidis N, Klein J, Nei M. Evolutionary redefinition of immunoglobulin light chain isotypes in tetrapods using molecular markers. Proc Natl Acad Sci USA. 2008;105(43):16647–16652. doi: 10.1073/pnas.0808800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolaidis N, Nei M. Concerted and nonconcerted evolution of the Hsp70 gene superfamily in two sibling species of nematodes. Mol Biol Evol. 2004;21(3):498–505. doi: 10.1093/molbev/msh041. [DOI] [PubMed] [Google Scholar]
- 31.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 36.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 37.Milanesi L, Muselli M, Arrigo P. Hamming-clustering method for signals prediction in 5′ and 3′ regions of eukaryotic genes. Comput Appl Biosci. 1996;12(5):399–404. doi: 10.1093/bioinformatics/12.5.399. [DOI] [PubMed] [Google Scholar]
- 38.Hiller K, Grote A, Scheer M, Münch R, Jahn D. PrediSi: Prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32(Web Server issue):W375-9. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 40.Gomi M, Sonoyama M, Mitaku S. High-performance system for signal peptide prediction: SOSUIsignal. Chem Bio Inform J. 2004;4:142–147. [Google Scholar]
- 41.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


