Abstract
We show that the M2 isoform of pyruvate kinase (M2PYK) exists in equilibrium between monomers and tetramers regulated by allosteric binding of naturally occurring small-molecule metabolites. Phenylalanine stabilizes an inactive T-state tetrameric conformer and inhibits M2PYK with an IC50 value of 0.24 mM, whereas thyroid hormone (triiodo-l-thyronine, T3) stabilizes an inactive monomeric form of M2PYK with an IC50 of 78 nM. The allosteric activator fructose-1,6-bisphosphate [F16BP, AC50 (concentration that gives 50% activation) of 7 μM] shifts the equilibrium to the tetrameric active R-state, which has a similar activity to that of the constitutively fully active isoform M1PYK. Proliferation assays using HCT-116 cells showed that addition of inhibitors phenylalanine and T3 both increased cell proliferation, whereas addition of the activator F16BP reduced proliferation. F16BP abrogates the inhibitory effect of both phenylalanine and T3, highlighting a dominant role of M2PYK allosteric activation in the regulation of cancer proliferation. X-ray structures show constitutively fully active M1PYK and F16BP-bound M2PYK in an R-state conformation with a lysine at the dimer-interface acting as a peg in a hole, locking the active tetramer conformation. Binding of phenylalanine in an allosteric pocket induces a 13° rotation of the protomers, destroying the peg-in-hole R-state interface. This distinct T-state tetramer is stabilized by flipped out Trp/Arg side chains that stack across the dimer interface. X-ray structures and biophysical binding data of M2PYK complexes explain how, at a molecular level, fluctuations in concentrations of amino acids, thyroid hormone, and glucose metabolites switch M2PYK on and off to provide the cell with a nutrient sensing and growth signaling mechanism.
Keywords: allosteric regulation, nutrient sensor, thyroid hormone T3, Warburg effect
The last of 10 enzymatic steps used to convert glucose to pyruvate is carried out by pyruvate kinase (PYK), which transfers a phosphate from phosphoenolpyruvate to ADP to generate ATP. There are four human PYK isoforms (1); RPYK is restricted to erythrocytes, LPYK is found predominantly in liver and kidney, M1PYK is in muscle and brain, and M2PYK is found in fetal tissues and in proliferating cells. All four isoforms are active as tetramers; M1PYK is constitutively fully active, whereas R-, L-, and M2PYKs are activated by the effector molecule fructose-1,6-bisphosphate (F16BP) (2). M2PYK is a splice variant of the nonallosteric M1PYK isoform and differs by 22 amino acid residues (3). Recent quantification of the concentrations of constitutively fully active M1PYK and allosterically regulated M2PYK isoforms in both cancerous and control tissue samples has revealed that M2PYK is almost always the most abundant isoform in cancer cells, although it can also be predominant in matched control tissues (4). The up-regulation of the M2PYK isoform plays a key role in cancer metabolism (3) and explains the Warburg effect, in which proliferating cancer cells metabolize increased amounts of glucose but with no increase in mitochondrial oxidative phosphorylation (5). Regulation of M2PYK activity by the allosteric effector F16BP provides a “metabolic budget system” (6) to balance the energy requirements of the cell against the requirements of a growing and dividing cell.
There are multiple allosteric feedback mechanisms at work controlling the balance between the forward enzyme reaction (to produce ATP and pyruvate) and M2PYK inhibition [resulting in accumulation of intermediate metabolites required for DNA and protein synthesis (7)]. There is a growing literature showing that M2PYK activity can be modulated by acetylation (8), phosphorylation (6, 9), cysteine oxidation (10), and proline hydroxylation (11), providing additional mechanisms for enzyme regulation as well as proffering recognition sites for a diverse range of protein partners (6), including for example HIF (11), HPV E7 (12, 13), and the peptide hormone somatostatin (14).
Switching on the production of the constitutively fully active M1PYK was shown to reverse the Warburg effect and to inhibit tumor growth in mouse xenograft models (3). This result suggests that allosteric activation of M2PYK by pharmacological agents could be of interest, and a number of potent activators have been identified with AC50 (concentration that gives 50% activation) values of approximately 30 nM (15, 16). Currently, the only examples of isoenzyme-specific inhibitors are three families of weak M2PYK inhibitors with IC50 values of 10–20 μM that show some selectivity over M1PYK (17).
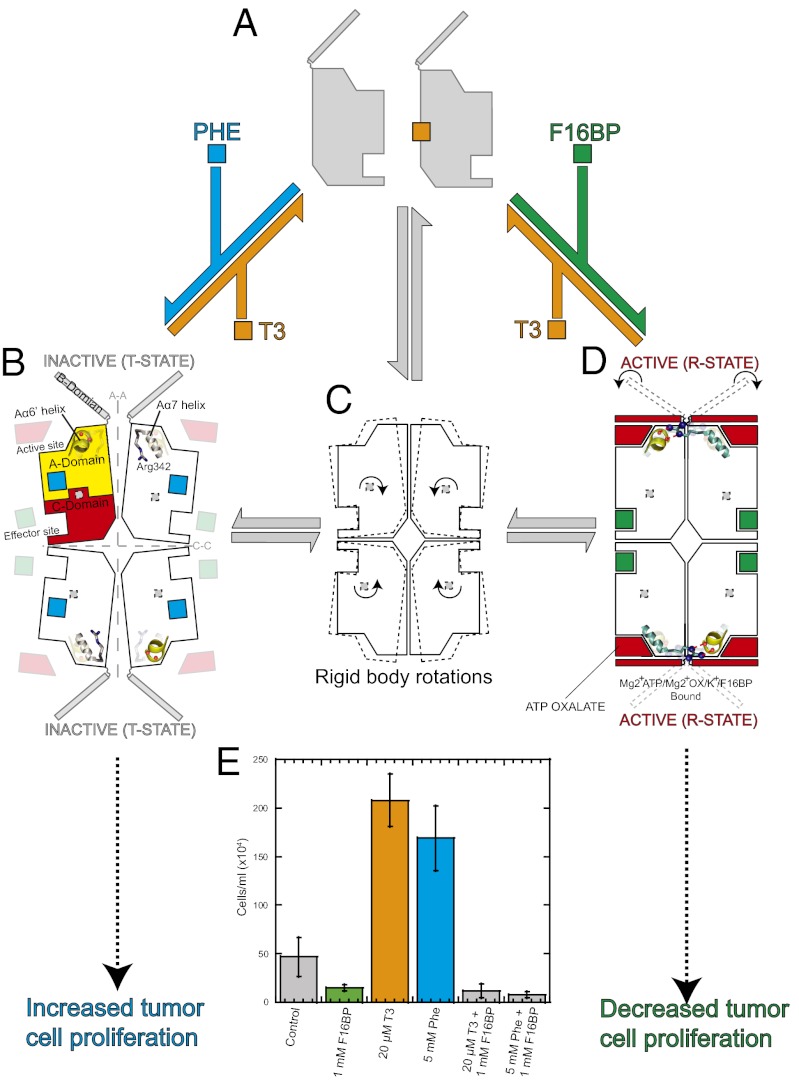
The results presented here show how M2PYK can be stabilized in three biologically relevant states: an inactive monomer, a distinct inactive T-state tetramer, and an active R-state tetramer (Fig. 1). X-ray structures of the different states show how the tetramer switches between the inactive T-state trapped as a phenylalanine complex and the fully active R-state, which is very similar in structure and enzyme activity to the constitutively fully active M1PYK isoform. From a screen of more than 50 natural metabolites, thyroid hormone triiodo-l-thyronine (T3) and phenylalanine were identified as the strongest enzyme inhibitors. In cellular assays we show that both T3 and phenylalanine increase cell proliferation, whereas the addition of the allosteric activator F16BP decreased cell proliferation. These enzymatic, structural, and cellular results provide plausible molecular mechanisms linking cell proliferation with allosteric regulation of M2PYK enzyme activity.
Fig. 1.
Allosteric nutrient sensing mechanism also regulates cellular proliferation. X-ray structures of the tetrameric Phe-bound T-state (B) and F16BP-activated R-state (D) are shown as cartoons, with each 50-kDa protomer represented by a rectangular shape showing the effector and active sites. M2PYK exists in equilibrium between tetrameric (C) and enzymatically inactive monomer (A) forms (gray arrows). Phenylalanine (Phe, cyan square) and the thyroid hormone T3 (orange) act as allosteric inhibitors and prevent the tetramer adopting an active R-state conformation. The activator F16BP (green square) clamps the tetramer in an enzymatically active conformation. (E) Addition of F16BP to HCT-116 cells inhibits proliferation, whereas both inhibitors of M2PYK (T3 and Phe) stimulate proliferation.
Results and Discussion
M2PYK Exists in Equilibrium Between Inactive Monomer and Active Tetramer.
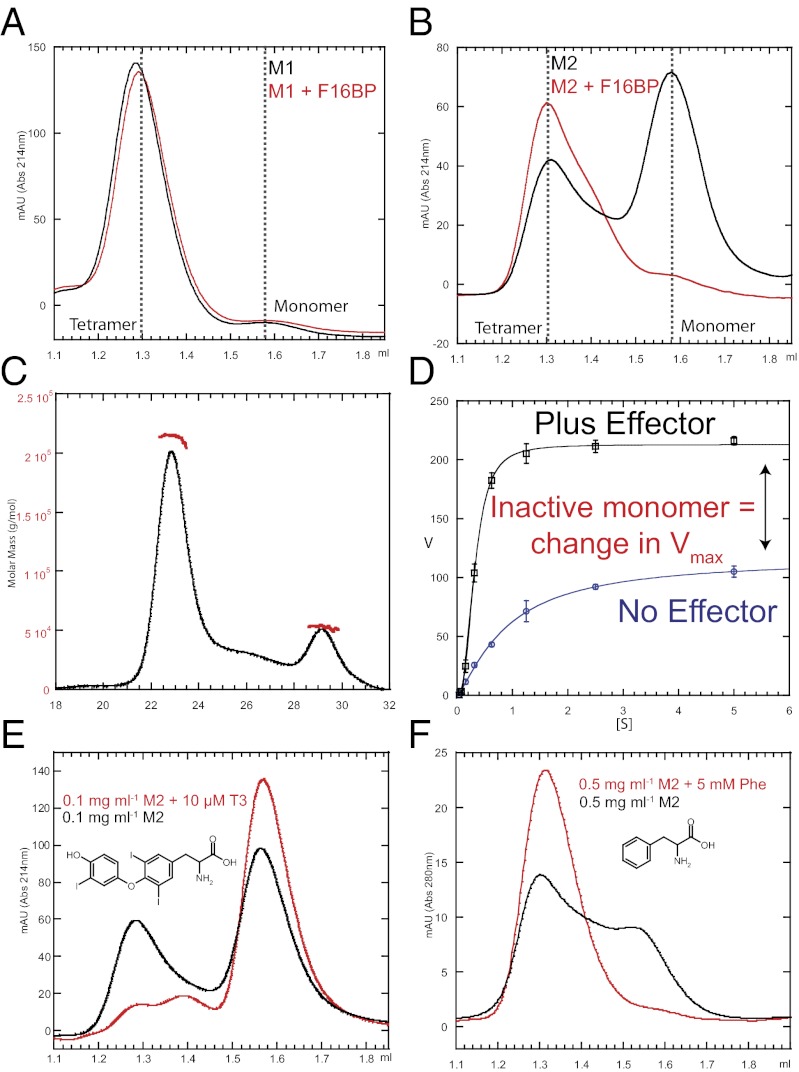
Analytical gel filtration was used to analyze the oligomeric natures of M1PYK and M2PYK. In the absence or presence of the allosteric effector F16BP, M1PYK eluted as a single species with a retention volume of 1.3 mL (Fig. 2A), consistent with M1PYK existing only as a tetramer. Under identical conditions (0.1 mg mL−1) M2PYK eluted as a mixed population of tetramer, dimer, and monomer in the absence of F16BP (Fig. 2B). Peaks corresponding to tetramer and monomer could be clearly assigned, whereas the putative dimer peak was poorly resolved. Deconvolution of the peaks gave an estimate of the molar ratios of tetramer (10%), dimer (10%), and monomer (80%). (Percentages correspond to molar ratios of the oligomers in the absence of F16BP.)
Fig. 2.
Oligomeric states of M1PYK and M2PYK. (A) Analytical gel-filtration elution profile observed for 10-µL sample injection of 0.1 mg mL−1 M1PYK in the absence (black) and presence (red) of 500 µM F16BP. (B) Same experiment as in A but for M2PYK. (C) Determination of the molar mass of M2PYK using SEC-MALS. Solid black line indicates the trace from the refractive index detector, and red dots are the weight-averaged molecular masses for each 0.5-s slice analyzed. A total of 200 µg of M2PYK was injected onto a Superdex 200 10/300 column. Flow rate was 0.5 mL min−1. (D) Concentration response curves observed for the titration of PEP against M2PYK in the presence (blue line) or absence (black line) of saturated F16BP. Error bars are derived from three independent repeat experiments. (E) Analytical gel-filtration elution profile observed for 10-µL sample injection of 0.1 mg mL−1 M2PYK in the absence (black) and presence (red) of 10 µM T3. (F) Analytical gel-filtration elution profiles observed for 10-µL sample injection of 0.5 mg mL−1 M2PYK in the absence (black) and presence (red) of 5 mM Phe. Experiments in which Phe was added to the running buffer could not be monitored at 214 nm because Phe saturated the absorbance. Monitoring M2PYK (that has a low extinction coefficient) at 280 nm required a higher M2PYK concentration, which increased lower limits from 0.1 mg/mL to 0.5 mg/mL. Thermal shift assays performed at 0.5 mg/mL M2PYK (Fig. S2) provide complementary results to those observed by gel filtration.
Size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS) was also used to measure molecular masses for M2PYK forms (Fig. 2C). The initial large peak corresponds to the tetramer with a molecular mass of 214 kDa, and a second smaller peak at 53 kDa is the monomeric form. The analytical gel filtration and SEC-MALS results thus provide independent confirmation of tetrameric and monomeric M2PYK forms. The gel chromatography trace from the latter technique showed a poorly resolved peak that eluted between the monomer and tetramer peaks, which is consistent with a small amount of the M2PYK dimer; however, the resolution of the column used for SEC-MALS was not sufficient to measure its molecular mass unambiguously.
Kinetic profiles of M1PYK and M2PYK were determined for phosphoenolpyruvate (PEP) in the presence or absence of the allosteric effector F16BP at 37 °C and are summarized in Table 1. Using PEP as a substrate (without effector) M1PYK has an apparent Vmax value of 346 µmol per min per mg and an S0.5(PEP) = 0.05 mM; addition of the effector F16BP made essentially no difference to the kinetics. By contrast, addition of effector F16BP to M2PYK resulted in a nearly twofold increase in apparent Vmax (Fig. 2D) and a marked shift in the S0.5(PEP) value from 0.9 to 0.1 mM, with a concomitant drop in cooperativity (from a nH of 1.2 to 1.0). PEP cellular concentrations are estimated to lie between 0.02 and 0.5 mM (18). Activation of M2PYK by F16BP would therefore increase reaction rates by between four- and 10-fold over this PEP concentration range. For a given protein concentration (∼0.03 µM), an increase in F16BP concentration (from ∼2 µM to 30 µM) results in marked changes in apparent Vmax (Fig. S1C). (Note that the cellular concentration of PYK is estimated to be in the region of 0.1 mg/mL or 2 µM (18), considerably higher than the concentrations used for enzymatic assays). The addition of F16BP stabilizes M2PYK in a predominantly enzymatically active tetrameric form (Fig. 2B), and the 45% increase in tetramer concentration observed in response to the addition of effector molecule correlates closely with the increase in observed apparent Vmax (Table 1 and Fig. 2D). M2PYK enzymatic activity seems therefore to be regulated in part by its ability to dissociate into inactive monomers. Such a regulatory mechanism is a feature of so called V-type allosteric enzymes (19), but there are few well-characterized V-type systems.
Table 1.
Kinetic parameters for PEP
| Protein | Apparent Vmax (µmol per min per mg) | S0.5(PEP) (mM) | nH |
| M2PYK-WT | 116.3 (3.3) | 0.86 (0.08) | 1.2 |
| M2PYK-WT + 500 µM F16BP | 212.9 (1.4) | 0.10 (0.10) | 1.0 |
| M1PYK-WT | 345.8 (12.8) | 0.05 (0.05) | 1.0 |
| M1PYK-WT + F16BP | 355.6 (11.5) | 0.05 (0.05) | 1.0 |
| M2PYK-R489A | 87.0 (3.9) | 0.77 (0.11) | 1.2 |
Errors shown in parentheses. SEs for nH are 0.1–0.2. Kinetic assays were carried out at 37 °C at pH 7.4. Assay conditions: PBS (8.1 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl, and 137 mM NaCl, pH 7.4) buffer with saturating [ADP] = 2 mM, [KCl] = 100 mM, and [MgCl2] = 10 mM in the presence or absence of 500 µM of F16BP. PEP was serially diluted for 5 mM to 0.04 mM. The corresponding kinetic data are shown in Fig. S1.
Monomer–Dimer–Tetramer Equilibrium and Enzyme Activity of M2PYK (but Not M1PYK) Are Regulated by Allosteric Effectors, Including T3, Phenylalanine, and F16BP.
A selection of more than 50 metabolites from the glycolytic, tricarboxylic acid cycle and pentose phosphate pathways and other potential PYK modulators (amino acids, oxalic acid, tartaric acid, and T3) were tested against both wildtype (WT) M2PYK and M1PYK (Table S1). Several activators were identified for M2PYK, which included F16BP (AC50 = 6.5 µM), serine, and histidine, but none were identified for M1PYK. Only phenylalanine, oxalic acid, and oxaloacetic acid were identified as weak inhibitors for M1PYK, whereas several inhibitors were observed for M2PYK, including phenylalanine, tryptophan, alanine, and more weakly, oxalic acid and ribose 5-phosphate. Phenylalanine has an apparent IC50 of 0.24 mM for M2PYK (Table 2) and exhibits significantly poorer affinity for M1PYK (IC50 >1 mM). Phenylalanine demonstrates an interesting specificity against M2PYK because similar molecules (tyrosine, dopamine, octopamine, 2-amino-1-phenylethanol, and tryamine) failed to elicit a response. SEC was used to examine the effect of phenylalanine on the oligomeric state of M2PYK, and it clearly stabilizes M2PYK in a tetrameric form (Fig. 2F).
Table 2.
Inhibition and thermal shift assays
| M2PYK-WT |
M1PYK-WT |
|||
| Ligand | AC/IC50 (µM) | ΔTm (°C)* | AC/IC50 (µM) | ΔTm (°C) |
| F16BP | 6.5 (0.3) | 7 | No effect | No shift |
| Ser | >1 mM | 0 | No effect | No shift |
| His | >1 mM | 0 | No effect | No shift |
| Phe | 240 (0.1) | 4 | >1 mM | No shift |
| Nor | 9,300 (1.9) | 3 | No effect | No shift |
| T3 | 0.072 | 3 | >4 µM | No shift |
| Trp | <1 mM | 2 | No effect | No shift |
| Ala | >1 mM | 3 | No effect | No shift |
Errors are in parentheses.
Thermal shift assays were performed at pH 7.4 in PBS buffer using 0.5 mg/mL of enzyme in the absence and presence of 1 mM ligand. The corresponding thermal melt data are shown in Fig. S2.
T3 contains the phenylalanine substructure and is known to inhibit the human cytosolic thyroid hormone-binding protein p58 (20), a mutant form of M2PYK found in human epidermoid carcinoma cells (20–22). We now show it to be a potent inhibitor of M2PYK enzyme activity, with an IC50 = 72 nM (Table 2 and Fig. S1). Again, T3 was selective for M2PYK over M1PYK. Intriguingly, SEC showed that in contrast to phenylalanine, T3 inhibits tetramer formation and stabilizes M2PYK monomers (Fig. 2E).
The stabilizing effects of PYK modulators were analyzed using a thermal denaturation assay (Table 2 and Fig. S2). An increase in melting temperature (Tm) reflects ligand binding and reduced conformational flexibility. The addition of the allosteric activator F16BP to M2PYK apoenzyme shows the most dramatic increase in the Tm, from 48 °C to 55 °C. The addition of inhibitory amino acids phenylalanine, alanine, and tryptophan resulted in significant increases in Tm values (2–4 °C). The addition of norphenylephrine and T3 to M2PYK resulted in Tm shifts of 3 °C. At identical concentrations, these ligands had no effect on the Tm of M1PYK, confirming the enzymatic results and the preference of these ligands for binding to M2PYK over M1PYK.
X-Ray Structural Studies of R-State M1PYK, R-State M2PYK, and T-State M2PYK.
Peg-in-hole geometry locks M1PYK and active M2PYK in identical conformations.
Both the human unligated M1PYK and M2PYK complexed with ATP, oxalic acid, and F16BP (M2PYK-ATP/OX/F16BP; active R-state) were crystallized at a physiologically relevant pH of 7.2 and the structures refined at 2.85 and 2.55 Å (Table S2), respectively. The R-state complex of M2PYK ATP/OX/F16BP provides an example of a structure of a mammalian allosteric PYK with ATP bound in the active site. The human M1PYK structure presented here differs by 16 amino acid residues from the previously published rabbit (2, 23) M1PYK structures.
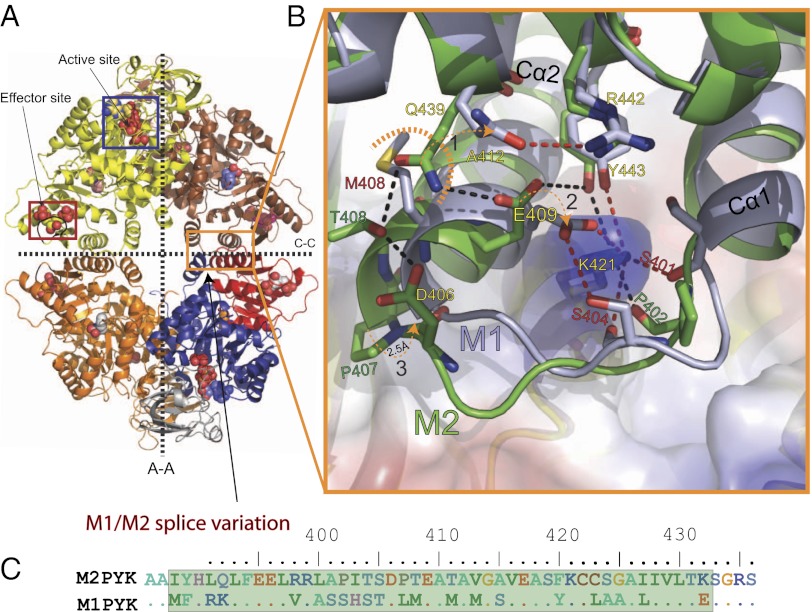
The M2PYK-ATP/OX/F16BP complex adopts a structure (Fig. 3A) similar to that of constitutively fully active M1PYK, with an rms fit for all Cα atoms (excluding the effector loops and the flexible B-domains) of ∼0.5 Å. The only region exhibiting any significant difference corresponds to the 22 amino acid splice difference (Fig. 3 B and C) between M1PYK and M2PYK, which makes up the “C-C” dimer interface between the C-domains in the tetramer. Key differences in hydrogen bonding across the C-C interface involve Lys421 (Fig. 3B). In the M1PYK structure, Cα1 and Cα2 helices form a tight clasp around Lys421, which makes a salt bridge with Glu409 and three additional hydrogen bonds with Ser401, Ser404, and Tyr443. In the M2PYK structure, the backbone of the linker between the Cα1 and Cα2 helices (402Pro-Ile-Thr-Ser-Asp-Pro407) adopts a significantly different conformation compared with the M1PYK linker (402Ser-His-Ser-Thr-Asp-Leu407), which pushes the helices further apart by 2.5 Å. This has the effect of relaxing the very tight peg-in-hole binding that was observed in the M1PYK structure, in which Lys421 acts as the peg and residues 390–420 form the hole (Fig. 3B).
Fig. 3.
M1PYK forms a tighter C-C interface than M2PYK. (A) The M2PYK tetramer is shown with the active and effector sites indicated. The bottom right chain has been colored to aid the identification of domains: domain-A (blue = residues 25–116 and 220–402), domain-B (gray = residues 117–219), and domain-C (red = residues 403–531). The large (A-A) and small (C-C) interfaces between monomers are shown as dashed lines. (B) Enlarged view of the dimer interface showing a comparison of the M1PYK and M2PYK side chains. M1PYK hydrogen bonds are shown in red and those of M2PYK in black (1). Gln439 is repelled by Met408 in M1PYK allowing Glu409 to form a salt bridge with Lys421 (2). (3) Cα2 of M1PYK is 2.5 Å closer to Lys421 than Cα2 of M2PYK. (C) Splice variant amino acids between M1PYK and M2PYK.
Allosteric inhibitor phenylalanine locks the M2PYK tetramer in the T-state.
To probe the allosteric mechanism of M2PYK we made a mutant, M2PYK-R489A. Replacement of Arg489 abolishes F16BP binding and prevents contamination by F16BP, which is commonly bound during purification of the protein from Escherichia coli culture. M2PYK-R489A has within experimental error the same S0.5(PEP) (0.77 ± 0.1 mM) as that of WT (0.86 ± 0.1 mM) (Table 1). A complex of M2PYK-R489A with phenylalanine was crystallized at pH 6.0 and the structure solved at a resolution of 2.9 Å (Table S2).
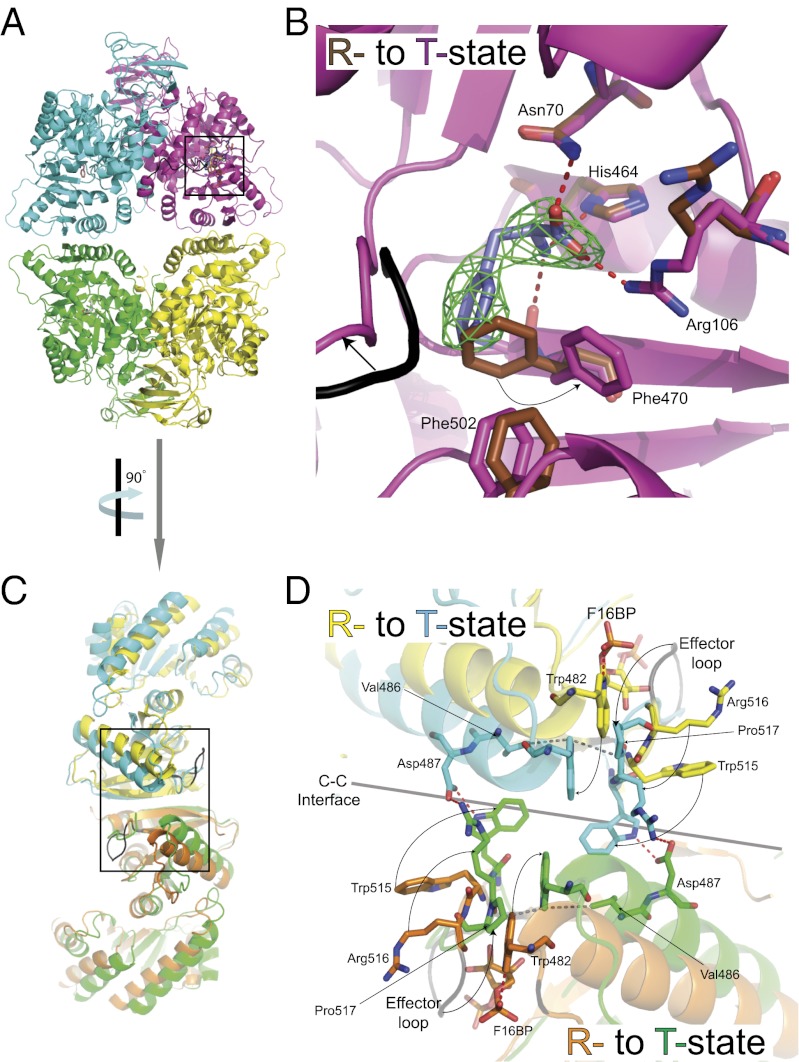
The phenylalanine complex adopts a T-state conformation in which each of the protomers (comprising the A and C domains) has rotated 13° from the active R-state M2PYK and M1PYK structures described above. Phenylalanine binds in an allosteric pocket located between the active and effector site (near the pivot point where the rigid body rotation occurs; Fig. 4 A and B). The T-state and R-state structures are shown in cartoon form in Fig. 1. Rotation of the protomers pulls Lys421 out of its binding pocket and destroys the peg-in-hole binding that stabilizes the C-C interface in the R-state M2PYK structure (Movie S1).
Fig. 4.
Allosteric inhibitor phenylalanine locks the M2PYK tetramer in the T-state. All ligands and interacting amino acids are shown as sticks; hydrogen bonds are shown as dashed red lines. The resolution of the electron density (Fo-Fc map, shown in green) map is 2.9 Å and is contoured at 3σ. (A) A cartoon representation of the T-state tetramer (M2PYK-Phe). (B) Enlargement of the Phe binding site. (C) Side view (rotated 90° to that of A) of the superposed R-state (M2PYK-ATP/OX/F16BP) and T-state structures (M2PYK-Phe). (D) Enlargement of the C-C interface highlighting conformational changes and side chain movements (arrows) as the protomers rotate from R- (effector bound) to T- (Phe bound) state.
The rigid body rotation of the protomers is similar to that observed for the T to R transition in Leishmania mexicana PYK (LmPYK) (24). In LmPYK the allosteric “rock-and-lock” regulation mechanism is controlled by a concerted rigid-body rocking motion of all four chains between an active (R) and inactive (T) state, with the effector (in this case F26BP) locking the active conformation by the formation of eight salt bridges across the C-C interface of the tetramer. The locking mechanisms that stabilize the R and T states are, however, very different between the two species. In M2PYK the rigid body rotation of the protomers allows the side chains of Trp515 and Arg516 of the (unoccupied) F16BP effector binding loop to swivel out and form salt bridges and hydrogen bonds across the C-C interface, stabilizing the tetramer in the inactive T-state. This conformation is further stabilized by the involvement of Trp515 and Trp482 in a series of stacking interactions across the interface (Fig. 4D and Movie S2).
Binding of the effector molecule F16BP causes the effector loop (514–523) to fold up round the effector, forcing the side chains of Trp515 and Arg516 to fold inward and breaking the T-state stabilizing Arg516…Asp487…Trp515 hydrogen bond network. The T-state-stabilizing hydrophobic stacking interactions are also disrupted by a unique role for the indole ring nitrogen of Trp482, which forms a hydrogen bond with the 1’-phospho group of FBP (Fig. 4D).
The inhibitory allosteric effect of phenylalanine on PYK activity has previously been reported, and a spectroscopic and biochemical study on rabbit kidney PYK showed the poor inhibition by phenylalanine of M1PYK relative to allosterically activated PYK (25). A high-resolution X-ray structure of a complex of alanine with rabbit M1PYK has been shown to adopt a standard R-state conformation that accommodates alanine bound in the same site as the Phe-humanM2PYK structure but without distorting the R-state conformation (26). It was also reported that amino acids with larger side chains could not be crystallized in complex with rabbit M1PYK, and this is consistent with the idea that the tight peg-in-hole binding across the dimer interface in the constitutively fully active M1 isoform is locking the R-state conformation, preventing entry of larger side chains into this newly defined phenylalanine allosteric pocket.
“Dock-rock-lock” model explains allosteric activation and inhibition of M2PYK.
The monomer-tetramer equilibrium of M2PYK, not observed in any other of the trypanosomatid or mammalian PYK isoforms, provides the first step in the activation in which the protomers “dock” together to form a tetramer (Fig. 1). The equilibria here are controlled by protein concentration and allosteric effector molecules. The initial step in forming an active complex is a docking across the C-C interface, which can be stabilized in the R-state by the peg-in-hole interaction or in the T-state by the Trp/Arg interactions as described in the phenylalanine complex. As the protomers rock 13° from T- to R-state in response to effector binding, the Trp-stabilized T-state interactions are broken and the disordered Lys421 (the peg) slots into the hole (formed by residues 390–420), thereby stabilizing the R-state (Fig. 3 A and B and Movies S1 and S2).
The concerted rocking motion of each of the protomers explains at the molecular level how the effector molecule F16BP can bind over 40 Å away from the active site and enhance enzyme activity. This motion is therefore similar in a number of ways to the rock-and-lock mechanism of LmPYK, although the residues involved in locking the protomers in a given T or R state are not conserved between the species. The 13° rigid body “rock” of M2PYK places Arg342 in position to prime the active site by stabilizing the short glycine-rich A6’ helix (295Gly-Asp-Leu-Gly-Ile-Glu-Pro301) of an adjacent protomer (Fig. S3). The involvement of adjacent protomers in shaping the active site provides a clear explanation for the inactivity of monomeric PYK and also therefore an explanation of the inhibitory effect of T3, which stabilizes monomeric M2PYK.
Inhibition of M2PYK by Phenylalanine and T3 Enhance Cell Proliferation.
M2PYK is frequently up-regulated in cancer cells (27), and it has been suggested that inactivating M2PYK would block metabolic flux, allowing build-up of glycolytic intermediates for macromolecular biosynthesis and tumor growth (28, 29). The structural and biochemical results described above provide mechanisms for phenylalanine, T3, and F16BP to act as allosteric inhibitors and activators with a high specificity for M2PYK over M1PYK. It was of interest therefore to determine whether these compounds would have an effect on cell growth.
We selected the cell line HCT-116 for this study because it is known to overexpress M2PYK (30). Addition of 5 mM Phe or 20 µM T3 to the cell culture media significantly increased cell proliferation compared with vehicle treated cells, whereas addition of 1 mM F16BP resulted in complete inhibition of cell growth (Fig. 1E). It has previously been shown that although F16BP is highly negatively charged it can enter the cell (31).
In vitro the enzymatic inhibition of p58 by T3 can be overcome by addition of high concentrations of F16BP (20). Another study showed a similar effect, with inhibition by phenylalanine (25). We were able to show comparable effects in cellular proliferation experiments, whereby the addition of 1 mM F16BP was able to abrogate the effects of both T3 and phenylalanine (Fig. 1E), providing strong evidence that M2PYK is indeed the cellular target. Together these data suggest that allosteric M2PYK activation can play a dominant role in regulating cancer cell growth.
There is increasing interest in M2PYK as a potential antiproliferative drug target, and stimulation of M2PYK activity (32–34) or inhibition (17) have both been explored. The reduction in size of tumor allografts observed using synthetic activators of M2PYK (34) fit with our observations that activation of M2PYK leads to the inhibition of proliferation of HCT-116 cells and supports the approach of developing small-molecule M2PYK activators for cancer therapy.
M2PYK as a Nutrient Sensor and Regulator of Cell Proliferation.
In this article we have described the detailed regulatory mechanisms of three small-molecule inhibitors and activators. The structural and biophysical data describe two molecular mechanisms for inhibiting M2PYK, as summarized in Fig. 1. T3 traps M2PYK in an inactive monomeric state, whereas phenylalanine traps M2PYK in an inactive tetrameric T-state. Despite these different inhibitory mechanisms each inhibitor has similar dramatic effects on cellular proliferation, suggesting that their interactions with M2PYK are biologically relevant.
There is a large body of published work on the biological roles of T3 (35). The most studied properties of thyroid hormones relate to their activities as gene regulators. However, there is a pool of unbound T3/T4 available at approximately nanomolar concentration (36), which regulates a number of nongenomic actions. In two breast cancer cell lines, increased T3 levels correlate with the degree of Warburg phenotype and increased level of M2PYK (36).
F16BP is effective in reducing the tissue damage associated with ischemia due to hypo-perfusion, and at concentrations in the millimolar range F16BP suppresses T-cell proliferation (37), which provides a potential therapy against inflammation and sepsis. Tyrosine phosphorylation has been shown to prevent binding of F16BP, thus preventing enzyme activation, and the ability of M2PYK to bind F16BP was inversely correlated with tumor growth in xenograft mice (38). In this context F16BP binding to M2PYK is acting as an inhibitor of cell proliferation, which correlates with our result.
Several reviews have mentioned that amino acids affect human M2PYK activity but without detailed kinetic data (6, 29), and one study on rabbit kidney PYK showed the poor inhibition by phenylalanine of rabbit M1PYK relative to allosterically activated PYK (25). The effect of phenylalanine on cancer cell proliferation has not been previously published, although in some early work a phenylalanine restricted dietary regimen has been shown to result in significant inhibition of tumor growth in mice (39, 40). It is compelling that phenylalanine and T3, which inhibit M2PYK enzyme activity, both show the same cellular phenotype in HCT-116 cells. Interestingly, earlier work showed that the effect of T3 on epithelial cell replication can be replaced by phenyalanine, and transformation strongly reduces both T3 and phenylalanine requirements for growth (41).
Phenylalanine is one of the essential amino acids and cannot be synthesized by mammals and requires constant monitoring. The allosteric pocket identified in the M2PYK-Phe crystal structure may therefore provide a potential feedback mechanism to block enzyme activity when phenylalanine is abundant in the cell, allowing metabolite build-up and cell proliferation. The various oligomeric states of M2PYK described here (Fig. 1) suggest how M2PYK may be acting as a complex nutrient sensor for the cell, responding directly or indirectly to fluctuations in oxygen (11), essential amino acids, hormones, and glucose.
Materials and Methods
M1PYK and M2PYK Production and Activity Measurements.
M1PYK and M2PYK were expressed and purified as described in SI Materials and Methods. Site-directed mutagenesis of M2PYK cDNA was performed using the QuikChange mutagenesis kit from Stratagene, according to the manufacturer’s instructions. Enzyme activity measurements in the absence and presence of modulators were performed using the standard lactate dehydrogenase coupled assay (SI Materials and Methods).
Crystallization and Structure Determination.
Single crystals of M1PYK and M2PYK were obtained as described in SI Materials and Methods, and diffraction data were collected at the Diamond synchrotron radiation facility in Oxfordshire, United Kingdom on beamline IO3 to a resolution of 2.55 Å (M2PYK-ATP/OX/F16BP), 2.9 Å (M2PYK-R489A-Phe), and 2.85 Å (M1PYK). All datasets were obtained from a single crystal flash-frozen in liquid nitrogen at 100 K. Structures were solved by molecular replacement as described in SI Materials and Methods. Atomic coordinates and the experimental structure factors for all structures have been deposited in the Protein Data Bank, with the following codes, M1PYK (3SRF), M2PYK-ATP/OX/F16BP (4FXF), and M2PYK-R489A-Phe (4FXJ).
Analytical Gel Chromatography.
M1PYK and M2PYK were purified under identical conditions, and highly purified samples of both isozymes were loaded independently onto a Superdex 200 PC 3.2/30 gel-filtration column. Unless stated otherwise, protein samples were analyzed at physiologically relevant concentrations (0.1 mg/mL) (18). Ten-microliter samples were injected, and the column flow rate was maintained at 0.1 µL min−1. Separations and equilibration steps were performed in PBS without calcium and magnesium (PBS-CM) or PBS-CM supplemented with the appropriate concentration of F16BP, T3, or phenylalanine at 26 °C. Protein peaks were monitored using absorbance at both 280 and 214 nm. All samples were incubated overnight at 26 °C before analysis.
SEC-MALS Analysis.
SEC was carried out at room temperature at a flow rate of 0.5 mL min−1 using a Superdex 200 10/300 GL. The Superdex column was connected in-line with the following detectors; UV detector, a light scattering detector (Wyatt Technology), and a refractometer.
Thermal Shift Assay.
Thermal shift assays were performed essentially as described previously (24), except PBS buffer was used throughout.
Cell Growth Experiments.
HCT-116 growing (37 °C and 5% CO2) exponentially in McCoy’s 5A media plus 10% FCS (MS) were suspended with trypsin EDTA (trypsin, 0.05%; EDTA, 0.02%), and ∼32,000 cells per well (1.6 mL of 20,000 cells per mL) were plated into sterile six-well (35-mm) plastic culture dishes. After sufficient time for the cells to become adherent had elapsed (12–16 h) an Alamar blue assay (Invitrogen) was performed to ensure equivalent cell density between wells as per the manufacturer's instructions. The medium was changed (1.6 mL of media per well) to McCoy’s 5A media plus 10% dialyzed FCS (Dundee Cell Products, catalog no. DS1002) supplemented with either T3 (a 25-mM stock solution was prepared in 100 mM NaOH), F16BP (a 100-mM stock solution was prepared in McCoy’s 5A media, and the pH adjusted to 7.4 with NaOH or phenylalanine (an 80-mM stock solution was prepared in McCoy’s 5A media). All ligand stocks were filtered through a 0.2-μm filter. Cells were then grown for a further 72 h and were collected by suspension in trypsin-EDTA and counted in a hemocytometer. Ligands used in this study interfere with metabolic pathways (TCA and glycolysis), leading to difficulties interpreting metabolic-based proliferation assays.
Supplementary Material
Acknowledgments
We thank Dr. J. Dornan for excellent advice and practical help, and the staff at the Synchrotron facilities at Diamond, United Kingdom. This project has been funded by the Medical Research Council. The Centre for Translational and Chemical Biology and the Edinburgh Protein Production Facility were funded by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council. J.R.O. was funded by a Wellcome Clinical Training Fellowship 094417/Z/10/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3SRF, 4FXF, and 4FXJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217157110/-/DCSupplemental.
References
- 1.Yamada K, Noguchi T. Nutrient and hormonal regulation of pyruvate kinase gene expression. Biochem J. 1999;337(Pt 1):1–11. [PMC free article] [PubMed] [Google Scholar]
- 2.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 3.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 4.Bluemlein K, et al. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2(5):393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011;14(4):443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv L, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42(6):719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2(97):ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anastasiou D, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazurek S, Zwerschke W, Jansen-Dürr P, Eigenbrodt E. Effects of the human papilloma virus HPV-16 E7 oncoprotein on glycolysis and glutaminolysis: Role of pyruvate kinase type M2 and the glycolytic-enzyme complex. Biochem J. 2001;356(Pt 1):247–256. doi: 10.1042/0264-6021:3560247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwerschke W, et al. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1999;96(4):1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steták A, et al. Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 2007;67(4):1602–1608. doi: 10.1158/0008-5472.CAN-06-2870. [DOI] [PubMed] [Google Scholar]
- 15.Boxer MB, et al. Evaluation of substituted N,N’-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. J Med Chem. 2010;53(3):1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang JK, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20(11):3387–3393. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, et al. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol. 2010;79(8):1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flory W, Peczon BD, Koeppe RE, Spivey HO. Kinetic properties of rat liver pyruvate kinase at cellular concentrations of enzyme, substrates and modifiers. Biochem J. 1974;141(1):127–131. doi: 10.1042/bj1410127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton RL, Chen S, Xu XL, Grant GA. Transient kinetic analysis of the interaction of L-serine with Escherichia coli D-3-phosphoglycerate dehydrogenase reveals the mechanism of V-type regulation and the order of effector binding. Biochemistry. 2009;48(51):12242–12251. doi: 10.1021/bi901489n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Fukuda T, Parkison C, McPhie P, Cheng SY. Cytosolic thyroid hormone-binding protein is a monomer of pyruvate kinase. Proc Natl Acad Sci USA. 1989;86(20):7861–7865. doi: 10.1073/pnas.86.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 1991;266(25):16842–16846. [PubMed] [Google Scholar]
- 22.Ashizawa K, McPhie P, Lin KH, Cheng SY. An in vitro novel mechanism of regulating the activity of pyruvate kinase M2 by thyroid hormone and fructose 1, 6-bisphosphate. Biochemistry. 1991;30(29):7105–7111. doi: 10.1021/bi00243a010. [DOI] [PubMed] [Google Scholar]
- 23.Larsen TM, Benning MM, Rayment I, Reed GH. Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel. Biochemistry. 1998;37(18):6247–6255. doi: 10.1021/bi980243s. [DOI] [PubMed] [Google Scholar]
- 24.Morgan HP, et al. Allosteric mechanism of pyruvate kinase from Leishmania mexicana uses a rock and lock model. J Biol Chem. 2010;285(17):12892–12898. doi: 10.1074/jbc.M109.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consler TG, Woodard SH, Lee JC. Effects of primary sequence differences on the global structure and function of an enzyme: A study of pyruvate kinase isozymes. Biochemistry. 1989;28(22):8756–8764. doi: 10.1021/bi00448a012. [DOI] [PubMed] [Google Scholar]
- 26.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Differentiating a ligand’s chemical requirements for allosteric interactions from those for protein binding. Phenylalanine inhibition of pyruvate kinase. Biochemistry. 2006;45(17):5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 27.Kumar Y, et al. In vivo factors influencing tumour M2-pyruvate kinase level in human pancreatic cancer cell lines. Tumour Biol. 2010;31(2):69–77. doi: 10.1007/s13277-009-0010-3. [DOI] [PubMed] [Google Scholar]
- 28.Mazurek S, Eigenbrodt E. The tumor metabolome. Anticancer Res. 2003;23(2A):1149–1154. [PubMed] [Google Scholar]
- 29.Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog. 1992;3(1-2):91–115. [PubMed] [Google Scholar]
- 30.Zhou CF, et al. Pyruvate kinase type M2 is upregulated in colorectal cancer and promotes proliferation and migration of colon cancer cells. IUBMB Life. 2012;64(9):775–782. doi: 10.1002/iub.1066. [DOI] [PubMed] [Google Scholar]
- 31.Hardin CD, Roberts TM. Metabolism of exogenously applied fructose 1,6-bisphosphate in hypoxic vascular smooth muscle. Am J Physiol. 1994;267(6 Pt 2):H2325–H2332. doi: 10.1152/ajpheart.1994.267.6.H2325. [DOI] [PubMed] [Google Scholar]
- 32.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491(7424):458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh MJ, et al. 2-Oxo-N-aryl-1,2,3,4-tetrahydroquinoline-6-sulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2011;21(21):6322–6327. doi: 10.1016/j.bmcl.2011.08.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8(10):839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31(2):139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suhane S, Ramanujan VK. Thyroid hormone differentially modulates Warburg phenotype in breast cancer cells. Biochem Biophys Res Commun. 2011;414(1):73–78. doi: 10.1016/j.bbrc.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopes RP, et al. The effects of fructose-1,6-bisphosphate and dexamethasone on acute inflammation and T-cell proliferation. Inflamm Res. 2006;55(8):354–358. doi: 10.1007/s00011-006-6044-8. [DOI] [PubMed] [Google Scholar]
- 38.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 39.Demopoulos HB. Effects of low phenylalanine-tyrosine diets on S91 mouse melanomas. J Natl Cancer Inst. 1966;37(2):185–190. [PubMed] [Google Scholar]
- 40.Mitamura A, Yuen T, Duke PS, Demopoulos HB. Ultrastructure of S-91 mouse melanomas subjected to phenylalanine and tyrosine restriction in vivo. Am J Pathol. 1966;49(2):309–324. [PMC free article] [PubMed] [Google Scholar]
- 41.Lechner JF. Interdependent regulation of epithelial cell replication by nutrients, hormones, growth factors, and cell density. Fed Proc. 1984;43(1):116–120. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.