Abstract
The bacterial antiporter GadC plays a central role in the glutamate (Glu)-dependent acid resistance system, which protects enteric bacteria against the extreme acidity of the human stomach. Upon acid shock, GadC imports Glu into the cytoplasm, where Glu decarboxylases consume a cytoplasmic proton, which ends up as a “virtual” proton in the decarboxylated product γ-aminobutyric acid (GABA) and is then exported via GadC. It was therefore proposed that GadC counters intracellular acidification by continually pumping out virtual protons. This scenario, however, is oversimplified. In gastric environments, GadC encounters substrates in multiple carboxyl protonation forms (outside: Glu−, Glu0, Glu+; inside: GABA0, GABA+). Of the six possible combinations of antiport partners, Glu+/GABA0 results in proton influx, Glu0/GABA0 and Glu+/GABA+ are proton neutral, and Glu−/GABA0, Glu−/GABA+, or Glu0/GABA+ lead to proton extrusion. Which of these exchanges does GadC catalyze? To attack this problem, we developed an oriented GadC liposome system holding a three-unit inward pH gradient to mimic the conditions facing bacteria in the stomach. By assessing the electrogenicity of substrate transport, we demonstrate that GadC selectively exchanges Glu− or Glu0 with GABA+, resulting in effective proton extrusion of >0.9 H+ per turnover to counter proton invasion into acid-challenged bacteria. We further show that GadC selects among protonated substrates using a charge-based mechanism, rather than directly recognizing the protonation status of the carboxyl groups. This result paves the way for future work to identify the molecular basis of GadC’s substrate selectivity.
Keywords: transporter, reconstitution, proton pump
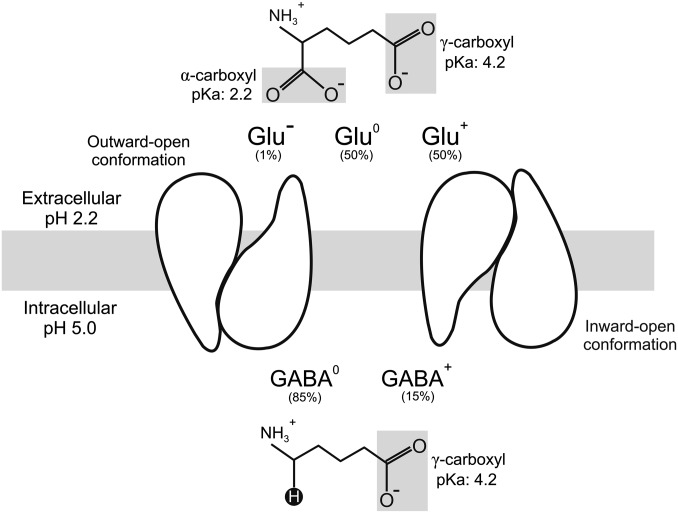
The companion article to this paper (1) showed that AdiC, the antiporter central to arginine-dependent extreme acid resistance in Escherichia coli and other enteric bacteria, acts as a decarboxylation-driven, outwardly directed virtual proton pump to counter intracellular acidification of bacteria in the stomach. The present work addresses the analogous issue for GadC, the key antiporter of the alternative Glu-dependent system (2, 3). We ask how the action of GadC produces acid resistance. It is known that upon acid shock, the antiporter delivers extracellular Glu to the acid-activated intracellular decarboxylases GadA and GadB (4) and expels the decarboxylated product GABA in a strict one-to-one exchange (5, 6). The decarboxylation reaction consumes a proton, which, as a “virtual proton,” ends up in a C–H bond of GABA and is exported from the cytoplasm (Fig. 1). It is widely accepted that removal of intracellular protons by this mechanism prevents cytoplasmic pH from falling to a dangerously low level below pH 5.
Fig. 1.
GadC in the gastric environment. GadC switches between outward- and inward-open conformations to import extracellular Glu while expelling cytoplasmic GABA, which carries the virtual proton in a C–H bond (black circle). The protonable carboxyl groups of Glu and GABA are highlighted.
This simple picture, however, requires additional experimental scrutiny. As with AdiC and arginine (1), GadC encounters its substrates in multiple protonation states under gastric acid-shock conditions (extracellular pH 1.5–3.5, intracellular pH 5). Specifically, Glu exists in three carboxyl-group forms (α,γ-doubly deprotonated Glu−, α-deprotonated, γ-protonated Glu0, and α,γ-doubly protonated Glu+); the last two represent the majority of extracellular Glu, and if imported would bring protons into the cell, exacerbating rather than resisting acid shock. Likewise, GABA presents two possible carboxyl-group forms for export (deprotonated GABA0 and protonated GABA+). Thus, GadC is faced with six possible combinations of antiport partners (Fig. 1). Among them, Glu+/GABA0 would produce proton influx, Glu0/GABA0 and Glu+/GABA+ would result in futile, proton-neutral antiport cycles, whereas virtual proton extrusion could be achieved by Glu−/GABA0, Glu−/GABA+, or Glu0/GABA+. It is not known which substrate forms are actually exchanged by GadC, and therefore the net proton flux coupled to GadC transport activity remains unclear. A simple calculation shows that if all substrate forms were transported equally well, at a typical gastric pH 2.2, GadC would import ∼0.4 H+ into the pH 5 cytoplasm of acid-resisting bacteria in each turnover.
Using a strategy as in the companion article (1), we attempt here to differentiate the six exchange options above by testing how substrate transport responds to an imposed negative-inside membrane potential set by adding the K+ ionophore valinomycin (Vln) to liposomes sustaining a 1,000-fold K+ gradient (1). This hyperpolarized potential would suppress electrogenic exchanges that generate outward positive current (Glu−/GABA0, Glu0/GABA+, or Glu−/GABA+), stimulate exchange moving net positive charge inward (Glu+/GABA0), and exert no effect on electroneutral exchanges (Glu0/GABA0 and Glu+/GABA+). Exploiting proteoliposomes reconstituted with functionally oriented antiporters facing a pH gradient mimicking acid-resistance conditions, we show that GadC catalyzes efficient virtual proton pumping by selecting against extracellular Glu+ for import while exclusively exporting GABA+. Further experiments suggest that GadC discriminates among the multiple protonation forms by recognizing net electric charge of substrates, rather than by close-up chemical scrutiny of carboxyl-group protonation.
Results
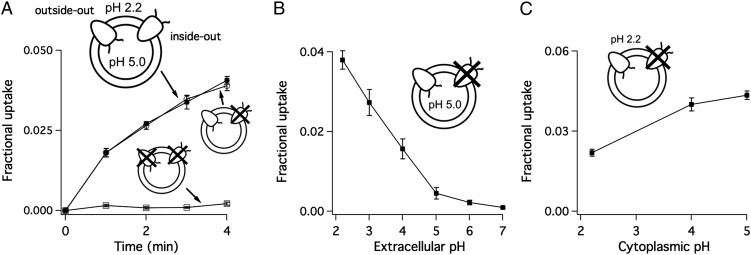
As in the companion article (1), examination of substrate transport under conditions resembling extreme acid shock requires two essential features of the reconstituted proteoliposome system. First, the liposomes must hold a three-unit inward pH gradient (outside pH 2.2; inside pH 5.0) to mimic pH conditions in acid-resisting bacteria. Second, all functional proteins must adopt the “outside-out” orientation, so that the extracellular side of the protein, as in the biological system, projects to the more acidic outside of the membrane (Fig. 2A). The first issue has been resolved (1), leaving us the task of establishing an oriented GadC system. To attack this problem, we introduced an innocuous cysteine mutation (I21C) in the cytoplasmic mouth of a cysteine-free GadC construct. Addition of a membrane-impermeant thiol reagent 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET) (7) to the extraliposomal solution completely inhibits the inside-out population while leaving the oppositely oriented, outside-out proteins functionally intact, thus establishing the sidedness of the reconstituted liposomes. Experimental documentation of the system’s full orientation, including the choice of the I21C cysteine substitution, is discussed in SI Methods and Figs. S1–S3.
Fig. 2.
Sidedness of GadC established by asymmetric pH. Icons represent the two orientations of GadC incorporated into liposomes, with the N- and C termini, which mark the transporter’s intracellular side, represented as wiggly lines. (A) The 3H-Gluex/GABAin exchange using I21C-GadC liposomes, with no treatments (filled square), MTSET in outside to silence inside-out GadC (open circle), or MTSET in both sides to react with all proteins (open square), in the presence of a three-unit inward pH gradient. (B) Effect of extracellular pH on 3H-Gluex/GABAin exchange. The extraliposomal pH of I21C-containing liposomes was varied from 2.2–7, with internal pH fixed at 5.0. Uptake of 3H-Gluex was recorded 4 min after initiation of transport. (C) Effect of cytoplasmic pH on 3H-Gluex/GABAin exchange. I21C liposomes, oriented outside-out (with external pH 2.2), were allowed to uptake Glu for 4 min in various inside pH ranging from 2.2 to 5.
In the AdiC reconstituted system (1), the inside-out protein is strongly inhibited by the pH conditions (outside pH 2.2/inside pH 5.0) used to mimic acid shock of bacteria in the stomach. Under the same pH setup, we found that I21C-containing liposomes pretreated with external MTSET to silence inside-out proteins, exhibit only a slight (<5%) reduction of external Glu/internal GABA (Gluex/GABAin) exchange compared with those receiving no treatment (Fig. 2A). In contrast, liposomes with the reagent on both sides show no substrate transport whatsoever (Fig. 2A). Thus, as in the AdiC system, the inside-out population of GadC contributes negligibly to overall transport in the presence of the acid-shock pH gradient. To understand this observation in more detail, we examine the separate effects of extracellular and cytoplasmic pH on GadC activity by using I21C-GadC oriented outside out. Gluex/GABAin exchange is greatly suppressed by extracellular-side pH above 4 (Fig. 2B) and slightly inhibited by intracellular-side pH lower than 4.0–5.0 (Fig. 2C). Therefore, the suppressed transport activity of inside-out proteins is due mainly to the exposure of GadC’s extracellular face to pH 5.0 inside the liposome, which can be considered in the context of acid shock to be “nonphysiologically basic.” The inhibitory effect of extraliposomal pH 2.2 on the transporter’s cytoplasmic face also contributes to silencing the inside-out population, but to a lesser extent. In the following experiments we will simply apply the three-unit inward pH gradient to orient the system using wild-type (WT) GadC, eliminating the need for chemical modification of the I21C mutant.
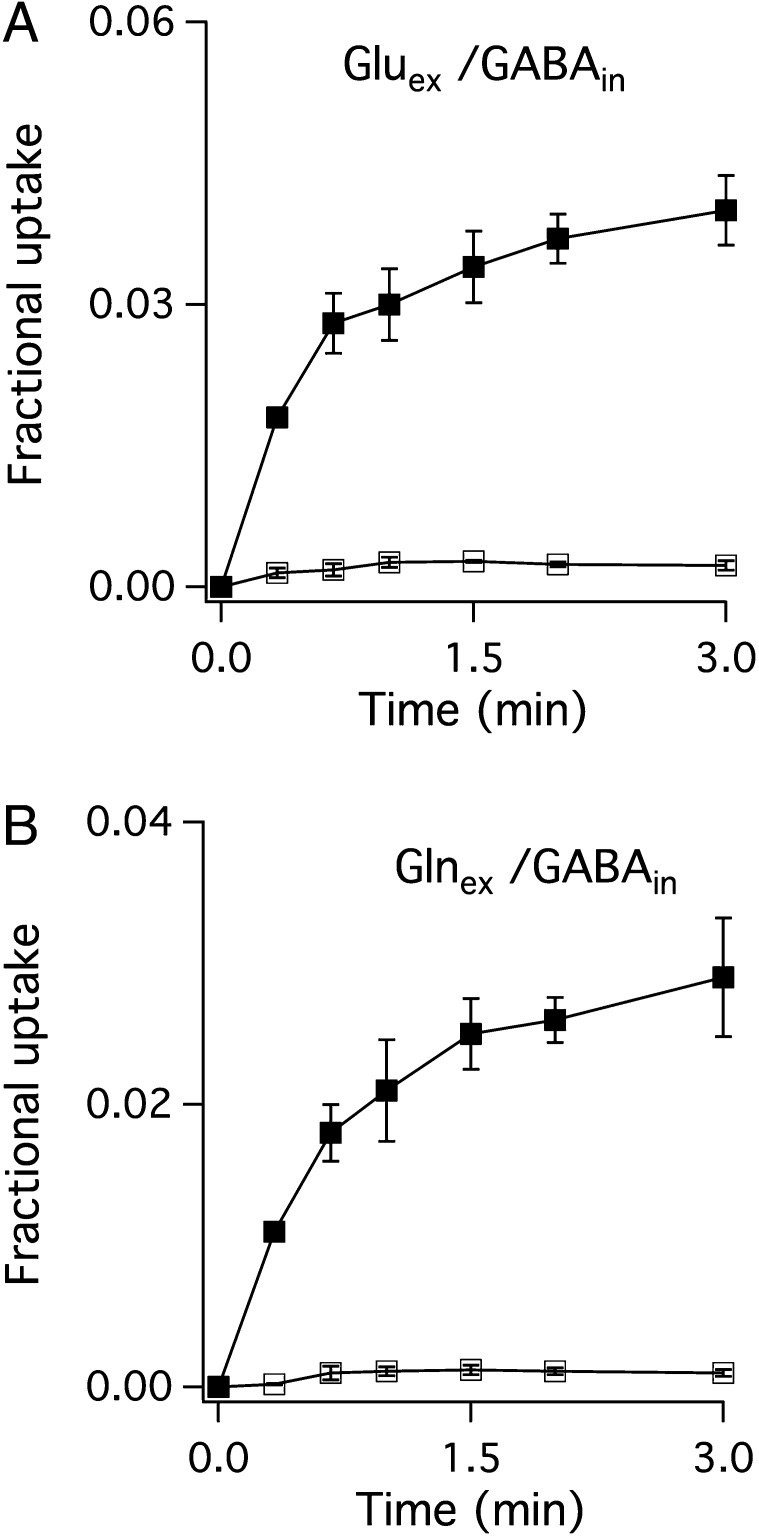
With these tools in hand, the stage is set to examine the electrogenicity of substrate antiport by GadC, a clue to the protonation states being transported (1). GadC catalyzes robust exchange of extraliposomal Glu (pH 2.2, 1% Glu−, 50% Glu0, 50% Glu+) and internal GABA (pH 5.0, 85% GABA0, 15% GABA+), as illustrated in Fig. 3A. Moreover, this transport activity is almost completely (>95%) inhibited by a large negative-inside membrane potential (Fig. 3A), a result indicating that nearly every turnover of GadC produces outward current at zero voltage. This observation limits the antiport partners to only three possibilities: 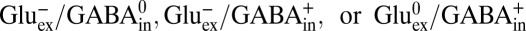 . Thus, we conclude that the outward-open conformation selectively excludes Glu+, whose α- and γ-carboxyls are both protonated.
. Thus, we conclude that the outward-open conformation selectively excludes Glu+, whose α- and γ-carboxyls are both protonated.
Fig. 3.
Substrate selectivity of GadC. WT GadC proteoliposomes (inside pH 5.0, outside pH 2.2) and a 1,000-fold outward K+ gradient, were used to quantify (A) 3H-Gluex/GABAin and (B) 3H-Glnex/GABAin exchanges, in the presence (open squares) or absence (filled squares) of Vln.
To test if it is GABA0 or GABA+ that serves as the internal exchange partner for Glu, we used glutamine (pH 2.2, 50%  , 50%
, 50% 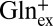 ), a robust substrate for GadC (6, 8), as a surrogate for external Glu. Again, a large negative voltage almost completely (>95%) abolishes transport (Fig. 3B), a result that cannot be explained unless GadC exclusively catalyzes
), a robust substrate for GadC (6, 8), as a surrogate for external Glu. Again, a large negative voltage almost completely (>95%) abolishes transport (Fig. 3B), a result that cannot be explained unless GadC exclusively catalyzes 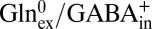 exchange, which moves net positive charge outward. Therefore, the inward-open conformation of GadC must selectively export GABA+ over the alternative GABA0 form.
exchange, which moves net positive charge outward. Therefore, the inward-open conformation of GadC must selectively export GABA+ over the alternative GABA0 form.
Taken together, the results above show that GadC mainly imports Glu− or Glu0 while exporting GABA+. However, it is difficult to quantify the selectivity from these experiments (Fig. 3 A and B), as the Vln-insensitive part of the transport is too small (∼5%) to be measured accurately. We can nevertheless obtain a conservative lower limit of substrate selectivity by setting the rare exchange cycles involving 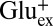 import or
import or 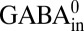 export at an overestimated 10% of total transport. This leads to the conclusion that the outward-open conformation of GadC must be at least 10-fold selective for Glu− and Glu0 over Glu+, and that the inward-open conformation is >100-fold selective for GABA+ over GABA0 (calculation described in Methods).
export at an overestimated 10% of total transport. This leads to the conclusion that the outward-open conformation of GadC must be at least 10-fold selective for Glu− and Glu0 over Glu+, and that the inward-open conformation is >100-fold selective for GABA+ over GABA0 (calculation described in Methods).
Before proceeding, three possible artifacts that would undermine the conclusions above must be eliminated. First, any nonspecific inhibitory effect of Vln on substrate exchange, irrelevant to the negative membrane potential, is negligible because the ionophore does not affect transport in the absence of a K+ gradient (Fig. S4). Second, the hyperpolarized membrane potential does not drive GadC into nonfunctional states, because it has a minimal effect on GABAex (pH 2.2, 1% GABA0, 99% GABA+)/GABAin (pH 5.0, 85% GABA0, 15% GABA+) exchange (Fig. S5), which would be primarily electroneutral (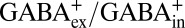 ). Third, to confirm that the asymmetric pH sufficiently suppresses contaminating signals from inside-out WT proteins as argued above, we repeated the voltage-inhibition experiments using the I21C-based strategy to fully orient the system outside out and obtained essentially identical results (Fig. S6).
). Third, to confirm that the asymmetric pH sufficiently suppresses contaminating signals from inside-out WT proteins as argued above, we repeated the voltage-inhibition experiments using the I21C-based strategy to fully orient the system outside out and obtained essentially identical results (Fig. S6).
To gain insight into the mechanism by which GadC distinguishes among protonation forms, we ask if the protein recognizes differences in carboxyl protonation status or merely the substrate net charge. For instance, the inward-open conformation of GadC selects GABA+, which differs from GABA0 in both the protonated γ-carboxyl and the overall charge. Which difference does the protein perceive in choosing the minor GABA+ over the predominant GABA0 form? If the protein recognizes the protonation status of the γ-carboxyl directly by close chemical interaction, we expect that it would not differentiate Gln+ and Gln0, which have the same neutral, unprotonatable side chain. This expectation, however, is contrary to fact. Exchange between Glnex (pH 2.2, 50% 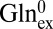 , 50%
, 50% 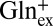 ) and Glnin (pH 5.0, 0.2%
) and Glnin (pH 5.0, 0.2% 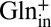 ) is almost completely inhibited by hyperpolarization (Fig. 4A), indicating that GadC selectively recruits Gln+ and rejects Gln0 from the intracellular side. This result strongly suggests that the inward-open conformation recognizes GABA+ by virtue of its +1 charge rather than by the protonation of its γ-carboxyl group.
) is almost completely inhibited by hyperpolarization (Fig. 4A), indicating that GadC selectively recruits Gln+ and rejects Gln0 from the intracellular side. This result strongly suggests that the inward-open conformation recognizes GABA+ by virtue of its +1 charge rather than by the protonation of its γ-carboxyl group.
Fig. 4.
Substrate selectivity of GadC. (A) 3H-Glnex, pH 2.2/Glnin, pH 5.0 and (B) 3H-GABAex, pH 3.0/GABAin, pH 5.0 exchanges were examined in WT GadC liposomes holding a 1,000-fold outward K+ gradient, in the presence (open squares) or absence (filled squares) of Vln.
Our analysis proceeds to the outward-facing conformation, which selects against extracellular Glu+. Glu+ differs from Glu0 and Glu– in three respects: the protonated α-carboxyl, the protonated γ-carboxyl, and the +1 valence. Which of these differences makes Glu+ a poor ligand for GadC? Two lines of evidence provide hints regarding this question. First, as shown above, GadC rarely imports 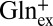 in exchange for
in exchange for  (Fig. 3B) or
(Fig. 3B) or 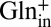 (Fig. 4A). Second, we performed GABAex/GABAin exchange experiments using a less steep pH gradient (outside pH 3.0, 5% GABA0, 95% GABA+; inside pH 5.0, 85% GABA0, 15% GABA+), which is still sufficient to maintain the sidedness of the system (Fig. S7). Under this condition, transport is suppressed ∼30% by negative voltage (Fig. 4B), indicating that
(Fig. 4A). Second, we performed GABAex/GABAin exchange experiments using a less steep pH gradient (outside pH 3.0, 5% GABA0, 95% GABA+; inside pH 5.0, 85% GABA0, 15% GABA+), which is still sufficient to maintain the sidedness of the system (Fig. S7). Under this condition, transport is suppressed ∼30% by negative voltage (Fig. 4B), indicating that 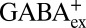 is ∼10-fold less preferred than
is ∼10-fold less preferred than 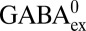 for import. Thus, the outward-open conformation of GadC appears to reject substrates with +1 valence, regardless of whether the charge is from the protonation of α-carboxyl (Gln+), γ-carboxyl (GABA+), or both (Glu+).
for import. Thus, the outward-open conformation of GadC appears to reject substrates with +1 valence, regardless of whether the charge is from the protonation of α-carboxyl (Gln+), γ-carboxyl (GABA+), or both (Glu+).
Discussion
AdiC and GadC, the two antiporters central to the extreme acid resistance response of enteric bacteria, are widely thought to counter intracellular acidification using a virtual proton pumping mechanism. In this picture, the extracellular substrate is imported and the decarboxylated product stoichiometrically expelled along with a virtual proton in the C–H bond formed in replacing the carboxyl group. In the extremely acidic gastric environment, however, a predominant fraction of extracellular substrates—arginine for AdiC and Glu for GadC—are protonated on carboxyl groups and could potentially carry protons into the cytosol, leading to futile exchange cycles or disastrous net proton influx. This issue, previously recognized as a serious threat to undermine the validity of the virtual proton pumping mechanism (2, 9), is addressed in these two studies.
The companion article (1) demonstrated that AdiC rejects extracellular Arg2+, greatly diminishing the possibility of proton entry via the protonated α-carboxyl. Here we show that GadC selectively imports Glu− or Glu0 while exporting GABA+. As a result, any dissociable proton smuggled into the cytosol by Glu0 is captured and returned to the extracellular side on the γ-carboxyl of GABA+. This ability of AdiC and GadC to discriminate substrate protonation forms ensures effective virtual proton pumping to defend bacteria against acid shock; we estimate that AdiC and GadC, with typical gastric pH 2.2 and cytoplasmic pH 5, remove at least 0.8 and 0.9 H+, respectively, from the cytosol in each antiport cycle. Our results contradict a recently proposed “proton influx” model (3, 10), which posits that these antiporters contribute to a positive membrane potential to produce an energy barrier for proton invasion. The substrate selectivity properties documented here show that AdiC and GadC should instead polarize the membrane in the negative direction.
In the studies of the present and the companion articles (1), we have sought to examine if AdiC and GadC discriminate among variously protonated substrates by recognizing carboxyl protonation states locally or by sensing the net charge of the substrate. These alternative scenarios have distinct mechanistic implications and would suggest different experimental approaches for future work. The former implies intimate chemical involvement of a few residues at the substrate binding site, whereas the latter invokes electrostatic effects, wherein charged residues, perhaps sitting on the wall of the translocation pathway, interact with substrates at a distance. Our results suggest that both of these antiporters use a charge-based mechanism, which, as also discussed in the companion article (1), makes biological sense. For instance, the outward-open conformation of GadC, in addition to selecting against Glu+ from the extracellular side, must also efficiently unload GABA+ recruited from the cytoplasm. The simplest way to achieve such a dual physiological role is for this conformation to be hostile to the +1 valence, rather than by recognizing the protonated α-carboxyl, which GABA+ lacks, to differentiate Glu+ from Glu0 and Glu−. The inward-open conformation, which needs to reject GABA0 and discharge Glu0 or Glu−, also adhere to this electrostatic picture.
We acknowledge, however, that a selectivity mechanism based on net charge is not as strongly supported with GadC as it is with AdiC. A “logic-gate” mechanism is possible wherein a single residue, or localized region of the outward-open conformation, monitors both α- and γ-carboxyls of Glu through H bonding and acts as a gatekeeper that prevents doubly protonated extracellular Glu+ from being transported. This alternative mechanism is far more complicated in molecular detail, however, as it requires intricate molecular logic in which a single gatekeeper rejects α-protonated Gln+ while selecting α-protonated Glu0, and rejects γ-protonated GABA+ while selecting γ-protonated Glu0.
A final issue is also worthy of note. We have no direct evidence that the outward-open conformation, which excludes Glu+, also selects between Glu− and Glu0. We suggest, however, that Glu0 might be the preferred substrate for import, because both Gln0 and GABA0 are robustly imported (Figs. 3 and 4). It follows that in the gastric environment, Glu0, which is ∼50-fold more abundant than Glu−, is likely to be the main species imported.
In light of the reports in the present and the companion articles (1), and many previous studies, we can now be confident that AdiC and GadC act as virtual proton pumps protecting enteric bacteria against extreme acid. However, many important questions remain unanswered. At the molecular level, a particular challenge is to identify the molecular basis of the charge-based substrate selectivity mechanism uncovered here. These antiporters are dormant at neutral pH and come into action only upon extreme acid shock; how do they sense the sudden pH drop? How does the cell dissipate the electrical imbalance arising from virtual proton pumping? The reconstituted systems developed here can serve as useful tools with which to attack these questions, as they permit analysis of the purified transporters in reduced, defined conditions reflecting acid shock in the stomach. Crystal structures of AdiC and GadC in multiple conformations are a great enhancement to experimental design, but caution must be taken in reading these structures, which, as we have shown for AdiC, can be mechanistically misleading in the absence of close functional analysis carried out in parallel.
Methods
Biochemical Procedures.
The gadC gene was cloned from E. coli BL21(DE3) and inserted into the pASK-IBA2 vector (9) between the XbaI and HindIII restriction sites. A thrombin-cleavable hexahistidine tag (HHHHHHSGGLVPRGSGT) was placed between the initiator methionine and the GadC sequence. The I21C mutant in a cysteine-less background (C60S/C247S/C380S) was constructed using standard two-step PCR and was confirmed with sequencing. To express GadC, transformed E. coli BL21(DE3) cells were grown in terrific broth at 37 °C to A600 of 1.0 and induced with 0.2 mg/L anhydrotetracycline for 1 h. Protein purification and reconstitution were exactly as in the companion article (1).
Functional Orientation of GadC in Reconsituted Liposomes.
The procedure for producing liposomes harboring fully oriented outside-out GadC is described in detail in SI Methods. In brief, a mutant containing a single cysteine (I21C) exposed near the substrate’s intracellular entryway was used for reconstitution. The liposomes were loaded with the desired substrate by freeze–thaw soniscation and treated with the membrane impermeant thiol reagent MTSET to inhibit all proteins in the inside-out orientation. The liposomes were then spun through G-50 columns to remove external substrates and MTSET. The resulting sample could then be adjusted to the desired extraliposomal composition for transport experiments.
Functional Analysis.
Flux assays were carried out similarly to those previously described (1). Proteoliposomes suspended in 100 mM K2SO4, 50 mM citric acid, KOH pH 5.0 were loaded with 5 mM substrate by freeze–thaw cycles and spun through Sephadex G-50 columns equilibrated with 100 mM Na2SO4, 1 mM citric acid, NaOH pH 5.0. The flow through was diluted into two-volume flux buffer [FB, 100 Na2SO4, 0.1 mM K2SO4, adjusted to pH 2.2–3 with 25 mM glycine, pH 4–5 with 25 mM citric acid, pH 6 with 25 mM 2-(N-morpholino)ethanesulfonic acid (MES), or pH 7 with 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS)] in the presence or absence of 1 μg/mL Vln, immediately before the flux experiment. For experiments requiring various cytoplasmic pH values, liposomes collected from the G-50 column were first dialyzed against 1,000 volumes pH buffer (100 Na2SO4, 0.1 mM K2SO4, pH 2.2, 1 mM glycine or pH 4–5, 1 mM citric acid) for 1 h to adjust internal pH, and then diluted into two-volume pH 2.2 FB to change external pH shortly before beginning the experiment. A total of 50 μM 3H-labeled Glu, GABA, or Gln (Perkin-Elmer, ∼2.5 μCi/mL) was added to liposome samples to initiate substrate exchange. At desired time points, the transport reaction was stopped by passing through G-50 columns. In all figures, each point represents the mean ± SE of three to five independent experiments.
The selectivity, SA, for substrate protonation form A preferred over form B is calculated as SA = TA/TB * [B]/[A], where TA/TB is the relative steady state uptake for the two protonation forms, obtained by comparing the Vln-sensitive and -insensitive parts of the transport, and [B], [A] is the concentration ratio of the protonation forms, given by the appropriate pKa values and pH.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant R01-GM089688.
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301444110/-/DCSupplemental.
References
- 1. Tsai, M-F, Miller C (2013) Substrate selectivity in arginine-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci USA 110:5893–5897. [DOI] [PMC free article] [PubMed]
- 2.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181(11):3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster JW. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat Rev Microbiol. 2004;2(11):898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 4.De Biase D, Pennacchietti E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol. 2012;86(4):770–786. doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- 5.Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419(6908):715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 6.Ma D, et al. Structure and mechanism of a glutamate-GABA antiporter. Nature. 2012;483(7391):632–636. doi: 10.1038/nature10917. [DOI] [PubMed] [Google Scholar]
- 7.Holmgren M, Liu Y, Xu Y, Yellen G. On the use of thiol-modifying agents to determine channel topology. Neuropharmacology. 1996;35(7):797–804. doi: 10.1016/0028-3908(96)00129-3. [DOI] [PubMed] [Google Scholar]
- 8. Lu P, et al. (2013) L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res, 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed]
- 9.Fang Y, Kolmakova-Partensky L, Miller C. A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J Biol Chem. 2007;282(1):176–182. doi: 10.1074/jbc.M610075200. [DOI] [PubMed] [Google Scholar]
- 10.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186(18):6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.