Abstract
Protein quality control systems protect cells against the accumulation of toxic misfolded proteins by promoting their selective degradation. Malfunctions of quality control systems are linked to aging and neurodegenerative disease. Folding of polypeptides is facilitated by the association of 70 kDa Heat shock protein (Hsp70) molecular chaperones. If folding cannot be achieved, Hsp70 interacts with ubiquitylation enzymes that promote the proteasomal degradation of the misfolded protein. However, the factors that direct Hsp70 substrates toward the degradation machinery have remained unknown. Here, we identify Fes1, an Hsp70 nucleotide exchange factor of hitherto unclear physiological function, as a cytosolic triaging factor that promotes proteasomal degradation of misfolded proteins. Fes1 selectively interacts with misfolded proteins bound by Hsp70 and triggers their release from the chaperone. In the absence of Fes1, misfolded proteins fail to undergo polyubiquitylation, aggregate, and induce a strong heat shock response. Our findings reveal that Hsp70 direct proteins toward either folding or degradation by using distinct nucleotide exchange factors.
Keywords: Hsp110, proteasome, triage, ubiquitin ligase, Ubr1
Cells use quality control (QC) mechanisms to counteract the cytotoxic accumulation of misfolded proteins triggered by internal damage and environmental stressors. QC mechanisms selectively recognize and target misfolded polypeptides to folding-promoting factors or remove them from the cell by degradation. Malfunction of QC mechanisms results in deleterious consequences and is linked to many pressing human maladies including neurological disorders as well as to aging (1, 2).
In eukaryotes, misfolded cytosolic or nuclear proteins are degraded by the ubiquitin-proteasome system (UPS) (3). Proteasomal degradation is induced by polyubiquitylation of the substrate, which labels the protein for degradation by the 26S proteasome. In this process, a set of enzymes is required to activate (E1 enzyme) and selectively conjugate (E2 and E3 enzymes) ubiquitin to the misfolded substrate or its growing polyubiquitin chain (4). E3 ligases involved in QC recognize structural characteristics of a variety of misfolded proteins. For example, the nuclear ubiquitin E3 ligase San1 targets misfolded proteins to proteasomal degradation by recognizing exposed hydrophobicity of misfolded proteins (5).
Molecular chaperones of the 70 kDa heat shock protein (Hsp70) class promote protein folding and prevent aggregation by binding and shielding hydrophobic peptide segments of client proteins (6, 7). The association of Hsp70 with substrate is controlled by its ATPase activity. When the Hsp70 carries ATP, substrate can bind and release with fast kinetics. ATP hydrolysis is stimulated by cochaperones of the Hsp40 class that directly associate with and deliver substrates to the Hsp70. Substrate remains associated with Hsp70 until ADP release is triggered by nucleotide exchange factors (NEFs) and ATP rebinds. As an outcome, NEFs trigger substrate release from Hsp70s.
Interaction of Hsp70 with substrates is coordinated with QC mechanisms so that misfolded proteins are triaged between folding and degradation (8–10). For example, in metazoan cells, the Hsp70-interacting E3 ligase Carboxyl-terminus of Hsp70 Interacting Protein (CHIP) induces ubiquitin-proteasome–mediated degradation of misfolded proteins (11, 12). In yeast, a functional Hsp70 system is required for ubiquitylation and proteasomal degradation of many misfolded proteins (13, 14). Three E3 ligases, Hul5, San1, and Ubr1, have been shown to ubiquitylate misfolded proteins that depend on Hsp70 for their degradation, but how Hsp70 and the E3 ligases coordinate their interactions is not known (15–19). NEFs are candidates that potentially could coordinate QC by releasing misfolded proteins from Hsp70 but up until now, their roles in this process have been undefined.
Here we report that the NEF Fes1 is a key triaging factor in protein QC that it is specifically required for targeting misfolded proteins recognized by Hsp70 to the UPS. Our results support a model in which Fes1 facilitates the selective release of misfolded proteins from the Hsp70 system, thereby promoting their destruction by the ubiquitin-proteasome machinery.
Results
Cells Lacking Fes1 Are Hypersensitive to Induced Protein Misfolding and Display a Strong and Constitutive Heat Shock Response.
Fes1 is a biochemically well-characterized yeast Hsp70 NEF of the Heat shock binding protein 1 (HspBP1)-class with no assigned cell biological function (20, 21). Using a reverse genetics approach, we set out to investigate the involvement of Fes1 in QC. We constructed a fes1∆ strain and could confirm the previously reported sensitivity to high temperature (Fig. S1A) (22). Importantly, fes1∆ cells were also found to be hypersensitive to protein misfolding induced either by the toxic proline ring analog azetidine-2-carboxylic acid or the toxic arginine analog canavanine (Fig. 1A). The sensitivity of fes1∆ cells to distinct conditions that induce protein misfolding implicates Fes1 in QC.
Fig. 1.
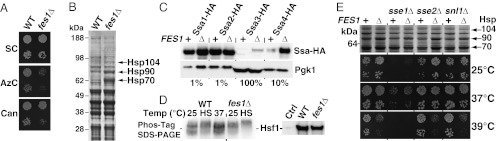
Inactivation of FES1 results in hypersensitivity to induced protein misfolding and strong activation of the heat shock response. (A) fes1Δ strains are hypersensitive to the amino acid analogs azetidine-2-carboxylic acid (AzC, 0.1 μM) and canavanine (Can, 3.6 mM). (B) Total protein extracts from WT and fes1Δ cells grown in YPD at 25 °C separated by SDS/PAGE and stained with Coomassie Brilliant Blue. The arrows mark the positions of induced Hsp70, Hsp90, and Hsp104 (Results). (C) Western analysis of HA-epitope tagged Hsp70s Ssa1, Ssa2, Ssa3, and Ssa4 expressed from their endogenous loci in WT and fes1Δ cells. Protein extract gel loading was adjusted as indicated (%). (D) Hsf1-Myc hypo- and hyperphosphorylation status in WT and fes1Δ cells was monitored using a gel shift assay based on Phos-Tag. Heat shocked (HS) cells were transferred from 25 °C to 37 °C for 30 min. Standard SDS/PAGE (Right). (E) Heat shock induction in Hsp70 NEF single- and double-mutant strains analyzed as in B and growth phenotypes at different temperatures.
To further investigate the physiological role of Fes1, we analyzed the overall protein profile of fes1∆ cells grown under nonstressful conditions (Yeast extract peptone dextrose, YPD, 25 °C) by SDS/PAGE. Three protein species migrating at positions between the 64- and 97-kDa size standards were markedly induced in samples from fes1∆ cells compared with the WT control (Fig. 1B). After purification, using denaturing anion exchange chromatography, these proteins were identified by MS as Hsp70 (70 kDa), Hsp90 (81 kDa), and Hsp104 (102 kDa), a finding that was confirmed by Western analysis (Fig. S1B). Further analysis of Stress-seventy class A (Ssa)-class Hsp70 proteins in fes1Δ cells revealed an expression pattern that was strikingly similar to that resulting from a strong and constitutive heat shock response in WT cells (Fig. 1C). Whereas SSA2 is a constitutively expressed gene that is unresponsive to stress, SSA1, SSA3, and SSA4 are all transcriptionally induced by heat shock factor (Hsf1) in response to cytoplasmic protein folding stress (23). This response is triggered by accumulation of misfolded proteins in the cytoplasm via a mechanism that relies on hyperphosphorylation of Hsf1 (24). We directly tested if Hsf1 was constitutively activated in fes1Δ cells under nonstressful growth conditions by assessing the phosphorylation status of Hsf1 using phosphoprotein gel shift analysis. This assay revealed that Hsf1 was hyperphosphorylated in the fes1Δ mutant in comparison with WT cells (Fig. 1D). Taken together, our data clearly show that the heat shock response is constitutively active in fes1Δ cells at nonstressful conditions, suggesting that the cells accumulate misfolded proteins in the cytoplasm.
As a supporting experiment to test whether fes1Δ mutants accumulate misfolded proteins, we used an aggregation reporter consisting of a fusion between highly stable GFP and thermolabile firefly luciferase that unfolds after heat shock and coaggregates with available misfolded proteins (25). Before heat shock, both WT and fes1Δ cells grown at 25 °C expressed luciferase-GFP evenly in the cytoplasm (Fig. S1C), with 9.3% and 25.0% of the cells containing luciferase-GFP aggregates, respectively. After heat shock at 37 °C for 30 min, 20.7% and 42.7% of the WT and fes1Δ cells, respectively, contained aggregates.
Because Fes1 represents only one of the three eukaryotic classes of Hsp70 NEFs present in the cytoplasm of yeast, we asked whether the Hsp110-class members Sse1 and Sse2 or the Bcl2-associated athanogene (BAG)-class member Snl1 have a similar or overlapping role in protecting cells against misfolded proteins (26, 27). Remarkably, we found a strong constitutive heat shock response to be a specific property of fes1Δ cells because none of the other mutants (sse1Δ, sse2Δ, and snl1Δ) displayed high-level expression of Hsp70, Hsp90, and Hsp104 (Fig. 1E). This is particularly striking in the light of the fact that the sse1Δ mutation causes a much more severe growth defect than fes1Δ (Fig. 1E). Moreover, NEF double mutants accumulated Hsp70, Hsp90, and Hsp104 proteins to similar extents as fes1Δ single mutants, and none of the double mutations exacerbated the growth phenotypes of the individual mutations.
The strong heat shock response in fes1Δ cells prompted us to perform a series of supporting experiments to investigate the importance of the induction of heat shock proteins for fes1Δ cells (Fig. S2). First, we partially attenuated the activity of the essential Hsf1 by removal of the C-terminal activation domain (hsf1ΔCT) (28). When fes1Δ was combined with this mutation, the hsf1ΔCT mutation attenuated the induction of Hsp70 and Hsp90, which went along with a strong synthetic phenotype; the double mutant displayed extremely poor growth at 30 °C (Fig. S2 A and B). Similarly, when fes1Δ was combined with deletions of genes encoding either cotranslationally acting Hsp70 proteins (ssb1Δ, ssb2Δ) or the inducible Hsp70 system (ssa1Δ, ssa3Δ, ssa4Δ) (29) as well as a deletion of HSP104, which encodes the main cytoplasmic disaggregation machine in Saccharomyces cerevisiae (30), all resulted in severe synthetic growth defects, in particular at higher temperature (Fig. S2C). Together, these findings demonstrate that fes1Δ cells depend on and induce multiple chaperone systems to cope with the defects associated with the lack of Fes1.
Fes1 Is Specifically Required for UPS-Dependent Degradation of Misfolded Proteins.
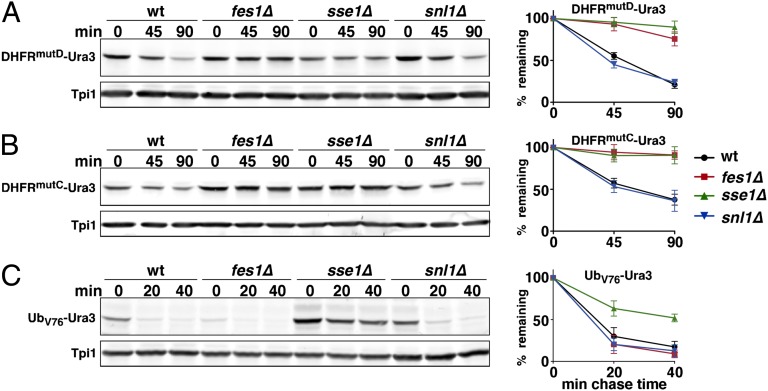
The observation that misfolded proteins accumulate in fes1∆ cells prompted us to ask if Fes1 is a general factor required for degradation of misfolded proteins associated with Hsp70. Initially, we took advantage of two mutant versions of the globular and compactly folded dihydrofolate reductase (DHFR) that are metabolically unstable at 30 °C (Fig. S3). The basic characterization of these DHFR mutants is presented in Figs. S3–S8. Briefly, degradation of reporter proteins (DHFR-Ura3) carrying the DHFRmutC or DHFRmutD modules was found to be dependent on the ubiquitin-proteasome pathway because their degradation was severely impaired by mutations in genes encoding UPS components (uba1-ts, ubc4∆ ubc5∆, ubr1∆, and ump1∆). Moreover, degradation of these proteins (DHFR-Ura3) requires the Ssa-class of Hsp70 and Hsp40 (Figs. S4–S6). Pull-down experiments performed with DHFRmutC-FLAG demonstrated that this folding-deficient protein is associated with Ssa1 and Ydj1, which are representatives of the Hsp70 and Hsp40 chaperone classes, respectively (Figs. S5 and S6). Failure to degrade DHFRmutC leads to its accumulation as intracellular aggregates (Fig. S7). We tested if Fes1 associates with DHFRmutC by performing immunoprecipitation. Fes1 copurified with DHFRmutC-FLAG but not with the natively folded control protein DHFR-FLAG (Fig. S8A), indicated that Fes1 interacts specifically with misfolded DHFR. We therefore asked whether Fes1 is required for degradation of DHFRmutC-Ura3 and DHFRmutD-Ura3 and found that the fes1Δ mutation caused a drastic stabilization of both proteins (Fig. 2), which was also reflected by growth of the cells on media lacking uracil (Fig. S8B).
Fig. 2.
Fes1 is required for UPS degradation of misfolded DHFR mutants. (A, B) Two folding-deficient variants of DHFR (DHFRMutC-Ura3, DHFRMutD-Ura3) fused to Ura3 were expressed from the PCUP1 promoter (Fig. S3) in isogenic WT, fes1∆, sse1∆, and snl1∆ strains. (C) A third substrate (Ub-V76-Ura3) is a noncleavable ubiquitin fusion protein that is targeted by the UFD pathway. Following cycloheximide (CH) addition, extracts were prepared by glass bead lysis of cells collected at the indicated time points and analyzed by quantitative Western blot. The data represent at least three experiments with independent transformants (error bars indicate SD).
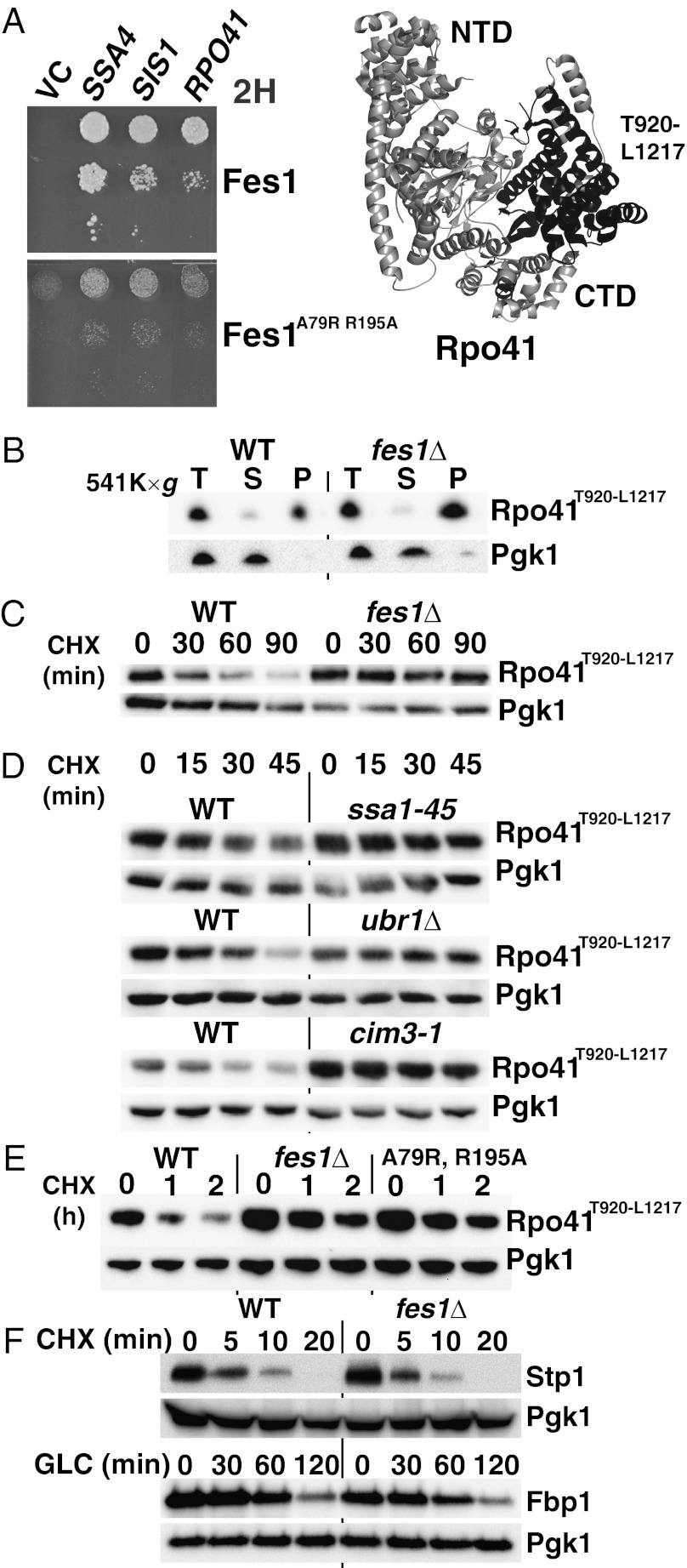
Independent evidence for a general role for Fes1 in the degradation of misfolded proteins came from the analysis of an interactor identified in a Fes1 two-hybrid screen. In this screen, we found Fes1 to interact with Hsp70 of both the Ssa class (nine isolates for Ssa1 and two for Ssa4) and the ribosome-associated Ssb class (one Ssb1 isolate) (31). Apart from Hsp70 proteins, a number of other interactors were found, including Sis1, an Hsp40 that binds directly to Hsp70 (32, 33), and an internal fragment of the mitochondrial RNA polymerase (Rpo41T920-L1217). Importantly, two well-characterized mutations in Fes1 substituting residues specifically required for the association with Hsp70 (Fes1A79R, R195A) abolished the interaction with both Sis1 and Rpo41T920-L1217 (Fig. 3A) (21). We conclude that Fes1 interaction with both of these proteins depends on Hsp70, most likely via formation of ternary complexes. A comparison of the interacting stretch of Rpo41T920-L1217 with the corresponding sequence (C897-I1179) in the crystal structure of human mitochondrial RNA polymerase (34) indicated that the identified segment does not represent a stable independently folding structural domain (Fig. 3A). Rpo41T920-L1217 is an internal fragment of the C-terminal polymerase domain taken out of structural context wherein it is normally stabilized by numerous interactions with both the N- and C-terminal domains. Direct measurement of solubility by ultracentrifugation showed that the bulk of the Rpo41T920-L1217 population was aggregated (Fig. 3B). These observations indicated that Rpo41T920-L1217 is a misfolded polypeptide with a tendency to aggregate and therefore a likely target for ubiquitin-dependent QC mechanisms. We tested this hypothesis by monitoring the stability of Rpo41T920-L1217 in WT and fes1Δ cells. Strikingly, although Rpo41T920-L1217 was degraded in WT cells, the protein was completely stable in fes1Δ cells over 90 min (Fig. 3C). Similar to what was observed for the misfolded DHFR derivatives described above, Hsp70 function and the proteasome system was required for the degradation of Rpo41T920-L1217 (Fig. 3D) (35). Inactivation of the E3 ubiquitin ligase Ubr1, which is required for the selective ubiquitylation of misfolded proteins, resulted in a stabilization of Rpo41T920-L1217 (16, 17). Cells expressing Fes1A79R, R195A that do not interact with Hsp70 did not support degradation of Rpo41T920-L1217 (Fig. 3E). We conclude that, similar to degradation of the misfolded DHFR derivatives described above, the proteolytic targeting of the aberrant polypeptide Rpo41T920-L1217 by the UPS depends on the Ssa1-class of Hsp70 and its NEF Fes1. Together, our findings obtained with multiple distinct test proteins identify Fes1 as a factor essential for the degradation of misfolded cytosolic proteins.
Fig. 3.
Fes1 interacts with and is required for UPS degradation of Hsp70-associated misfolded protein Rpo41T920-L1217. (A) Growth on medium selective for interaction of a yeast two-hybrid reporter strain expressing Fes1 or the Hsp70-binding mutant Fes1A79R, R195A as baits and Hsp70 (SSA4), Hsp40 (SIS1), or a fragment of RPO41 as preys Vector control (VC). The isolated RPO41 expresses a truncated polypeptide (T920-L1217) that corresponds to an internal region of the C-terminal domain (CTD) of mitochondrial RNA polymerase (marked in black in the crystal structure of human homolog). The N-terminal domain (NTD) is indicated. (B) Western analysis of total lysate (T) and supernatant (S) and pellet (P) fractions following ultracentrifugation (541,000 × g). Pgk1 functions as a soluble control protein. (C) Degradation of Rpo41T920-L1217 in WT and fes1Δ cells after translation arrest by cycloheximide (CHX). (D) Degradation at a restrictive temperature (37 °C) as in C in strains carrying mutations in Hsp70 (ssa1-45 ssa2Δ ssa3Δ ssa4Δ), the proteasome (cim3-1) or the ubiquitin E3 ligase Ubr1 (ubr1Δ). (E) Degradation as in C in strains carrying Hsp70 binding (A79R, R195A) mutations in the chromosomal FES1 locus. (F) UPS degradation (CHX) of transcription factor Stp1-HA (Stp1). Induced Hsp70-dependent ubiquitin-proteasome degradation of fructose-1,6-bisphosphatase (Fbp1-HA) followed after addition of 2% glucose to Yeast extract peptone-ethanol medium.
We found that the role of Fes1 in ubiquitin-dependent proteolysis is specific for misfolded proteins because the degradation of several other known ubiquitin-dependent substrates was unaffected. Specifically, the artificial ubiquitin (Gly76Val)-fusion degradation (UFD) pathway substrate UbV76-Ura3 (36) is degraded normally in fes1Δ cells (Fig. 2C and Fig. S8). Similarly, the extremely fast degradation of two natural UPS substrates, the latent transcription factor Stp1 (37) and the glucose-triggered and Hsp70-dependent degradation of the gluconeogenic enzyme fructose-1,6-bisphosphatase 1 (38) proceeded at WT rates (Fig. 3F). In contrast to the specific effects of the fes1∆ mutation toward misfolded proteins, a severe reduction of the total NEF activity in the cytoplasm by the sse1Δ mutation (Sse1 is sevenfold more abundant than Fes1; ref. 39) resulted not only in an increased stability of folding-impaired substrates, but also of the UFD pathway substrate UbV76-Ura3 (Fig. 2C and Fig. S8). Deletion of SNL1 had no effects on any of the substrates tested (Fig. 2 and Fig. S8). Thus, in contrast to Sse1 or Snl1, Fes1 has a dedicated and specific role in misfolded protein degradation.
Fes1 Releases Misfolded Proteins from Hsp70, Thereby Targeting Them for Ubiquitylation.
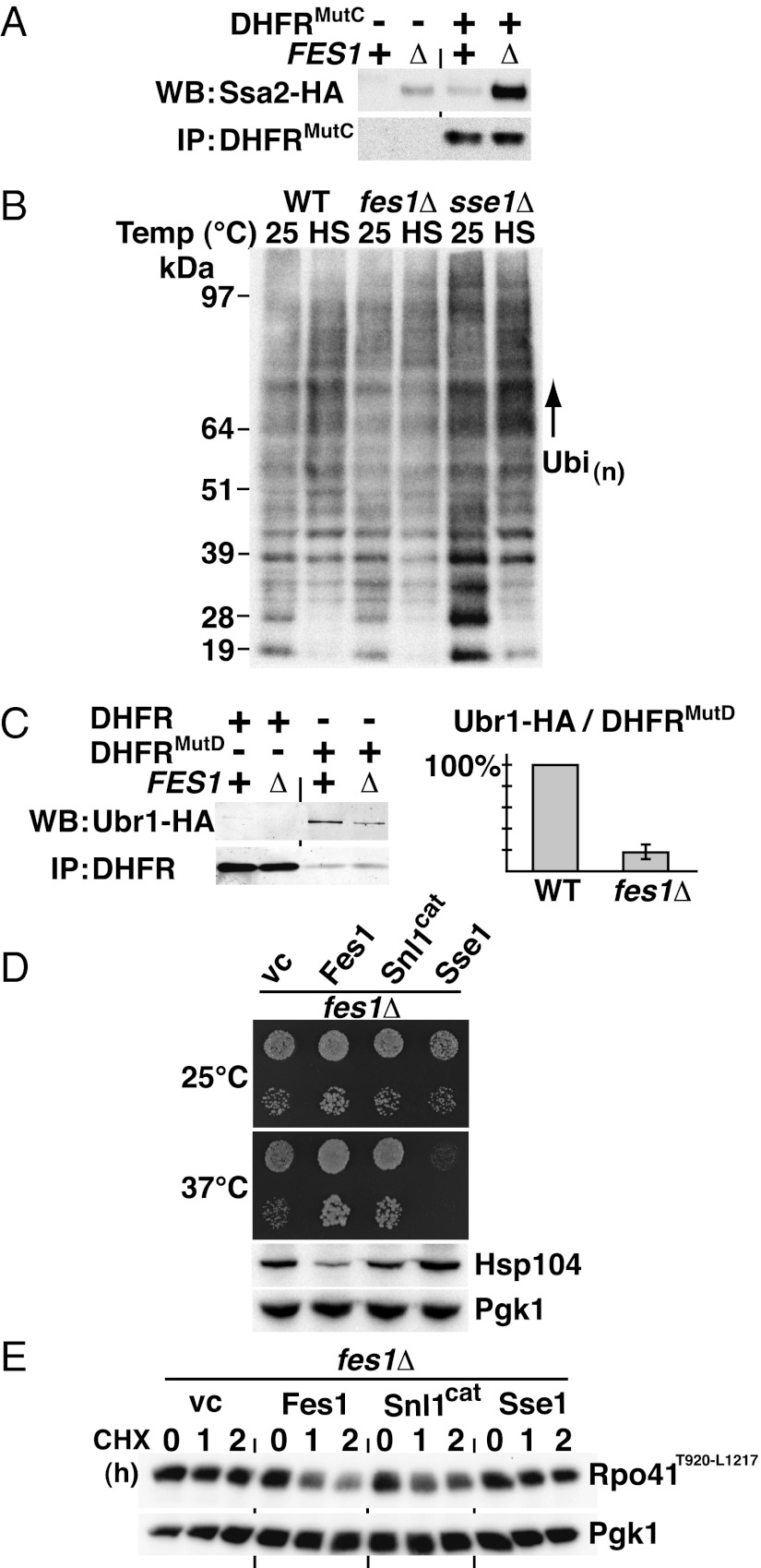
An established biochemical function of NEFs is to stimulate nucleotide exchange in Hsp70, a process that results in substrate release (22, 31). We tested if Fes1 promotes the release of misfolded proteins from Hsp70 by monitoring the amount of Hsp70 that binds misfolded DHFRMutC in fes1Δ cells and WT cells. We choose the Hsp70 Ssa2 for detection because it is expressed at the same constitutive level in both cell types (Fig. 1C). It is worthy to note that Ssa2 is competing with the higher induced levels of Ssa1, Ssa3, and Ssa4 in the fes1Δ strain. Strikingly, when immunoprecipitating DHFRMutC from WT and fes1Δ cells and analyzing the amount of Ssa2 that copurified, we found that substantially more Ssa2 copurified with DHFRMutC immunoprecipitated from fes1Δ cells than from WT cells (Fig. 4A) despite the higher levels of competing Ssa chaperones. Next, we investigated the involvement of Fes1 in heat shock–induced ubiquitylation (15). We predicted that misfolded proteins induced by heat treatment require Fes1 for efficient ubiquitylation. Confirming our prediction, heat shock–induced ubiquitylation was severely impaired in fes1Δ cells (Fig. 4B). In sse1Δ cells, in contrast, the levels of ubiquitylated proteins were already increased at the lower temperature and further increased upon heat stress. The increased amounts of ubiquitin-modified proteins in these mutants even in the absence of external stress argue strongly against a specific function of Sse1 in promoting ubiquitylation of misfolded proteins and rather point to a general role of Sse1 in protein folding. Finally, we directly investigated if Fes1 promotes interactions between ubiquitin ligases and their misfolded protein substrates. The ubiquitin ligase Ubr1-HA specifically copurified with misfolded DHFRMutD but not with native DHFR (Fig. 4C). Importantly, approximately fivefold more Ubr1-HA bound the misfolded protein in purifications from WT compared with fes1Δ extracts. We conclude that Fes1 has an important general role in releasing misfolded proteins from Hsp70 and targeting them for ubiquitylation and proteasomal degradation.
Fig. 4.
Fes1 is required for the release of misfolded proteins from Hsp70 and their targeting to the UPS. (A) Western blot (WB) analysis of amounts of Hsp70 Ssa2-HA copurifying with DHFRMutC-FLAG immunoprecipitated (IP) from WT and fes1Δ protein extracts. (B) Western analysis of total polyubiquitin levels (Ubi(n)) in WT, fes1Δ, and sse1Δ cells grown at 25 °C or heat-shocked (HS) at 42 °C for 30 min. Loading was normalized to amount of cells used for denaturing protein extraction. (C) WB analysis of amounts of Ubr1-HA copurifying with DHFRMutD-FLAG IP from WT and fes1Δ protein extracts. Quantification of the amount of Ubr1-HA copurifying with DHFRMutD-FLAG normalized to WT (100%) (error bars represent SD of experiments performed with five independent transformants). (D) Growth of cells (fes1Δ) transformed with vector control (vc) or plasmids expressing Fes1, Snl1cat, or Sse1. Heat shock response of each transformant was assessed by immunoblotting (Hsp104). (E) UPS degradation after cycloheximide (CHX) of Rpo41T920-L1217 followed in fes1Δ cells expressing Fes1, Snl1cat or Sse1.
Structurally Unrelated NEF but Not Sse1 Can Partially Replace Fes1 Function.
We tested if Fes1 function could be executed by a structurally unrelated Hsp70 NEF by expressing the soluble C-terminal catalytic BAG domain of Snl1 (Snl1Cat) (40) and Sse1. Expression of the Snl1Cat in fes1Δ cells resulted in partial suppression of the temperature-sensitive growth phenotype (Fig. 4D) as well as partial restoration of Rpo41T920-L1217 degradation (Fig. 4E). In contrast, Sse1 overexpression exacerbated the temperature-sensitive growth phenotype and did not suppress the defect in Rpo41T920-L1217 degradation. Expression of Snl1Cat or Sse1 did not suppress the induced heat shock response of fes1Δ cells (Fig. 4D). The observation that Snl1Cat can partially replace the function of Fes1 suggests that Fes1 supports protein degradation by a simple Hsp70 release mechanism, whereas Sse1 functions in a competing pathway.
Discussion
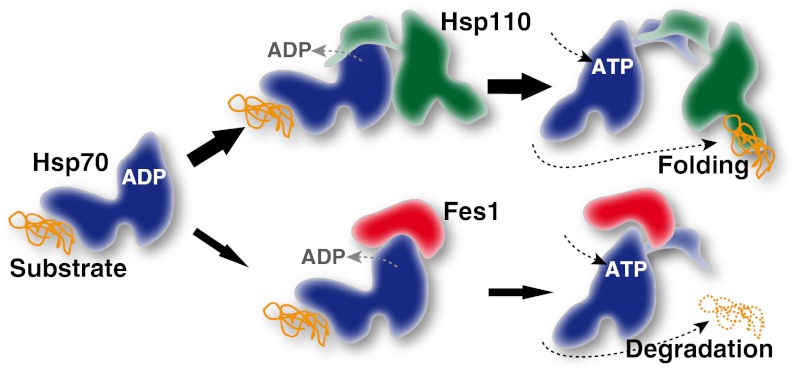
Based on the findings reported here, we propose a model for the role of Fes1 in cytoplasmic protein QC that integrates all available data (Fig. 5). Abundant Hsp70 associates transiently with exposed hydrophophic patches of misfolded proteins and maintains these aggregation-prone proteins soluble. Hsp70 can refold misfolded proteins in ATPase-driven cycles catalyzed by the dominant NEFs of the Hsp110 class (Sse1/Sse2). Hsp110 contributes directly to the folding process by binding hydrophobic peptide stretches of the substrate protein. Misfolded proteins resilient to Hsp70-Hsp110 refolding will undergo repetitive and futile interactions with Hsp70-Hsp110. Eventually the Hsp70-substrate complexes will encounter the NEF Fes1. Fes1 triggers nucleotide exchange of the Hsp70, which results in the release of the misfolded protein from Hsp70 and facilitates the interaction with downstream ubiquitin E3 ligases that target misfolded proteins for degradation by the UPS.
Fig. 5.
Model for Hsp70 substrate triage regulated by NEFs (Discussion).
In our model, we propose that NEFs of the Hsp110 class mainly support protein folding activities of Hsp70s, whereas Fes1 targets Hsp70-associated misfolded proteins for degradation. This notion is supported by comparisons of the phenotypes of sse1Δ and fes1Δ strains (Fig. 1E) as well as by the observation that overexpression of the catalytic domain of Snl1 suppresses fes1Δ, whereas that of Sse1 does not (Fig. 4 D and E). We find that sse1Δ cells, in contrast to fes1Δ cells, are very slow growing at all temperatures, likely because removal of this abundant NEF induces a general Hsp70 activity defect. Consistent with this understanding, sse1Δ mutants expressing folding-impaired Ura3 reporter remain uracil auxotrophs despite the observed stabilization of these reporter proteins, indicating that they accumulate as nonnatively folded proteins (Fig. S8). Furthermore, sse1Δ cells do not exhibit a strong heat shock phenotype as do fes1Δ cells (Fig. 1E), but do accumulate larger amounts of ubiquitylated proteins at normal growth temperature as well as under heat stress conditions (Fig. 4B). In conclusion, sse1Δ degradation phenotypes likely reflect general malfunction of the Hsp70 system (i.e., that misfolded proteins become stabilized in sse1Δ cells because of extensive aggregation or as a consequence of abundant production of misfolded polypeptides competing for the degradation pathways).
Biochemical differences between Hsp110 and Fes1 might explain how these two NEFs can induce such diverse fates of Hsp70 substrates as folding and degradation. In contrast to Fes1, Hsp110 is a chaperone that recently has been shown to directly bind hydrophobic peptides (41–43). Thus, direct binding of hydrophobic patches might help to maintain the misfolded protein soluble, help the protein to fold, and shield it from interactions with the ubiquitylation machinery. In support of the latter concept, the E3 ligase San1 directly interacts with hydrophobic patches of its misfolded protein substrates (5). Indeed, Sse1 inhibits San1-dependent ubiquitylation in vitro, corroborating the notion of direct competition for substrate binding (14). Further support for the hypothesis of qualitative activity differences between Hsp110 and Fes1 is provided by the observation that purified Hsp70-Hsp40-Sse1 can extract and refold luciferase from aggregates, whereas Hsp70-Hsp40-Fes1 cannot (42). When all these data are considered, a simple hypothesis emerges: Sse1 has substrate binding activities and promotes protein folding probably by mediating rebinding to Hsp70, whereas Fes1 is devoid of the same activities and therefore triggers release of Hsp70 substrates that are to be ubiquitylated and degraded. Our model provides the conceptual foundation for how distinct classes of NEFs regulate Hsp70 function in protein folding and degradation.
Materials and Methods
Strains, Plasmids, and Growth Media.
Details on constructions of strains and plasmids are available upon request. Plasmid inserts expressing various DHFR-derived test proteins used in this study are depicted in Fig. S1A. Constructs expressing DHFR-Ura3 and DHFRmutC-Ura3 were generated and kindly provided by Jörg Höckendorff. Standard media include YPD medium and ammonia-based synthetic complete dextrose (SC).
Heat Shock Protein Purification and MS Analysis.
Strains were expanded in YPD medium at 25 °C and harvested at logarithmic growth phase by centrifugation. Cells were lysed in denaturing lysis buffer (40 mM Tris⋅HCl pH 7.5 1 mM EDTA 8 M urea) by bead beating. After preclearing by centrifugation (21,100 × g), the supernatant was applied onto a gravity-flow Q-Sepharose (GE Healthcare Life Sciences) column. The column was extensively washed with denaturing lysis buffer, and bound proteins were eluted using an NaCl step gradient (50-mM increments up to 600 mM NaCl) prepared in denaturing lysis buffer. Eluted proteins were separated by SDS/PAGE followed by Coomassie Brilliant Blue staining. Protein bands were excised and subjected to tryptic extraction and MS analysis.
Yeast Two-Hybrid Assay.
A genomic yeast plasmid library (pJG4-5) was screened for interactors (growth on SC-galactose media lacking lysine and leucine) with full-length Fes1 fused to the N terminus of the DNA-binding domain of LexA in yeast strain CAY235 (44).
Analytical Ultracentrifugation of Rpo41T920-L1217.
Cells expressing Rpo41T920-L1217-HA were grown at 30 °C to logarithmic growth phase, harvested by centrifugation, and lysed in 50 mM Tris⋅HCl pH 7.5, 5 mM MgCl2, 10 mM DTT, 1 mM PMSF, and 1 mM N-ethylmaleimide by bead beating. Lysates were cleared from cell debris by low-speed centrifugation (1500 × g) and detergents were added to the following final concentrations, 1% (vol/vol) Nonidet P-40, 1% (vol/vol) Tween 20, and 0.1% SB3-14. The solubilized lysates were subjected to 30 min of ultracentrifugation in a TLA 100.3 rotor (Beckman) at 100,000 rpm (541,000 × g) and analyzed by Western blotting.
Western Blot Analysis and Determination of Protein Stability.
Cells were grown at 30 °C to logarithmic growth phase, and cycloheximide was added to the cultures at a final concentration of 100 mg/L. Strains carrying temperature-sensitive alleles were expanded at permissive 25 °C and incubated at restrictive 37 °C for 30 min before cycloheximide addition. Protein extracts were prepared from samples taken at the time points indicated either by glass bead lysis in ice cold Native lysis buffer (50 mM Na⋅Hepes pH 7.5, 150 mM NaCl, 5 mM EDTA, and 1% (vol/vol) Triton-X100 containing Complete Protease Inhibitor Mixture (Roche)) or by NaOH and trichloroacetic acid as described previously (45). Equal amounts of proteins were separated by SDS/PAGE and analyzed by quantitative Western blotting (46) or by chemiluminescence detection (SuperSignal West Dura Extended-Duration Substrate; Pierce) and quantified using an LAS1000 system (Fuji Photo Film Co.).
Coimmunoprecipitation of Mutant DHFR and Associated Proteins.
Cells were grown at 30 °C to logarithmic growth phase in SC medium supplemented with 100 or 400 μM CuSO4 to induce the expression of the DHFR constructs. After harvest by centrifugation, cells were broken by glass bead lysis in either LWB150 [40 mm Hepes-KOH, pH 7.4, 150 mm KCl, 5 mm MgCl2, 5% (vol/vol) glycerol], 0.1% (vol/vol) Triton X-100) or Native lysis buffer (see previous paragraph). The lysates, precleared by centrifugation, were incubated with ANTI-FLAG M2 Affinity Gel (Sigma) for 60 min. After washing extensively with buffer, bound proteins were eluted with sample buffer or Native lysis buffer containing 3× FLAG-Peptide (200 mg/L). Coimmunoprecipitated proteins were detected by Western blotting.
Supplementary Material
Acknowledgments
We thank Andreas Bachmair, Stefan Jentsch, Erica Johnson, Nils Johnsson, Jörg Höckendorff, Mark Hochstrasser, Kiran Madura, Thomas Sommer, and Alexander Varshavsky for yeast strains and plasmids. This work was supported by Deutsche Forschungsgemeinschaft Grant Do 649/2 (to R.J.D.) and grants from the Swedish Science Council, Carl Tryggers stiftelse för vetenskaplig forskning, and Jeanssons Stiftelser (to C.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216778110/-/DCSupplemental.
References
- 1.Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol. 2005;6(11):891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 3.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22(13):2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goloubinoff P, De Los Rios P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem Sci. 2007;32(8):372–380. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arndt V, Rogon C, Höhfeld J. To be, or not to be—molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64(19-20):2525–2541. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kettern N, Dreiseidler M, Tawo R, Höhfeld J. Chaperone-assisted degradation: Multiple paths to destruction. Biol Chem. 2010;391(5):481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 10.McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: Chaperones culling corrupt conformations. Nat Cell Biol. 2005;7(8):736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- 11.Jiang J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 12.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121(5):739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Park SH, et al. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18(1):153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13(11):1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107(3):1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582(30):4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21(13):2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nillegoda NB, et al. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21(13):2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andréasson C, Fiaux J, Rampelt H, Druffel-Augustin S, Bukau B. Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc Natl Acad Sci USA. 2008;105(43):16519–16524. doi: 10.1073/pnas.0804187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shomura Y, et al. Regulation of Hsp70 function by HspBP1: Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17(3):367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002;22(13):4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingolia TD, Slater MR, Craig EA. Saccharomyces cerevisiae contains a complex multigene family related to the major heat shock-inducible gene of Drosophila. Mol Cell Biol. 1982;2(11):1388–1398. doi: 10.1128/mcb.2.11.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54(6):855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 25.Gupta R, et al. Firefly luciferase mutants as sensors of proteome stress. Nat Methods. 2011;8(10):879–884. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]
- 26.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25(11):2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25(11):2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorger PK. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62(4):793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- 29.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71(1):97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 30.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372(6505):475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 31.Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A. Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol Chem. 2006;387(12):1593–1600. doi: 10.1515/BC.2006.198. [DOI] [PubMed] [Google Scholar]
- 32.Aron R, Lopez N, Walter W, Craig EA, Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169(4):1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Wu Y, Qian X, Sha B. Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem J. 2006;398(3):353–360. doi: 10.1042/BJ20060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringel R, et al. Structure of human mitochondrial RNA polymerase. Nature. 2011;478(7368):269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 35.Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16(8):4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270(29):17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 37.Pfirrmann T, Heessen S, Omnus DJ, Andréasson C, Ljungdahl PO. The prodomain of Ssy5 protease controls receptor-activated proteolysis of transcription factor Stp1. Mol Cell Biol. 2010;30(13):3299–3309. doi: 10.1128/MCB.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juretschke J, Menssen R, Sickmann A, Wolf DH. The Hsp70 chaperone Ssa1 is essential for catabolite induced degradation of the gluconeogenic enzyme fructose-1,6-bisphosphatase. Biochem Biophys Res Commun. 2010;397(3):447–452. doi: 10.1016/j.bbrc.2010.05.123. [DOI] [PubMed] [Google Scholar]
- 39.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 40.Sondermann H, et al. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J Biol Chem. 2002;277(36):33220–33227. doi: 10.1074/jbc.M204624200. [DOI] [PubMed] [Google Scholar]
- 41.Goeckeler JL, et al. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 2008;582(16):2393–2396. doi: 10.1016/j.febslet.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6(10):e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X, et al. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J Biol Chem. 2012;287(8):5661–5672. doi: 10.1074/jbc.M111.275057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omnus DJ, Pfirrmann T, Andréasson C, Ljungdahl PO. A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol Biol Cell. 2011;22(15):2754–2765. doi: 10.1091/mbc.E11-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silve S, et al. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11(2):1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurian L, Palanimurugan R, Gödderz D, Dohmen RJ. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature. 2011;477(7365):490–494. doi: 10.1038/nature10393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.