Abstract
Terminase enzymes are viral motors that package DNA into a preformed capsid and are of interest both therapeutically and as potential nano-machines. The enzymes excise a single genome from a concatemeric precursor (genome maturation) and then package the duplex to near-crystalline density (genome packaging). The functional motors are oligomers of protomeric subunits and are the most powerful motors currently known. Here, we present mechanistic studies on the terminase motor from bacteriophage λ. We identify a mutant (K76R) that is specifically deficient in packaging activity. Biochemical analysis of this enzyme provides insight into the linkage between ATP hydrolysis and motor translocation. We further use this mutant to assemble chimeric motors with WT enzyme and characterize the catalytic activity of the complexes. The data demonstrate that strong coordination between the motor protomers is required for DNA packaging and that incorporation of even a single mutant protomer poisons motor activity. Significant coordination is similarly observed in the genome maturation reaction; however, although the motor is composed of a symmetric tetramer of protomers, the maturation complex is better described as a “dimer-of-dimers” with half-site reactivity. We describe a model for how the motor alternates between a stable genome maturation complex and a dynamic genome packaging complex. The fundamental features of coordinated ATP hydrolysis, DNA movement, and tight association between the motor and the duplex during translocation are recapitulated in all of the viral motors. This work is thus of relevance to all terminase enzymes, both prokaryotic and eukaryotic.
Keywords: AAA+ ATPases, bacteriophage lambda, molecular motors, virus assembly, Walker A
The assembly pathway of double-stranded DNA (dsDNA) viruses is conserved from bacteriophage to eukaryotic viruses of therapeutic interest, including the adenovirus and herpesvirus groups (1, 2). A key step is the translocation of genomic DNA into a capsid shell by a packaging motor, fueled by ATP hydrolysis (3–6). These motors are among the most powerful discovered to date; they package DNA to near-crystalline density and can generate up to 50 atmospheres of internal capsid pressure (7, 8). A mechanistic characterization of viral DNA packaging is of interest not only in a therapeutic sense, but also toward a fundamental understanding of complex biological motors and in the development of powerful nanomachines.
Terminase enzymes catalyze viral genome packaging. All characterized terminases are composed of large (TerL) and small (TerS) subunits in hetero-oligomeric complexes, although the subunit stoichiometry in the functional motors remains ill defined in most cases (3, 6). Most of the enzymes also possess a DNA “maturation” activity that excises a single genome from a concatemeric precursor in preparation for packaging (3, 6). Bacteriophage λ terminase is prototypical of these viral enzymes and has been extensively characterized. The enzyme may be isolated as a pure and homogenous species and a variety of physical and kinetic assays have been established to interrogate the maturation and packaging activities of the motor. Here we use the λ system to elucidate the mechanistic features of DNA packaging by these viral motors.
λ terminase is composed of a large (gpA; TerL) and a small subunit (gpNu1; TerS) in a well-defined TerL1:TerS2 heterotrimer complex (6, 9, 10). Biochemical studies indicate that this heterotrimer likely represents the native state of the enzyme in vivo, and we refer to it as the terminase protomer (11, 12). Although the protomer possesses little to no catalytic activity, it self-assembles into a tetrameric ring [(TerL1:TerS2)4] that possesses full maturation and packaging activities (11, 12). The λ genome maturation and packaging pathway has been reviewed (5, 9, 13) and is summarized in Fig. 1.
(i) Maturation complex assembly: The protomer site specifically assembles at the cohesive-end-site (cos); this ∼200-bp sequence represents the junction between two genomes in the immature DNA concatemer. Escherichia coli integration host factor (IHF) is necessary for optimal virus yield in vivo, and we demonstrated cooperative assembly of terminase and IHF at the cos site in vitro (14).
(ii) Genome end maturation: Terminase introduces symmetric nicks into the cosN subsite (cos-cleavage) to afford a nicked, annealed intermediate. The enzyme then separates the strands to afford the mature 12 base single-stranded “sticky” end of the genome tightly bound by the terminase complex. Both intermediates are extremely stable and collectively represent “complex I” isolated from infected cells in vivo (12).
(iii) Transition to the packaging motor: Terminase next binds to the portal, a ring-like structure that resides at a unique vertex of a procapsid shell. Minimally, the portal serves as a conduit through which DNA is translocated into the shell during packaging and out of the shell during infection. Binding to the portal triggers the transition of terminase from a stable maturation complex to a mobile packaging motor complex; however, the role that the portal proteins play in mechano-chemical translocation of DNA remains uncertain.
(iv) Genome packaging. Procapsid binding activates the packaging ATPase activity of terminase, release of the motor from cos, and translocation of DNA into the shell. A series of events follows that ultimately afford an infectious virus particle containing a “matured” unit-length genome tightly packaged into the capsid shell. The ejected terminase•DNA complex binds a second procapsid to initiate another round of packaging and the process is repeated with three to four genomes packaged per terminase binding event.
Fig. 1.
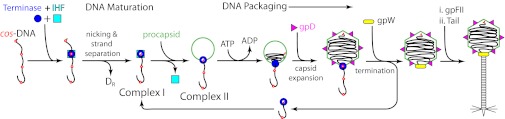
Maturation and packaging of a λ genome by the terminase enzyme. Details are provided in the main text.
Terminase enzymes function as genome maturation and packaging motors that alternate between a stable, site specifically–bound maturation complex and a dynamic, powerful DNA packaging motor. Single-molecule (7) and ensemble biochemical studies (15) indicate that λ terminase packages DNA at a rate of ∼600 bp/s, is highly processive, and can generate the significant force required to package the entire genome. Although these studies have provided significant mechanistic insight, little is known about the ensemble biochemical features of mechano-chemical coupling and force generation by these viral motors in solution. The physio-kinetic features of packaging imply significant and tight coordination between the protomers assembled into the motor complex, but there are scant biochemical data to directly support this presumption.
Here we characterize the genome maturation and DNA packaging reactions of λ terminase using chimeric motor complexes assembled from WT and packaging-deficient protomers. We demonstrate that there is exceptionally strong coordination between the protomers during translocation of DNA by the motor complex in solution. These ensemble biochemical studies further provide insight into the stoichiometry of the maturation and packaging motor complexes, and we propose a unified model coupling the two reactions in virus assembly. The relevance of these results with respect to the general features of viral packaging motors is discussed.
Results
The goal of this work is to evaluate the coordination between protomers assembled into a viral genome packaging motor. Our approach is to incorporate packaging-deficient protomers into the complex and to quantify their effect on motor function. This approach requires a mutant enzyme that is selectively defective in DNA translocation but is otherwise fully WT. Thus, we first identify an appropriate mutation in the enzyme.
Purification and Physical Characterization of Terminase K76R.
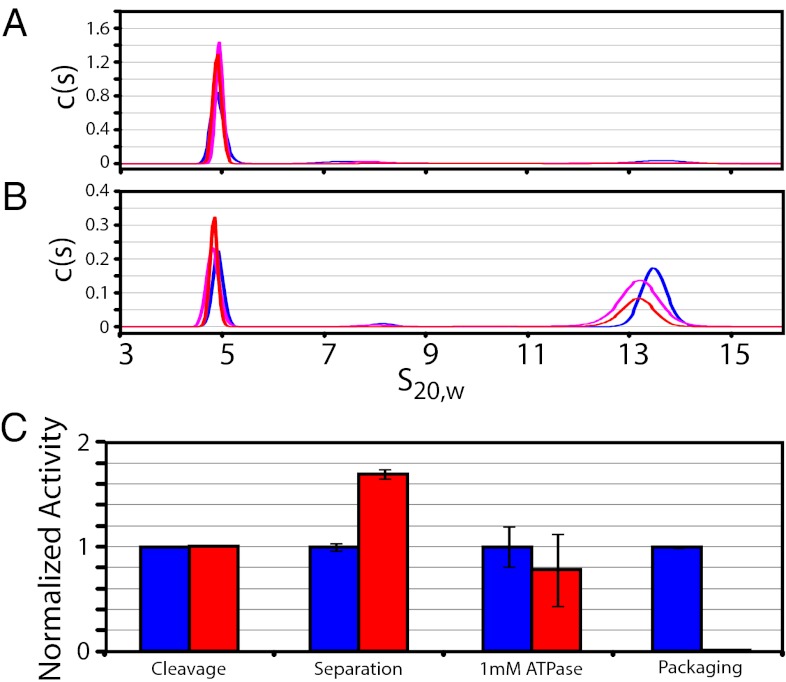
Duffy and Feiss (16) identified a number of terminase mutations that disrupt genome packaging in vivo; five classes were observed, including those that slow the packaging rate, those that affect packaging processivity, and those that have a severe packaging defect. To simplify the interpretation of results obtained in the present study, we focused on mutations that abolish packaging completely, and three were chosen for further study based on this criteria (SI Text). All three enzymes showed WT chromatographic behavior and were purified to homogeneity. Their self-association characteristics were evaluated by sedimentation velocity analytical ultracentrifugation (AUC), which identifies terminase gpA-K76R as the most WT protomer in this respect (Fig. 2 A and B; Fig. S1). This mutant was chosen for further study, and we refer to this enzyme as K76R for simplicity.
Fig. 2.
The gpA-K76R mutation specifically abrogates DNA packaging activity. Sedimentation distribution profiles for the purified protomers in low salt buffer (A) and the assembled tetramer species in high salt buffer (B). WT, K76R, and a 50:50 mix of the two protomers are shown in blue, red, and violet, respectively. (C) Catalytic activity of the WT (blue) and K76R (red) terminase protomers. Error bars represent SD (n = 3).
An essential requirement for the mutant protomer is that it must assemble with the WT enzyme in a stochastic manner to afford a chimeric motor of defined composition. AUC analysis confirms that the K76R protomer is structurally homogenous with a S20,w identical to that of the WT protomer and that it self-assembles into a native-like tetramer of protomers (Fig. 2 A and B). Importantly, mixtures of WT and mutant protomers similarly assemble into native-like tetramers, with no evidence for attenuated assembly or aberrant oligomerization (Fig. 2B). These data, in combination with the kinetic data presented below, demonstrate that chimeric motors can be assembled with both WT and K76R mutant protomers in a defined composition.
Biochemical Characterization of K76R.
λ terminase possesses multiple catalytic activities that are required to first mature and then package the viral genome into a procapsid. An idealized mutant enzyme requires specific deletion of packaging activity without affecting the maturation activities of the enzyme. We first examined the cos cleavage activity of K76R, which is the first step in genome maturation (Fig. 1). The data demonstrate that cos cleavage by K76R is essentially identical to that of the WT enzyme (Fig. 2C). Thus, neither protomer assembly at cos nor catalytic activity has been compromised by the mutation. We next examined the second step of maturation: separation of the nicked ends. Unexpectedly, the strand separation activity of K76R is slightly greater than that of the WT enzyme (Fig. 2C). This observation is discussed further below.
Maturation of the duplex end is followed by recruitment of a procapsid and activation of the packaging activities of terminase. Activation includes up-regulation of the packaging ATPase in TerL, release of the motor from the cos site, and translocation of the duplex into the procapsid shell (Fig. 1). Although the K76R mutation has little to no effect on ATPase activity, the mutant enzyme is essentially devoid of detectable DNA packaging activity (Fig. 2C). In sum, K76R possesses WT assembly properties and retains near-native maturation and ATPase activities but is profoundly deficient in DNA packaging activity. Thus, K76R is ideally suited to interrogate coordination between protomers in the DNA packaging motor.
Model for Ensemble Catalytic Activity of Chimeric Packaging Motors in Solution.
The activity of a chimeric motor will depend on the total number of protomers in the functional motor (n), the number of mutant protomers incorporated into the motor (k), the relative activities of the WT (AWT) and mutant (AMT) protomers, and the degree of coordination between them; we consider two extremes of coordination in the motor. If the protomers act completely independently (i.e., zero coordination), then motor activity can be calculated by simply adding the fractional activity of each protomer in the complex
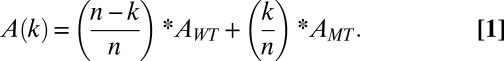 |
For instance, K76R is devoid of packaging activity (AMT = 0; Fig. 2C), and a tetrameric motor (n= 4) with two WT and two mutant protomers (k = 2) will possess a normalized activity A(2) = 0.5. In contrast, if one considers “infinite” coordination (100% coupling), then incorporation of even a single K76R protomer completely poisons motor activity. In this case, only the fully WT motor, A(0), possesses activity, and all of the chimeric motors are packaging deficient.
Although the activity of an individual chimeric motor can be calculated as described above, the activity of an ensemble of chimeric motors requires consideration of the population distribution of the motors in solution, which can be obtained from a binomial distribution model:
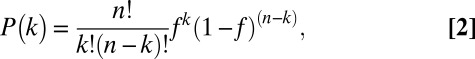 |
where P(k) is the probability that a motor contains k mutant subunits, and f is the probability that a mutant protomer is incorporated into the motor. An example of the probability distribution of a n= 4 motor containing k subunits as a function of f is shown in Fig. S2A.
Finally, the observed activity of an ensemble of chimeric motors in solution is determined by summing the activities of each individual chimera multiplied by its probability distribution in solution
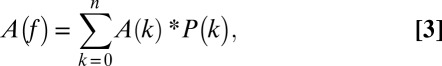 |
where A(f) is the observed packaging activity as a function of f, which varies between 0 and 1; in the limit, A(0) = AWT (fully WT motor) and A(1) = AMT (fully mutant motor). This analysis assumes that WT and mutant protomers assemble stochastically; that is, the mutation neither affects homo- nor hetero-association interactions. Stochastic assembly appears to be the case for K76R, as the AUC and packaging data (see below) indicate that bulk assembly of the WT and the K76R protomers at cos is unperturbed by the mutation and that the mixtures coassemble in a stochastic manner. Thus, f equals the fraction of K76R enzyme added to the reaction mixture.
Nature and Catalytic Activity of Chimeric Packaging Motors.
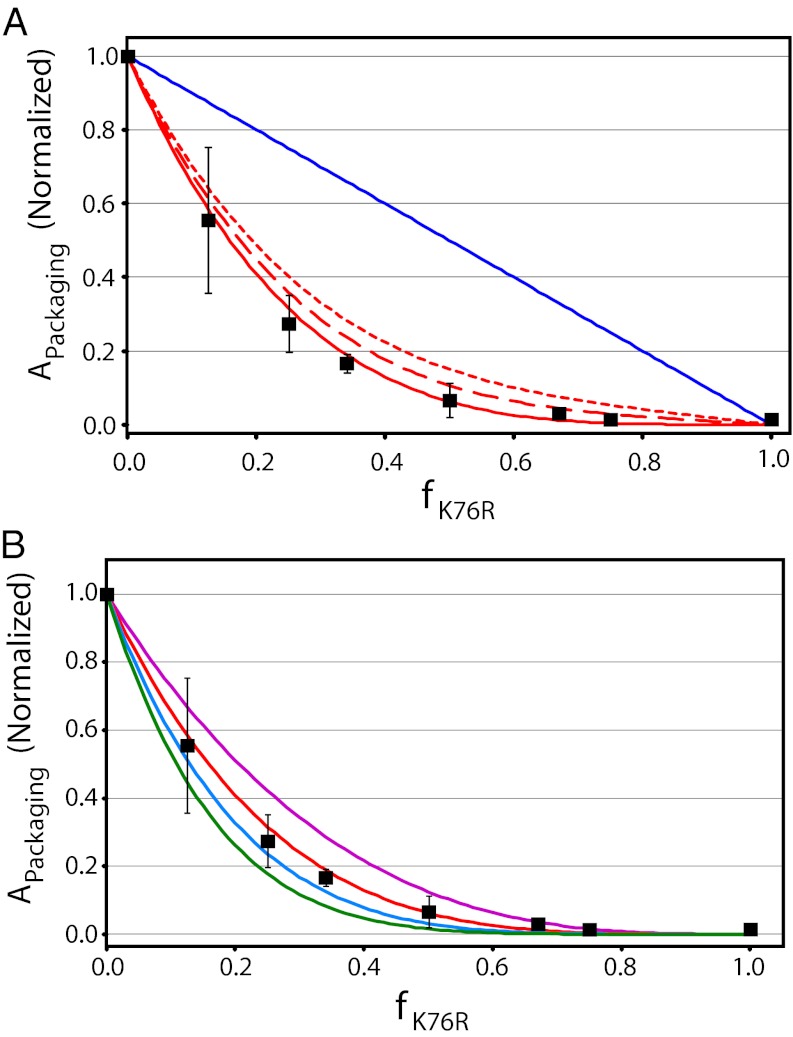
As shown above, the WT protomer packages DNA efficiently, whereas the K76R mutant enzyme is devoid of packaging activity (Fig. 2C). Incremental addition of K76R to the WT protomer keeping the total enzyme concentration constant (increasing f) strongly and significantly poisons the packaging activity of the chimeric motors (Fig. 3A). These data were evaluated using the model described above.
Fig. 3.
The DNA packaging motor shows strong coordination between the protomers. (A) Relative packaging activity as a function of f is indicated in black squares. Error bars represent SD (n = 3). The predicted ensemble activity of the motors in solution was calculated from Eq. 3 incorporating zero (blue line; SSE = 0.91), 100% (red line; SSE = 4.55 × 10−3), 90% (red dashed line; SSE = 1.98 x10−2), and 80% (red dotted line; SSE = 5.10 × 10−2) coordination in the individual motors (Fig. S2B). (B) The experimental data from A is redisplayed, and the predicted ensemble activity of motors containing three (purple; SSE = 5.65 × 10−2), four (red; SSE = 4.55 × 10−3), five (blue; SSE = 6.49 × 10−3), and six (green; SSE = 3.03 × 10−2) protomers was calculated from Eq. 3 assuming 100% coordination.
We demonstrated that four terminase protomers assemble into a functional packaging motor complex (11). Moreover, kinetic interrogation of DNA packaging suggests that the catalytically competent packaging motor is composed of at least four protomers, each of which hydrolyzes ATP in a cooperative manner (12, 17). Thus, we initially analyzed the packaging data assuming a tetrameric packaging motor (n = 4). The calculated activity of individual chimeric motors assuming zero coordination between the protomers is presented in Fig. S2B. These values were used in Eq. 3 to predict the ensemble activity of chimeric motors in solution, which is shown as a blue line in Fig. 3A; clearly, this model does not describe the data [sum of squared error (SSE) = 0.92]. In contrast, the predicted ensemble activity of chimeric motors with 100% coupling describes the data exceptionally well (SSE = 4.55 × 10−3; Fig. 3A, red line). We also considered relaxed coordination between the protomers, but relaxing coupling by as little as 10% results in a decrease in the quality of the fit and relaxing coupling by 20% poorly describes the data (4- and 11-fold increase in SSE, respectively; Fig. 3A; Fig. S2B). Finally, we considered models that incorporate strong coordination between the protomers (100%) but different motor stoichiometries (n = 3–6); the experimental data are best described by a model that incorporates four or five protomers in a tightly coupled symmetric motor complex (Fig. 3B). Importantly, these results confirm that the WT and mutant protomers coassemble in a stochastic manner. If this was not the case, WT and mutant motors would act independently, and the kinetic data would resemble the zero coordination model (Fig. 3, blue line), which is clearly not the case.
Nature and Catalytic Activity of the DNA Maturation Complex.
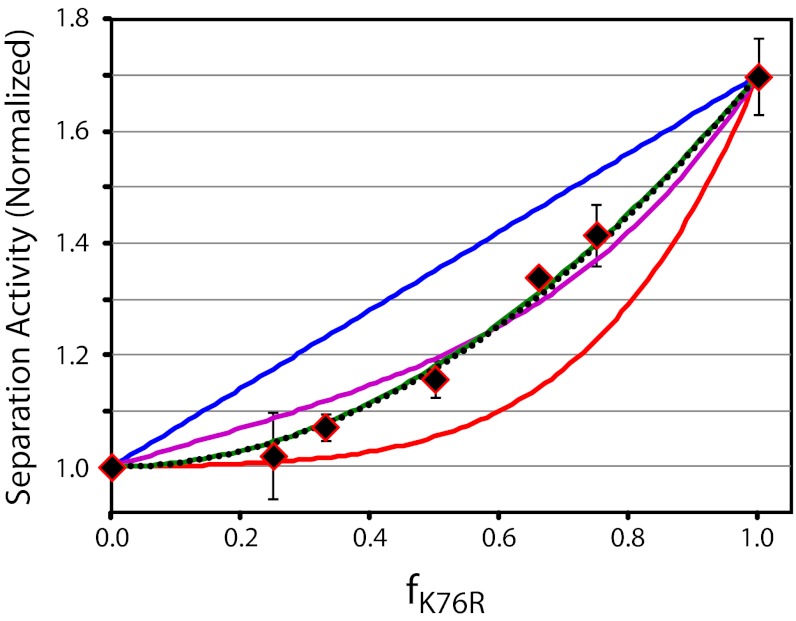
K76R exhibits increased strand separation activity relative to the WT enzyme (Fig. 2C), and this feature allows an interrogation of DNA maturation by chimeric complexes. The observed strand separation activity of WT terminase, K76R, and chimeric maturation complexes assembled from mixtures of the two is displayed in Fig. 4. The activity of the complex is strongly influenced by the fraction of mutant protomer added to the reaction mixture, and we evaluated the data assuming a symmetric maturation complex composed of four protomers, as described above for the motor complex. This model clearly fails to recapitulate the experimental data in both the zero and 100% coordination limits (Fig. 4, blue and red lines, respectively). Models that incorporate relaxed coordination and/or alternate motor stoichiometries afford a better fit, but systematically deviate from the experimental data (Fig. 4). We therefore considered alternate models for the genome maturation complex.
Fig. 4.
The genome maturation complex shows complex coordination. Relative strand separation activity as a function of f is indicated in black diamonds. Error bars represent SD (n = 3). The predicted ensemble activity of a symmetric tetrameric complex in solution was calculated from Eq. 3 assuming zero (blue line; SSE = 2.84 × 10−2), 50% (purple line; SSE = 2.99 × 10−3), or 100% (red line; SSE = 2.51 × 10−2) coordination in the individual motors (Fig. S3). The predicted ensemble activities of a simple dimer model (black dots; SSE = 5.71 × 10−4) and a dimer of dimers model (green line; SSE = 5.65 × 10−4) are shown assuming 100% coordination (Figs. S4 and S5B).
Early studies suggested that a terminase dimer assembles at the symmetric cosN sequence to mature the genome end (9, 13) in analogy to the classical type II restriction enzymes (see Fig. S4A) (18). This model was evaluated as described in SI Text and affords an exceptional fit to the experimental data when strong coordination (100%) between the protomers is incorporated (SSE = 5.71 × 10−4; Fig. 4, black dotted line). Although this simple model is adequate, we recently proposed that the maturation complex is actually composed of a ring tetramer assembled from a “dimer of dimers” (12, 19). This model is analogous to the type IIF restriction endonucleases where two symmetrically disposed subunits within a tetrameric ring are required to nick the DNA; the other two subunits are required for optimal binding activity but are catalytically silent (half-site reactivity; Fig. S5A) (18, 20). In this case, the predicted activity of a chimeric machine is more complex; although a mutant protomer may be incorporated into any of the four positions, only those incorporated into the active dimer will directly affect the reaction. The calculated activity of individual chimeric dimer of dimer complexes is shown in Fig. S5B, and these values were used in Eq. 3 to predict the ensemble activity of chimeric maturation complexes in solution. This model similarly describes the experimental data exceptionally well when 100% coordination is incorporated into the model (SSE = 5.65 × 10−4; Fig. 4, green line). This observation is consistent with biochemical studies demonstrating significant communication between the protomers that nick the duplex in the cos cleavage reaction (21).
Discussion
The terminase TerL subunits possess all of the catalytic activities required to mature and package DNA, whereas the TerS subunits are required for specific recognition of viral DNA (3, 5, 6). Although there is limited sequence similarity among the terminase enzymes, genetic, biochemical, and sequence alignment studies have shown that the TerL subunits possess a similar domain organization, where DNA packaging and maturation activities reside in N-terminal and C-terminal domains, respectively (5, 9). The K76R mutation in λ terminase lies within the TerL motor domain and specifically abrogates packaging activity. The study of this mutant enzyme provides insight into the nature of the nucleoprotein complexes involved in first maturation and then packaging a viral genome.
K76R Mutation Disrupts Mechano-Chemical Coupling.
Structural and phylogenetic analysis suggests that the terminase motor domains are most closely related to the SF2 family of helicases (5). Consistently, the λ motor domain contains the conserved signature motifs associated with these enzymes, including the adenine binding motif, the Walker A and B sequences, the catalytic carboxylate, and the mechano-chemical coupling C-motif (Fig. S6) (5).
The Walker A phosphate binding loop (a.k.a., P-loop) provides an essential lysine that coordinates the β-γ phosphate and a (T/S) residue that interacts with the Mg•ATP chelate; both interactions are required for nucleotide binding and/or hydrolysis (22). Consistently, mutation of K166 in the Walker A sequence of T4 terminase abrogates ATPase activity and the coupled packaging activity (23). The putative Walker A sequence of λ (76KSARVGYS83) diverges from the canonical sequence (GXXXXGK[T/S]), where the putative essential Lys residue is indicated in boldface type. It has been proposed that K76 represents a variant P-loop Lys. We thus anticipated that the K76R mutation would abolish ATP hydrolysis and in turn DNA packaging; however, although the K76R mutation indeed abrogates DNA packaging, it affects ATPase activity very little. We consider several possible roles for this residue in DNA packaging.
First, the mutation could affect terminase assembly into a motor complex. We do not favor this possibility based on the native self-association and cos cleavage activities of the enzyme. Second, it is feasible that the mutation alters terminase binding to the portal vertex to complete the packaging motor (Fig. 1). Genetic and biochemical studies indicate that the extreme C terminus of λ TerL is involved in portal binding (24). This region is quite distant from the K76 mutation in both primary sequence and tertiary structure (Fig. S6). Although we cannot rigorously exclude the possibility that this mutation affects the global architecture of the motor, the native-like properties of K76R in all other respects mitigate this possibility. Finally, the K76R mutation could affect mechano-chemical coupling between ATP hydrolysis and motor movement; the data favor the latter explanation.
We constructed a structural model for λ TerL (Fig. S6) that shows that K76 is proximate to the putative C-motif (a.k.a., helicase motif III; 212GST214) that is conserved in terminase enzymes. C-motif residues, which were first identified in the SF2 superfamily of helicase enzymes, form a network of hydrogen bonds that connect the γ-phosphate of ATP to DNA and play an intimate role in coupling ATP hydrolysis to motor movement (5). It appears that, although the conservative K→R mutation is insufficient to affect nucleotide hydrolysis, it uncouples these chemical events from interaction with the C-motif and the associated mechanical movement. Current studies in our laboratory seek to define the mechanistic features of this critical coupling in the λ packaging motor.
Strong Coordination Between Protomers in the λ Packaging Motor.
Translocating motors couple ATP hydrolysis to motor movement (25–27). The motors act as oligomeric complexes, and varying degrees of coordination between the protomers have been reported. For instance, the ClpB protease that translocates and degrades polypeptide substrates is composed of six subunits that exhibit little coordination during movement (28). In contrast, tight protomer coupling has been reported in the dynein and hexameric helicase motors (26, 29). Structural models of the T4 TerL subunit bound to the procapsid suggest that there is minimal contact between the protomers, and it was suggested that they might work independently of each other during translocation (30). In contrast, single molecule studies suggest that the protomers are tightly coordinated and in constant contact with the DNA in the T4 and ϕ29 packaging motor systems (31, 32).
The λ motor is highly processive and packages the entire 48.5-kb genome with minimal slips or stalls (7, 15). In addition, the motor generates extremely high forces as required to package dsDNA to near crystalline density (7). These features suggest that the motor protomers must be in contact with each other and with the DNA at all times. Indeed, we previously demonstrated that λ terminase maintains a tight grip on the duplex to prevent leakage of the highly pressurized DNA even at the end of the packaging reaction (33). The biochemical data presented here further demonstrate that strong coordination and communication between the protomers is essential to the packaging reaction.
Genome Maturation Complex.
The first step in DNA packaging is assembly of terminase protomers at cos to engender a genome maturation complex. Early models presumed this was a dimeric complex that introduced symmetric nicks into the cosN sequence (9). In contrast, we recently proposed that the maturation complex is actually comprised of a dimer of dimers with half-site activity (12). The kinetic interrogation of chimeric maturation complexes described here is consistent with either of these two models; however, we prefer the latter based on a variety of physical and biochemical data. First, AUC studies indicate that the protomer is in slow equilibrium with a ring tetramer, and we have not seen evidence for a significant population of dimers in solution (10–12). Second, kinetic studies indicate that the catalytically competent maturation complex is composed of four to five protomers (12). Finally, a unified complex involved in both maturation and packaging is mechanistically pleasing. Terminase alternates between a stationary maturation complex and a dynamic packaging motor to processively package monomeric genomes from the concatemer (Fig. 1). A singular complex avoids recruitment and ejection of protomers during each cycle and requires only conformational adjustments to alternate between the distinct complexes. For parsimony, we favor a model where a single complex, the ring tetramer, is responsible for the entirety of the maturation and packaging processes.
Conclusions
We propose that the catalytically competent λ packaging motor is a symmetric complex composed of four protomers, each of which hydrolyzes ATP and translocates DNA in a tightly coordinated manner. This motor stoichiometry differs from the pentameric motors proposed in the ϕ29 and T4 systems (34, 35); however, the operative feature of tight coordination in the motor protomers appears to be common, and it is likely that the fundamental features of coordinated ATP hydrolysis, DNA movement, and tight association between the motor and the duplex during translocation are recapitulated in all of the viral motors. We further propose that the λ maturation complex is similarly composed of four protomers that are assembled as a dimer of dimers with half-site reactivity. This unified model obviates cyclic recruitment and ejection of protomers from the maturation and packaging complexes during viral assembly. The essential features of coupled genome maturation and packaging are likely recapitulated in all of the dsDNA viruses that package DNA from a concatemeric precursor, from phage to herpesviruses. All of these viruses face similar challenges in particle assembly: specific recognition of viral DNA, preparation of the genome for packaging, assembly of a processive and powerful packaging motor, and cyclic alternation between the complexes to allow for multiple packaging events from a single DNA encounter.
Materials and Methods
WT and mutant terminase enzymes were expressed and purified as homogenous TerL1•TerS2 protomers as previously described (12). Sedimentation velocity analytical ultracentrifugation (AUC) analysis of the purified preparations was performed as described previously (11, 12). DNA packaging and maturation reactions were performed and quantified as described previously (12). The experimental data were evaluated according to discrete molecular models as described in the text, and the sum of squared error (SSE) for each model is presented in the figure legends and in SI Text.
Supplementary Material
Acknowledgments
We thank Dr. Michael Feiss for providing the gpA-K76R expression vector. We also thank the members of the C.E.C. laboratory for spirited discussions. This work was supported by National Institutes of Health Grants GM088186 (to C.E.C.) and F32GM-90565 (to B.T.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222820110/-/DCSupplemental.
References
- 1.Calendar R, Abedon ST. The Bacteriophages. New York: Oxford Univ Press; 2006. [Google Scholar]
- 2.Knipe DM, Howley PM. Fields Virology. 5th Ed. New York: Lippincott-Williams, and Wilkins; 2007. [Google Scholar]
- 3.Catalano CE. Viral genome packaging machines: An overview. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 1–4. [Google Scholar]
- 4.Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. New York, NY: Lippincott, Williams, and Wilkins; 2007. pp. 2501–2602. [Google Scholar]
- 5.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 6.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9(9):647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 7.Fuller DN, et al. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373(5):1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurmemmedov E, Castelnovo M, Catalano CE, Evilevitch A. Biophysics of viral infectivity: Matching genome length with capsid size. Q Rev Biophys. 2007;40(4):327–356. doi: 10.1017/S0033583508004666. [DOI] [PubMed] [Google Scholar]
- 9.Feiss M, Catalano CE. Bacteriophage lambda terminase and the mechanism of viral DNA packaging. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 5–39. [Google Scholar]
- 10.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: Implications for the DNA packaging motor. J Mol Biol. 2005;347(3):523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45(51):15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 12.Andrews BT, Catalano CE. The enzymology of a viral genome packaging motor is influenced by the assembly state of the motor subunits. Biochemistry. 2012;51(46):9342–9353. doi: 10.1021/bi300890y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catalano CE. The terminase enzyme from bacteriophage lambda: A DNA-packaging machine. Cell Mol Life Sci. 2000;57(1):128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: Implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45(16):5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- 15.Yang Q, Catalano CE, Maluf NK. Kinetic analysis of the genome packaging reaction in bacteriophage lambda. Biochemistry. 2009;48(45):10705–10715. doi: 10.1021/bi901016n. [DOI] [PubMed] [Google Scholar]
- 16.Duffy C, Feiss M. The large subunit of bacteriophage lambda’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316(3):547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 17.Yang Q, Catalano CE. Biochemical characterization of bacteriophage lambda genome packaging in vitro. Virology. 2003;305(2):276–287. doi: 10.1006/viro.2002.1602. [DOI] [PubMed] [Google Scholar]
- 18.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: Structure and mechanism. Cell Mol Life Sci. 2005;62(6):685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 19.Chang JR, Andrews BT, Catalano CE. Energy-independent helicase activity of a viral genome packaging motor. Biochemistry. 2012;51(1):391–400. doi: 10.1021/bi201604b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan S-H, Stoddard BL, Xu SY. Natural and engineered nicking endonucleases—from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res. 2011;39(1):1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang JQ, Catalano CE, Feiss M. The functional asymmetry of cosN, the nicking site for bacteriophage lambda DNA packaging, is dependent on the terminase binding site, cosB. Biochemistry. 2001;40(44):13370–13377. doi: 10.1021/bi011126r. [DOI] [PubMed] [Google Scholar]
- 22.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao VB, Mitchell MS. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J Mol Biol. 2001;314(3):401–411. doi: 10.1006/jmbi.2001.5169. [DOI] [PubMed] [Google Scholar]
- 24.Yeo A, Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J Mol Biol. 1995;245(2):141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 25.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18(2):243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gennerich A, Vale RD. Walking the walk: How kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21(1):59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajendar B, Lucius AL. Molecular mechanism of polypeptide translocation catalyzed by the Escherichia coli ClpA protein translocase. J Mol Biol. 2010;399(5):665–679. doi: 10.1016/j.jmb.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 28.Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J Mol Biol. 2008;378(1):178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 29.Patel SS, Pandey M, Nandakumar D. Dynamic coupling between the motors of DNA replication: Hexameric helicase, DNA polymerase, and primase. Curr Opin Chem Biol. 2011;15(5):595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feiss M, Rao VB. The Bacteriophage DNA Packaging Machine. In: Rossmann MG, Rao VB, editors. Viral Molecular Machines. New York: Springer; 2012. pp. 498–509. [Google Scholar]
- 31.Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochemistry of a viral DNA packaging motor. J Mol Biol. 2010;400(2):186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Chistol G, et al. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151(5):1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Q, Maluf NK, Catalano CE. Packaging of a unit-length viral genome: The role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage lambda. J Mol Biol. 2008;383(5):1037–1048. doi: 10.1016/j.jmb.2008.08.063. [DOI] [PubMed] [Google Scholar]
- 34.Morais MC, et al. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16(8):1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135(7):1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.