Abstract
We performed a population genomics study of the aye-aye, a highly specialized nocturnal lemur from Madagascar. Aye-ayes have low population densities and extensive range requirements that could make this flagship species particularly susceptible to extinction. Therefore, knowledge of genetic diversity and differentiation among aye-aye populations is critical for conservation planning. Such information may also advance our general understanding of Malagasy biogeography, as aye-ayes have the largest species distribution of any lemur. We generated and analyzed whole-genome sequence data for 12 aye-ayes from three regions of Madagascar (North, West, and East). We found that the North population is genetically distinct, with strong differentiation from other aye-ayes over relatively short geographic distances. For comparison, the average FST value between the North and East aye-aye populations—separated by only 248 km—is over 2.1-times greater than that observed between human Africans and Europeans. This finding is consistent with prior watershed- and climate-based hypotheses of a center of endemism in northern Madagascar. Taken together, these results suggest a strong and long-term biogeographical barrier to gene flow. Thus, the specific attention that should be directed toward preserving large, contiguous aye-aye habitats in northern Madagascar may also benefit the conservation of other distinct taxonomic units. To help facilitate future ecological- and conservation-motivated population genomic analyses by noncomputational biologists, the analytical toolkit used in this study is available on the Galaxy Web site.
Keywords: conservation genomics, landscape species concept, genomics of non-model species
Madagascar maintains one of the highest levels of unique biodiversity—coupled with imminent extinction risk—in the world (1–4). The endemic primates, lemurs, are among the most diverse faunal groups on Madagascar, with ∼100 distinct extant taxa (5). Because of a unique ecological and demographic profile, the lemur species with the largest geographical distribution, the aye-aye (Daubentonia madagascariensis) (6), may also be among the most sensitive to continuing degradation of Madagascar’s forests. Specifically, aye-ayes have very large individual home-range size requirements relative to other lemurs (7–11), population densities that are inferred to be very low (12), a relatively slow life history (13), and the lowest nuclear genetic diversity of any primate yet studied (14). Therefore, their ability to maintain sufficient individual numbers for long-term population viability in remaining forest patches may be at risk.
Aye-ayes are highly specialized extractive foragers, with relatively large, continuously growing incisors that are used to gnaw through decaying tree bark (deadwood) or bamboo to access wood-boring insect larvae and through the endocarp of seeds from the ramy tree (Canarium) to access endosperm (7, 15–17). A slender, flexible, probing third digit is used to extract these foods and bring them to the mouth (18, 19). Limitations in the availability of either deadwood or Canarium resources may explain the large individual home-range requirements, but this has not yet been shown. Aye-ayes are also nocturnal, cryptic, and primarily solitary, making them difficult to study and sample in the wild. As a result, no comparative population studies of this species have been published previously.
Adequate conservation planning requires knowledge of both long-term landscape dynamics and patterns of species distribution in suitable habitats (20). Wider geographic scale assessments are particularly important for species with large-range requirements (e.g., refs. 21 and 22), such as aye-ayes. Thus, it is important to understand the patterns of genetic differentiation that exist among surviving aye-aye populations. Because of their extensive individual home-range sizes and low population densities, conservation efforts relevant to aye-ayes will need to prioritize the preservation of large and contiguous forests. Although such protected areas do exist in Madagascar [albeit many of them currently under stress (23)], prior to this study we have not been able to assess whether current protected areas and conservation strategies maximize the preservation of distinct aye-aye populations and overall genetic diversity, because the genetic relationships among aye-aye populations have been unknown.
The analysis of population-level genome sequence data offers potentially powerful insights into both demographic and evolutionary processes. Although such analyses could thus benefit behavior, conservation, and ecological research across many taxa (24–27), large-scale whole-genome sequencing population studies conducted to date have typically focused on humans and model organisms (e.g., refs. 28–30). With continued increases in sequencing capacity, genomic-scale population studies of nonhuman, nonmodel organisms are increasingly feasible. Indeed, transcriptome sequencing, other reduced representation methods, and whole-genome sequencing approaches have been used in multiple recently published studies (14, 31–35).
In this study, we have generated and analyzed intermediate-coverage whole-genome sequence data for 12 aye-aye individuals from three regions of Madagascar (Fig. 1). We investigated population structure and quantified population differentiation using an analytical toolkit that we have made available through the Galaxy Web site (36, 37) to facilitate similar, future studies of other species. Although the analyses in this study benefitted from our previous assembly of an aye-aye reference genome (38) and high-quality DNA samples isolated from blood and tissue, the absence of such resources would not preclude the application of a similar pipeline to other nonmodel species (SI Text). For example, for this study we collected a total of more than twice as much genome sequence data than we had used to assemble the aye-aye reference genome (38). As an initial step in analyzing other species, a genome assembly could be constructed, benefitting from continually improving genome-assembly algorithms. For analyses like those reported in this article, long-range contiguity of the assembly is immaterial and thus relatively simple assembly methods are adequate. Alternatively, analyses can use the available genome sequence and gene annotations of a related species, such as analyzing bear sequences using the dog assembly and genes (34), possibly restricting attention to gene-coding regions (39) and focusing on synonymous sites. Finally, once sequence data are available from one individual of a species, genomic-scale population analyses can then be accurately performed with noninvasive samples using a DNA capture approach (40).
Fig. 1.
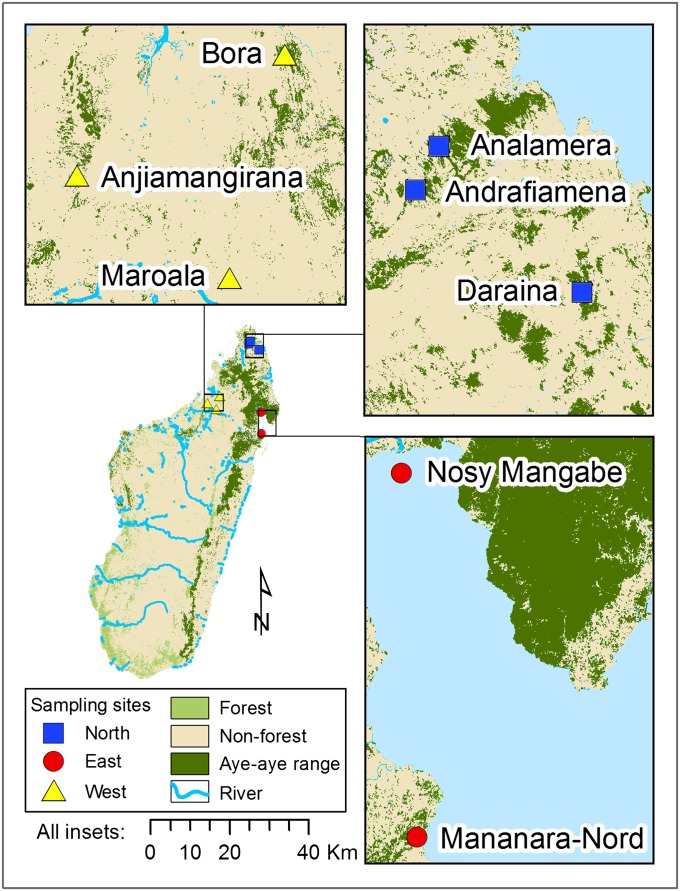
Aye-aye geographical species range and sampling locations. Remaining forests (light green) and presumed current species distribution of aye-ayes (dark green) in Madagascar. Capture locations for the North (blue squares), West (yellow triangles), and East (red circles) aye-aye population samples included in this study are indicated. Species distribution is based on the selection of remaining forest [identified from classified satellite imagery, deforestation data from Harper, et al (2007), courtesy of Cambridge University Press (51)] using a vector polygon of aye-aye distribution from Andrainarivo et al. (71), which was then further modified according to our field observations of aye-aye feeding traces and occasional sightings. Nonforested areas were not represented in the aye-aye distribution.
Results
We analyzed intermediate-coverage whole-genome sequence data for 12 aye-aye individuals from three regions of Madagascar: North (n = 4 individuals), West (n = 3), and East (n = 5) (Fig. 1). We identified SNPs following sequence-read alignment to an aye-aye reference genome (38). To limit the incorporation of erroneous genotypes into our analyses, we focused on the genotypes of 666,256 SNPs (of a total of 4,555,737 SNPs) that were covered by a minimum of four sequence reads per individual (see Materials and Methods).
Aye-Aye Population Structure.
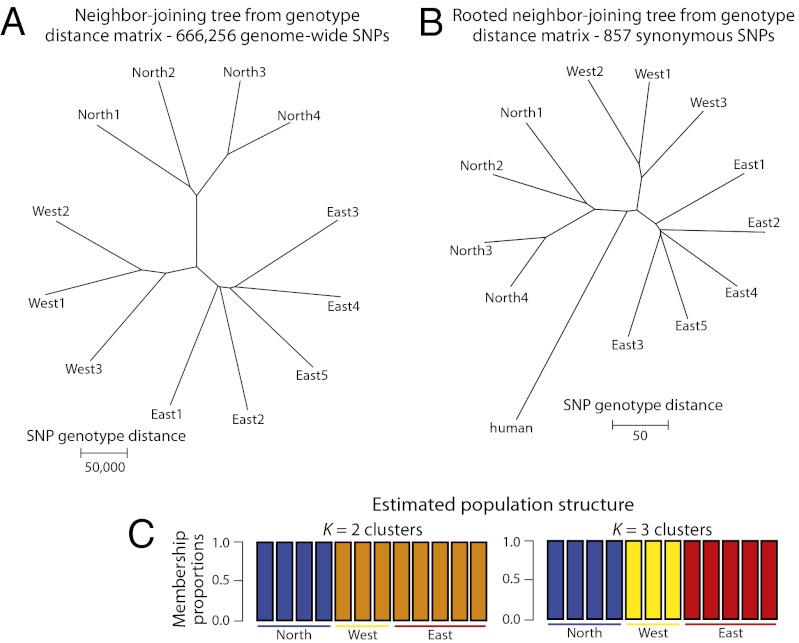
To examine relationships among the aye-aye individuals from the three regions of Madagascar, we constructed a neighbor-joining tree based on genome-wide SNP genotype distances. Individuals from each of the three populations cluster, and the East and West population clusters are more similar to each other than either is to the North (Fig. 2A). In addition, we constructed a rooted neighbor-joining tree by first aligning the aye-aye and human genome sequences (hg19). Given the relatively high levels of sequence divergence in introns and intergenic regions, to ensure orthology it was necessary to focus on gene coding regions. We then analyzed only autosomal synonymous SNPs from the gene-coding regions, because such SNPs do not affect amino acid sequences and are thus presumably neutral. Although the resulting tree is based on a distance matrix constructed from magnitudes of fewer SNPs than that of the genome-wide dataset (857 vs. 666,256), the patterns of aye-aye population structure are the same and the root of the tree separates the North from the East and West populations (Fig. 2B). A population structure analysis produced consistent results, with the North individuals distinguished from all others at k = 2 populations, and individuals from each of the three regions completely separated at k = 3 (Fig. 2C). Results from a principal component analysis are also similar. The first principal component clearly separates the North individuals from all others, and the second principal component separates West and East individuals (Fig. S1).
Fig. 2.
Aye-aye population structure. Analyses of estimated genotype SNPs with minimum 4× sequence coverage in each of the 12 individuals studied, and maximum 120× coverage in those individuals combined (Materials and Methods). (A) Neighbor-joining tree estimated from a genotype distance matrix based on all 666,256 genome-wide SNPs. Pairwise distances were calculated as total SNP genotype distance, with distance for an individual SNP the difference between two individuals’ genotypes scored as 0, 0.5, and 1 (e.g., AA, AT, and TT, respectively). (B) Rooted neighbor-joining tree estimated from a distance matrix based on 857 autosomal synonymous SNPs from gene coding regions that could be aligned to the human genome (hg19). Pairwise distances were calculated as described above. The nucleotide of the human reference sequence was different from both aye-aye alleles for 73 of the 857 SNPs; in these cases the human genotype was scored as 0.5. (C) Population structure analyses based on all 666,256 genome-wide SNPs. Cluster membership proportions for each individual are depicted for both k = 2 and k = 3 populations. Each individual is represented as a vertical bar with population origins indicated below the bars.
Population Differentiation.
To quantify the level of genetic differentiation between aye-aye populations, or the amount of total genetic variation that can be explained by population structure, we estimated FST for each SNP that was not fixed for the same allele in each of the two populations being compared. We calculated FST values using an unbiased estimator from Reich et al. (41) that is not adversely affected by small population sample sizes (42). The average FST values were 0.169 for the North vs. East populations (596,785 SNPs), 0.194 for the North vs. West populations (517,323 SNPs), and 0.129 for the East vs. West populations (536,734 SNPs).
We next assessed the level of observed genetic differentiation between aye-aye populations in a comparative context. To do so, we created an equivalent dataset for humans based on publicly available data. Specifically, we obtained genome sequence data for a total of 12 human individuals from three populations that were, as were our aye-aye data, generated using Illumina sequencing technology. The sampled human populations were sub-Saharan African agriculturalists (n = 4 individuals), European (n = 5 individuals), and Southeast Asian (n = 3 individuals). We matched sequence coverage levels to the aye-aye data at both the individual and population levels (Fig. S2), and used the same pipeline for sequence alignment, SNP genotype estimation, data filtering (e.g., minimum fourfold sequence coverage per individual), and FST analysis.
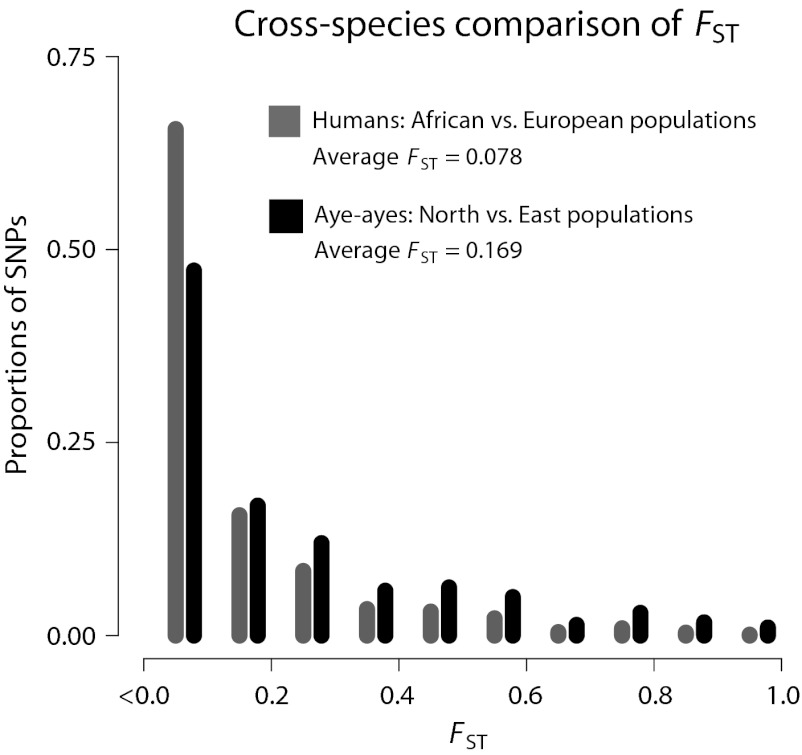
The average FST values were 0.078 for the African vs. European populations (1,061,671 SNPs), 0.091 for the African vs. Asian populations (988,646 SNPs), and 0.069 for the European vs. Asian populations (748,232 SNPs). Thus, the level of genetic differentiation between aye-aye populations from the North and East regions of Madagascar is more than 2.1-times greater than that between human Africans and Europeans based on an equivalent dataset (Fig. 3). The relative level of aye-aye versus human population differentiation was similar for each of the other equivalent comparisons. Furthermore, the two least-differentiated aye-aye populations, East and West (average FST = 0.129), are likely more genetically differentiated than Africans and Asians (average FST = 0.091), the two most differentiated human populations in our analysis. We obtained consistent results from SNP subsets generated using sequence coverage cut-offs of five-, six- and sevenfold per individual, and when using Weir and Cockerham’s unbiased estimator for FST (43) or Wright’s original FST definition (44) instead of Reich et al.’s unbiased FST estimator (41) (Fig. S3).
Fig. 3.
Cross-species FST frequency distribution comparison. Frequency distributions of FST values for the North vs. East aye-aye populations and the African vs. European human populations for SNPs meeting sequence coverage requirements (Materials and Methods). SNPs that were fixed for the same allele in both of the two compared populations were excluded (i.e., some SNPs were variable only among or the East aye-aye population sample or the Asian human population sample, or between these populations and the analyzed populations). There were 596,785 and 1,061,671 SNPs analyzed for aye-ayes and humans, respectively. FST values were computed using the unbiased estimator from Reich et al. (41). For the figure, SNPs with negative FST values were included in the first bin (FST ≤ 0.1). The average FST value for the two aye-aye populations (0.169) is 2.17-times greater than the average human FST (0.078).
Neutral Genetic Diversity.
Aye-ayes have the lowest level of genetic diversity of any studied primate species (14, 38, 45), which is likely a function of large individual range requirements and low population densities throughout the aye-aye species distribution. Alternatively, because the previous genetic diversity estimates were based primarily on individuals with ancestry from only one region of Madagascar (equivalent to our East population), they could reflect population-specific rather than species-wide demographic processes. We sought to address this issue by estimating neutral genetic diversity separately for each of the three aye-aye populations in our study. Although we expect to underestimate true genetic diversity with the intermediate coverage sequence data (because of the undercalling of rare SNPs and heterozygous sites), these estimates should be generally comparable both across aye-aye populations and against humans (using our sample size and sequence coverage-matched human dataset).
We identified the total number of autosomal synonymous sites covered by a minimum of four sequence reads per individual for each species (aye-aye = 368,675 synonymous sites; human = 915,245 sites) (SI Materials and Methods) and computed average pairwise genetic diversity (π) for each population: aye-aye North π = 0.054%, aye-aye East π = 0.057%, aye-aye West π = 0.049%, human African π = 0.093%, human European π = 0.070%, human Asian π = 0.066%. As expected, these estimates are lower than those from previous studies that used higher coverage sequence data (14), but the magnitude of the between-species differences are similar. Moreover, in contrast to the variability observed among human populations [as expected, with higher genetic diversity in Africa (46)], genetic diversity levels are similar among aye-aye populations suggesting that relatively low genetic diversity is a species-wide characteristic.
Discussion
We conducted this study to characterize patterns and levels of genetic differentiation among aye-aye populations for conservation planning purposes as well as to contribute to the general understanding of biogeographical processes in Madagascar, as this species has the widest geographical distribution of any lemur (6, 12). The aye-aye’s demographic profile suggests a particular sensitivity of this species to the rapid degradation and fragmentation of Madagascar’s forests. Specifically, aye-aye home-range sizes of 120–215 ha for males and 30–40 ha for females [with travel distances up to 4.4 km/night (47)] are very large for a solitary animal only ∼2.5 kg in size and considerably larger than those of other lemurs across a diversity of activity patterns, social systems, and body sizes [for example, 1–2 ha for woolly lemurs (8), 5 ha for fork-marked lemurs (9), 5.3 ± 5.2 ha for blue-eyed black lemurs (10), and 5.7–10.1 ha for Verreaux’s sifakas (11)]. Because there is minimal same-sex overlap in female home ranges (47), aye-aye population densities are inferred to be relatively very low (12). Perhaps unsurprisingly, the level of estimated nuclear genomic diversity in aye-ayes is the lowest of any primate yet studied (14). Thus, the ability of aye-ayes to maintain sufficient individual numbers for long-term population viability in remaining forest patches may be at risk.
Our analyses revealed that although aye-ayes from the East and West coasts of Madagascar are distinguished readily by their genome sequences, divergence between either of these populations and the northern Madagascar population is greater. Although connected forests between the habitats of the North and East sampled populations no longer exist (Fig. 1), the level of genetic differentiation between the populations in these regions implies a longer-term reproductive barrier than that which could be attributed to the human-mediated habitat loss that began only within the past 2,300 y (48, 49) before accelerating rapidly over the past century (50, 51). We found that the level of genetic differentiation between the North and East populations is substantially greater than that between, for example, human African and European populations, based on the analysis of an equivalently curated human SNP database. Although the relative level of aye-aye genetic differentiation—across only a relatively small geographic distance (248 km), representing a small part of the total aye-aye range (Fig. 1)—is intriguing, what relevance does this result have for conservation planning? In particular, the level of genetic differentiation among human populations is not typically considered high among primates (52). In addition, the observed level of aye-aye population differentiation may not be unusual for a species with low population sizes and geographical barriers across its range. Future expectations of an expanded population genomic database that will include many additional endangered taxa will help us to better contextualize this result.
As a starting point, we also computed the average FST between populations of Alaskan and Norwegian polar bears using data from a recent genomics study (34). Although the polar bear sequence coverage levels could not be matched precisely to our aye-aye and human datasets, the average North vs. East aye-aye FST was more than five-times greater than that for polar bears (SI Materials and Methods), strengthening the belief that the observed aye-aye FST values may be unusual for a wide-ranging animal over such a small geographic distance. For now, we are not suggesting that the North aye-aye population should necessarily be considered a distinct taxonomic unit. However, if general goals of conservation planning include preserving distinct populations and maximizing overall species-level genetic diversity, then this population merits particular protection, especially for a species with a demographic profile that suggests high extinction risk.
Because aye-ayes have a geographical species distribution that only excludes the central highlands and the southwest of Madagascar [the largest distribution of any lemur (6, 12)], these results are also particularly valuable for our broader understanding of the island’s biogeography. Indeed, the distinctiveness of the North aye-aye population accords well with 1 of 12 proposed cross-taxa centers of endemism, based on an analysis of elevation, the location of watersheds, and Quaternary climatic shifts (53). Multiple river systems, such as the Manambato, Bemarivo, and Ankavanana, have the potential to limit the dispersal of aye-ayes and other species between northern Madagascar and adjacent regions. Furthermore, the Tsaratanana Massif, which includes the highest peak in Madagascar, rises above the known elevational limits of aye-ayes (54), and thus may form at least a partial barrier to gene flow. We have sampled from only a portion of the total aye-aye species distribution. The cryptic, nocturnal nature of this species and low population densities constrain sampling efforts, but future population genomic studies that include populations from further south along the west and east coasts of Madagascar would likely contribute further to biogeographic knowledge and to aye-aye conservation efforts.
Preservation of the distinctive aye-aye populations in northern Madagascar would likely have indirect benefits for the conservation of other taxa within this center of endemism, the genetic diversity and structure of which are not yet fully characterized. The geographical correspondence of this northern region with patterns of species turnover in other lemur taxa (53) suggests similar underlying biogeographic processes and responses to landscape variation. Although the use of surrogate species for conservation management is strongly debated (55, 56), carefully selected individual species or groups may serve as appropriate indicators of overall biodiversity, although these applications are often highly context-dependent (57). Similarly, protecting species at the highest trophic levels and with large home ranges (e.g., apex predators) can serve ecosystem-level conservation goals and preserve disproportionate amounts of diversity in other taxa (58). Although not strictly predatory, aye-ayes maintain an ecological niche that necessitates large individual ranges, which can incorporate entire communities and populations of other endemic fauna, including other lemurs. Thus, any efforts to preserve contiguous forests large and diverse enough to support a viable population of aye-ayes would be very likely to also meet or exceed the space requirements for other taxa. Additionally, because of their relatively slow life history, aye-ayes may be particularly sensitive to rapid habitat changes. The combination of these factors would make aye-ayes a strong candidate for a focal species in a “landscape species” approach to conservation planning (59, 60).
Conclusion
In this study, we generated and analyzed complete genome sequences from 12 aye-aye individuals to characterize levels and patterns of genetic differentiation and to highlight a distinct population in northern Madagascar. This work serves as a potential model for future conservation- and ecology-motivated population genomic studies of nonmodel species; such research is expected to become more feasible with continuing advances in sequencing technology and capacity. Thus, we have made the analytical toolkit used for our analyses available on the Galaxy Web site. We argue that conservation attention should be directed toward an important center of endemism in northern Madagascar. Such efforts would preserve distinct populations of a species that is one of the world’s most unusual and highly specialized mammals, as well as other potentially distinct populations and taxa in the region.
Materials and Methods
DNA Samples.
DNA samples from 13 wild-caught aye-aye individuals were initially included in this study. Genomic DNA was extracted from liver-tissue samples collected at necropsy or whole venous blood from wild-born founders at the Duke Lemur Center (Durham, NC) and from whole venous blood samples collected from free-ranging individuals in Madagascar. In Madagascar, aye-ayes were immobilized with a CO2 projection rifle or blowgun with 10 mg/kg of Telazol (Fort Dodge Animal Health), and 1.0 cc/kg whole blood was collected and placed in storage buffer [0.1 M Tris, 0.1 M NA2EDTA, 2% (vol/vol) SDS] at room temperature until transferred to the laboratory for storage at −80 °C. All collection and export permits were obtained from Madagascar National Parks, formerly Association Nationale pour la Gestion des Aires Protégées, the Ministère des Eaux et Forêts of Madagascar. All rules and regulations were followed according to Malagasy law. Samples were imported to the United States under Convention for International Trade in Endangered Species permits 08US121039/9, 08US121040/9, and 08US121041/9 from the US Fish and Wildlife Service. Capture and sampling procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Omaha’s Henry Doorly Zoo and Aquarium under IACUC #12–101. Genomic DNA was isolated from the samples using a standard Phenol-chloroform extraction protocol (61).
Sequencing, Sequence Alignment, and SNP Identification.
For detailed descriptions of the library preparation, sequencing, alignment, and SNP identification methods see SI Materials and Methods. Each sample was pair-end sequenced for 101 bp from each end using one lane of the Illumina HiSEq. 2000 sequencing system. We obtained an average of 204,202,246 total reads per lane (SD = 40,714,120) (Dataset S1), or ∼20 Gb of raw sequence data per individual. Sequence data have been deposited in the National Center for Biotechnology Institute short read archive under accession no. SRA066444. Sequence reads were aligned to the aye-aye reference genome sequence (38) using the Burrows–Wheeler Aligner (62). On average, we mapped 16 Gb of sequence data per individual (SD 4.1 Gb) (Dataset S1), corresponding to an average of ∼5.6-fold coverage of the 2.9-Gb aye-aye reference genome sequence. We used SAMtools (63) to identify the locations of SNPs and estimate genotypes at all SNPs for each individual, regardless of sequence coverage for that SNP and individual.
Quality Control and SNP Filtering.
We selected an intermediate-coverage population genomics study design, using one HiSeq lane per individual (resulting in an average of ∼5.6× per-individual mapped sequence coverage). For analyses of these data, our approach was to focus on the subset of identified SNPs with a minimum of 4× sequence coverage for each individual in the study. In doing so, we ignored the majority of our data, but we have more confidence in the accuracy of the estimated genotypes of the SNPs that we do analyze. The number of SNPs remaining after this filtering step is still large (see below), sufficient for accurate population genetics inference (e.g., ref. 64). In future population genomics studies, it may become possible to extend the analysis to a larger proportion of variable sites in the genome by generating high-coverage sequence data for each individual. However, such a design would have been economically and computationally inefficient for our study, given current output of the Illumina HiSEq. 2000 sequencing system and data storage and analysis computing resource needs.
With only 4× coverage, some error in the estimated genotypes is expected (e.g., the probability of observing both SNP alleles among four reads for a heterozygous individual = 0.875). However, the 4× threshold is the minimum coverage for each individual; thus for any given SNP, the coverage levels for most individuals are higher. At this coverage threshold, overall genotype estimate qualities are expected to be high.
Before applying the 4× minimum coverage filter, we considered the coverage distributions (for the 4,555,737 identified SNPs) for each individual (Fig. S2A). One individual, North5, was a coverage outlier, with a mode of only twofold coverage and 70% of the SNP sites covered by fewer than four reads. North5 also had the fewest mapped reads of any individual in the study (Dataset S1). Accordingly, we excluded North5 from further analyses. We also examined the sum total coverage distribution for the remaining 12 individuals (North1 to -4, West1 to -3, East1 to -5) (Fig. S2B). SNPs with relatively low sum total coverage will be filtered because they would not meet the 4× per-individual threshold. Some proportion of the SNPs with relatively high sum total coverage could be located in duplicated regions of the genome, and thus potential false-positives. Therefore, we chose to filter all SNPs with >120× sum total coverage. With this step, we likely excluded many true SNPs, but the remaining number of SNPs was large. Specifically, there were 666,256 SNPs with minimum 4× coverage in each of the 12 individuals, and not more than 120× coverage in those individuals combined. These SNPs were the focus of our population genomic analyses.
Galaxy Tools.
We created tools on the Galaxy Web site usegalaxy.org (36, 37) to facilitate ecological- and conservation-motivated analyses of population genomics datasets such as ours, by noncomputational biologists. The user uploads SNP genotype calls with coverage and genotype quality information (alternatively, SAMtools functionality is also available through Galaxy). From the uploaded SNP table, the user may specify populations, compute and display coverage distributions at the individual and population levels, filter SNPs based on individual and population minimum and maximum sequence coverage levels, as well as minimum genotype quality, examine population structure, and perform analyses based on FST. Several popular population genetics and genomics analysis programs, including SMARTPCA (65) and ADMIXTURE (66), have been integrated into the Galaxy functionality. For detailed descriptions of the population genomic analyses used in this study, see SI Materials and Methods.
Human SNP Comparative Data.
To understand the significance of the average FST value between the North and East aye-aye populations, we created a similar dataset of human sequence data. Specifically, given the relatively small sample sizes for each population in our study and the use of intermediate-coverage sequence data, unknown false-positive and false-negative SNP call error rates and genotyping errors may have affected the accuracy of our aye-aye average FST estimate. Therefore, it was inappropriate to compare our result for the two aye-aye populations to published reports of average FST from other species that were based on microarray-based SNP genotyping data or Sanger sequencing data (or high-coverage massively-parallel sequencing data, but to our knowledge such datasets have not yet been published). To address this issue, we downloaded Illumina sequence reads for four African agriculturalist (e.g., Bantu-speaking) individuals, five individuals of European descent, and three Southeast Asian individuals from published and publicly available genome-sequencing studies (28, 29, 67–70) (Dataset S2). For our comparative purposes, we considered the African sample equivalent to the aye-aye North population sample (as described in Results, the aye-aye North population is more distinct from the West and East populations than either of the West and East populations are from each other; this is equivalent to the relationship among human African versus European and Southeast Asian populations), the European sample equivalent to the East, and the Southeast Asian sample to the West. We included varying numbers of reads for each individual (Dataset S3) to match the aye-aye coverage distributions, at both the individual and population levels (Fig. S2C).
We aligned reads, identified SNPs, and estimated genotypes in a manner identical to that used for the aye-aye data, except using the human genome (hg19) as a reference. A total of 8,598,051 SNPs were identified. There are at least two reasons why the number of identified human SNPs was greater than the number of identified aye-aye SNPs. First, nuclear genetic diversity is lower in aye-ayes than in humans (14). Second, although the total size (∼2.9 Gb) of the aye-aye reference genome sequence (38) is similar to that of humans, it is comprised of ∼2.6 million scaffolds. Thus, the effective size of the genome for SNP analysis is lower for aye-ayes because of expected reductions in mapability and coverage levels near scaffold ends. We filtered the human SNPs to a subset of 1,146,658 SNPs with a minimum of 4× sequence coverage per individual and a maximum of 120× sum total coverage for all individuals. Population structure analyses performed on these genotype data produced the expected results (Fig. S4).
Supplementary Material
Acknowledgments
We thank Sarah Zehr and the Duke Lemur Center for providing captive aye-aye samples; Shannon Engberg and Carolyn Bailey for sample preparation; Lynn Tomsho and John McGraw for library preparation and sequencing; Tracy Wyman for assistance with Fig. 1; Emily Davenport and Kate Thompson for comments on an earlier draft of the manuscript; the Madagascar Biodiversity Partnership for assistance in sample collection and field logistics in Madagascar; and the Madagascar National Parks, formerly Association Nationale pour la Gestion des Aires Protégées, and the Ministère des Eaux et Forêts of Madagascar for sampling permission. Funding for aye-aye sample collection was provided by Conservation International, the Primate Action Fund and the Margot Marsh Biodiversity Foundation (E.E.L.), along with logistical support from the Ahmanson Foundation and the Theodore F. and Claire M. Hubbard Family Foundation. Genome sequencing was funded by the College of the Liberal Arts, Pennsylvania State University (G.H.P.). The development of the Galaxy tools to analyze intermediate-coverage sequence data from multiple individuals was supported by the National Institutes of Health Grant UL1 RR033184-01 (to the Pennsylvania State Clinical and Translational Science Institute), with additional funding from National Science Foundation Award DEB 0733029 and a grant from the Pennsylvania Department of Health using Tobacco Commonwealth Universal Research Enhancement Funds. This is Duke Lemur Center publication #1241.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences have been deposited in the National Center for Biotechnology Information short read archive, www.ncbi.nlm.nih.gov/Traces/sra (accession no. SRA066444). Analysis workflows and the full input table of 4,555,737 aye-aye SNPs are available on the Galaxy Web site (usegalaxy.org; usegalaxy.org/r/aye-aye).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211990110/-/DCSupplemental.
References
- 1.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 2.Vieites DR, et al. Vast underestimation of Madagascar’s biodiversity evidenced by an integrative amphibian inventory. Proc Natl Acad Sci USA. 2009;106(20):8267–8272. doi: 10.1073/pnas.0810821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allnutt TF, et al. A method for quantifying biodiversity loss and its application to a 50-year record of deforestation across Madagascar. Conserv Lett. 2008;1(4):173–181. [Google Scholar]
- 4.Godfrey LR, Irwin MT. The evolution of extinction risk: Past and present anthropogenic impacts on the primate communities of Madagascar. Folia Primatol (Basel) 2007;78(5–6):405–419. doi: 10.1159/000105152. [DOI] [PubMed] [Google Scholar]
- 5.Mittermeier RA, et al. Lemur diversity in Madagascar. Int J Primatol. 2008;29(6):1607–1656. [Google Scholar]
- 6.Sterling EJ. Taxonomy and distribution of Daubentonia: A historical perspective. Folia Primatol (Basel) 1994;62(1–3):8–13. doi: 10.1159/000156758. [DOI] [PubMed] [Google Scholar]
- 7.Sterling EJ. 1993. The behavioral ecology of the aye-aye on Nosy Mangabe, Madagascar. PhD dissertation (Yale University, New Haven, CT)
- 8.Ganzhorn JU, Abraham JP, Razanahoera-Rakotomalala M. Some aspects of the natural history and food selection of Avahi laniger. Primates. 1985;26(4):452–463. [Google Scholar]
- 9.Schulke O. Evolution of pair-living in Phaner furcifer. Int J Primatol. 2005;26(4):903–919. [Google Scholar]
- 10.Volampeno MSN, Masters JC, Downs CT. Home range size in the blue-eyed black lemur (Eulemur flavifrons): Between dry and wet seasons. Mamm Biol. 2011;76(2):157–164. [Google Scholar]
- 11.Benadi G, Fichtel C, Kappeler P. Intergroup relations and home range use in Verreaux’s sifaka (Propithecus verreauxi) Am J Primatol. 2008;70(10):956–965. doi: 10.1002/ajp.20588. [DOI] [PubMed] [Google Scholar]
- 12.Mittermeier RA, et al. Lemurs of Madagascar. 3rd Ed. Arlington, VA: Conservation International; 2010. [Google Scholar]
- 13.Catlett KK, Schwartz GT, Godfrey LR, Jungers WL. “Life history space”: A multivariate analysis of life history variation in extant and extinct Malagasy lemurs. Am J Phys Anthropol. 2010;142(3):391–404. doi: 10.1002/ajpa.21236. [DOI] [PubMed] [Google Scholar]
- 14.Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22(4):602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterling EJ. Aye-ayes: Specialists on structurally defended resources. Folia Primatol (Basel) 1994;62(1–3):142–154. doi: 10.1159/000156771. [DOI] [PubMed] [Google Scholar]
- 16.Erickson CJ. Tap-scanning and extractive foraging in aye-ayes, Daubentonia madagascariensis. Folia Primatol (Basel) 1994;62(1–3):125–135. doi: 10.1159/000156769. [DOI] [PubMed] [Google Scholar]
- 17.Goodman SM, Sterling EJ. The utilization of Canarium (Burseraceae) seeds by vertebrates in the Reserve Naturelle Integrale d'Andringitra, Madagascar. Fieldiana Zoology. 1996;85:83–92. [Google Scholar]
- 18.Milliken GW, Ward JP, Erickson CJ. Independent digit control in foraging by the aye-aye (Daubentonia madagascariensis) Folia Primatol (Basel) 1991;56(4):219–224. doi: 10.1159/000156551. [DOI] [PubMed] [Google Scholar]
- 19.Lhota S, Jůnek T, Bartos L, Kubĕna AA. Specialized use of two fingers in free-ranging aye-ayes (Daubentonia madagascariensis) Am J Primatol. 2008;70(8):786–795. doi: 10.1002/ajp.20548. [DOI] [PubMed] [Google Scholar]
- 20.Franklin J. Moving beyond static species distribution models in support of conservation biogeography. Divers Distrib. 2010;16(3):321–330. [Google Scholar]
- 21.Carroll C. Interacting effects of climate change, landscape conversion, and harvest on carnivore populations at the range margin: Marten and lynx in the northern Appalachians. Conserv Biol. 2007;21(4):1092–1104. doi: 10.1111/j.1523-1739.2007.00719.x. [DOI] [PubMed] [Google Scholar]
- 22.Gregory SD, et al. Long-term field data and climate-habitat models show that orangutan persistence depends on effective forest management and greenhouse gas mitigation. PLoS ONE. 2012;7(9):e43846. doi: 10.1371/journal.pone.0043846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner CJ. IUCN management categories fail to represent new, multiple-use protected areas in Madagascar. Oryx. 2011;45(3):336–346. [Google Scholar]
- 24.Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma RK, Hedrick PW. Conservation genetics in transition to conservation genomics. Trends Genet. 2010;26(4):177–187. doi: 10.1016/j.tig.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nat Rev Genet. 2010;11(10):697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- 26.Frankham R. Challenges and opportunities of genetic approaches to biological conservation. Biol Conserv. 2010;143(9):1919–1927. [Google Scholar]
- 27.Ekblom R, Galindo J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity (Edinb) 2011;107(1):1–15. doi: 10.1038/hdy.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. The 1000 Genomes Project Consortium; Abecasis GR, et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073; Erratum in: Nature (2011) 473(7348):544. [DOI] [PMC free article] [PubMed]
- 29.Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463(7283):943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones FC, et al. Broad Institute Genome Sequencing Platform and Whole Genome Assembly Team The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everett MV, Grau ED, Seeb JE. Short reads and nonmodel species: Exploring the complexities of next-generation sequence assembly and SNP discovery in the absence of a reference genome. Mol Ecol Resour. 2011;11(Suppl 1):93–108. doi: 10.1111/j.1755-0998.2010.02969.x. [DOI] [PubMed] [Google Scholar]
- 32.Geraldes A, et al. SNP discovery in black cottonwood (Populus trichocarpa) by population transcriptome resequencing. Mol Ecol Resour. 2011;11(Suppl 1):81–92. doi: 10.1111/j.1755-0998.2010.02960.x. [DOI] [PubMed] [Google Scholar]
- 33.Miller MR, et al. A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol Ecol. 2012;21(2):237–249. doi: 10.1111/j.1365-294X.2011.05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller W, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci USA. 2012;109(36):E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller W, et al. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil) Proc Natl Acad Sci USA. 2011;108(30):12348–12353. doi: 10.1073/pnas.1102838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giardine B, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goecks J, Nekrutenko A, Taylor J. Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry GH, et al. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 2012;4(2):126–135. doi: 10.1093/gbe/evr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratan A, Zhang Y, Hayes VM, Schuster SC, Miller W. Calling SNPs without a reference sequence. BMC Bioinformatics. 2010;11:130. doi: 10.1186/1471-2105-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry GH, Marioni JC, Melsted P, Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Mol Ecol. 2010;19(24):5332–5344. doi: 10.1111/j.1365-294X.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willing EM, Dreyer C, van Oosterhout C. Estimates of genetic differentiation measured by F(ST) do not necessarily require large sample sizes when using many SNP markers. PLoS ONE. 2012;7(8):e42649. doi: 10.1371/journal.pone.0042649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 44.Wright S. The genetical structure of populations. Ann Eugen. 1951;15(4):323–353. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 45.Perry GH, Martin RD, Verrelli BC. Signatures of functional constraint at aye-aye opsin genes: The potential of adaptive color vision in a nocturnal primate. Mol Biol Evol. 2007;24(9):1963–1970. doi: 10.1093/molbev/msm124. [DOI] [PubMed] [Google Scholar]
- 46.Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc Natl Acad Sci USA. 2012;109(44):17758–17764. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sterling EJ. Patterns of range use and social organization in aye-ayes (Daubentonia madagascariensis) on Nosy Mangabe. In: Kappeler PM, Ganzhorn JU, editors. Lemur Social Systems and Their Ecological Basis. New York: Plenum; 1993. pp. 1–10. [Google Scholar]
- 48.Burney DA, et al. A chronology for late prehistoric Madagascar. J Hum Evol. 2004;47(1–2):25–63. doi: 10.1016/j.jhevol.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Dewar RE, Richard AF. Madagascar: A history of arrivals, what happened, and what will happen next. Annu Rev Anthropol. 2012;41:495–517. [Google Scholar]
- 50.Green GM, Sussman RW. Deforestation history of the eastern rain forests of madagascar from satellite images. Science. 1990;248(4952):212–215. doi: 10.1126/science.248.4952.212. [DOI] [PubMed] [Google Scholar]
- 51.Harper GJ, Steininger MK, Tucker CJ, Juhn D, Hawkins F. Fifty years of deforestation and forest fragmentation in Madagascar. Environ Conserv. 2007;34(4):325–333. [Google Scholar]
- 52.Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16(11):1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 53.Wilmé L, Goodman SM, Ganzhorn JU. Biogeographic evolution of Madagascar’s microendemic biota. Science. 2006;312(5776):1063–1065. doi: 10.1126/science.1122806. [DOI] [PubMed] [Google Scholar]
- 54.Goodman SM, Ganzhorn JU. Elevational ranges of lemurs in the humid forests of Madagascar. Int J Primatol. 2004;25(2):331–350. [Google Scholar]
- 55.Simberloff D. Flagships, umbrellas, and keystones: Is single-species management passe in the landscape era? Biol Conserv. 1998;83(3):247–257. [Google Scholar]
- 56.Caro T. Conservation by Proxy: Indicator, Umbrella, Keystone, Flagship and Other Surrogate Species. Washington, DC: Island Press; 2010. [Google Scholar]
- 57.Hess GR, et al. Effectiveness of biodiversity indicators varies with extent, grain, and region. Biol Conserv. 2006;132(4):448–457. [Google Scholar]
- 58.Sergio F, Newton I, Marchesi L, Pedrini P. Ecologically justified charisma: Preservation of top predators delivers biodiversity conservation. J Appl Ecol. 2006;43(6):1049–1055. [Google Scholar]
- 59.Sanderson EW, Redford KH, Vedder A, Coppolillo PB, Ward SE. A conceptual model for conservation planning based on landscape species requirements. Landsc Urban Plan. 2002;58(1):41–56. [Google Scholar]
- 60.Didier KA, et al. The Landscape Species Approach: Spatially-explicit conservation planning applied in the Adirondacks, USA, and San Guillermo-Laguna Brava, Argentina, landscapes. Oryx. 2009;43(4):476–487. [Google Scholar]
- 61.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 65.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahn SM, et al. The first Korean genome sequence and analysis: Full genome sequencing for a socio-ethnic group. Genome Res. 2009;19(9):1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujimoto A, et al. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat Genet. 2010;42(11):931–936. doi: 10.1038/ng.691. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andrainarivo C, et al. 2012. Daubentonia madagascariensis, 2008. International Union for the Conservation of Nature 2012: IUCN Red List of Threatened Species. Available at http://www.iucnredlist.org/details/6302/0. Accessed on March 01, 2012.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.