Abstract
The multiprotein exon junction complex (EJC), deposited by the splicing machinery, is an important constituent of messenger ribonucleoprotein particles because it participates to numerous steps of the mRNA lifecycle from splicing to surveillance via nonsense-mediated mRNA decay pathway. By an unknown mechanism, the EJC also stimulates translation efficiency of newly synthesized mRNAs. Here, we show that among the four EJC core components, the RNA-binding protein metastatic lymph node 51 (MLN51) is a translation enhancer. Overexpression of MLN51 preferentially increased the translation of intron-containing reporters via the EJC, whereas silencing MLN51 decreased translation. In addition, modulation of the MLN51 level in cell-free translational extracts confirmed its direct role in protein synthesis. Immunoprecipitations indicated that MLN51 associates with translation-initiating factors and ribosomal subunits, and in vitro binding assays revealed that MLN51, alone or as part of the EJC, interacts directly with the pivotal eukaryotic translation initiation factor eIF3. Taken together, our data define MLN51 as a translation activator linking the EJC and the translation machinery.
The information relayed by mRNAs is modulated by a set of proteins that together form ribonucleoprotein particles (mRNPs). These particles are unique because their composition depends on both mRNA sequence and processing history (1). Understanding mRNPs’ dynamics along their journey is a challenge in determining their importance for gene expression in eukaryotes. In this intricate network, the exon junction complex (EJC) plays a central role in coordinating posttranscriptional events in metazoans (2). This multiprotein complex is assembled onto mRNA as a consequence of pre-mRNA splicing upstream of exon–exon junctions (3, 4). The EJC remains associated with mRNAs and is exported to the cytoplasm until it is stripped off by translating ribosomes (5). Hence, the EJC transmits mRNA history to subsequent posttranscriptional events as a molecular signature of splicing.
The EJC is a dynamic structure organized around four core proteins: the DEAD-box RNA helicase, eukaryotic initiation factor 4A3 (eIF4A3), metastatic lymph node 51 (MLN51, also known as “CASC3” or “Barentsz”), and the heterodimer Magoh/Y14 (4). Its structure revealed an atypical RNA-binding mechanism (6). In the presence of ATP, the two RecA domains of eIF4A3 form a large RNA clamp without sequence preference. The most conserved domain of MLN51, named “SELOR” for “SpEckle LOcalizer and RNA-binding module” (7), binds each RecA domain of eIF4A3 and also contacts RNA. Finally, Magoh/Y14 prevents conformational changes of eIF4A3, stabilizing this core complex. Once clamped onto mRNA, the tetrameric core acts as a platform to recruit multiple factors conferring different functions to the EJC as mRNA progresses from splicing to translation. Notably, the EJC contributes in regulating the splicing of specific transcripts (8–10) and participates in mRNA transport (11). The EJC also plays a major role in nonsense-mediated mRNA decay (NMD) that triggers the decay of aberrant mRNAs containing a premature termination codon (12). In humans, efficient NMD requires the stepwise assembly of a surveillance complex originating from the EJC core. When translation terminates prematurely upstream of a remaining EJC, the NMD up-frameshift factors 3, 2, and 1 (Upf3, Upf2, and Upf1) successively join the remaining EJC core to trigger mRNA degradation.
Finally, the EJC also is involved in mRNA translation (13). Numerous studies reported that splicing contributes to the up-regulation of mRNAs and notably to their translation (14–16). Different approaches, including the use of reporters in cultured cells and direct microinjection of synthetic RNAs in Xenopus oocytes, demonstrated that the EJC per se is responsible in part for enhancing translation by splicing (17, 18). In addition, artificial tethering of Magoh, Y14, and MLN51 to luciferase reporter mRNAs showed that these proteins enhance the efficiency of mRNA translation without affecting mRNA levels (19). Independently, the EJC peripheral factor S6K1 Aly/REF-like substrate (SKAR) also contributes to the increase in translation by recruiting the mammalian target of rapamycin (mTOR)-related S6K1 kinase that phosphorylates numerous translation factors (20). Hence, it has been proposed that SKAR relays signals from the mTOR signaling pathway to mRNPs. However, it still is not clear whether SKAR’s action is sufficient to account for the positive effect of EJC on translation or whether other EJC components participate directly in this function (13).
We used a combination of experimental approaches to show that the EJC core component MLN51 is a translation enhancer. MLN51 preferentially increases the translation of intron-containing mRNAs via its incorporation into the EJC core. MLN51 interacts physically with the pivotal initiation complex eIF3 and associates with translation factors. Our study demonstrates a direct communication between the EJC core and the translation machinery in human cells.
Results
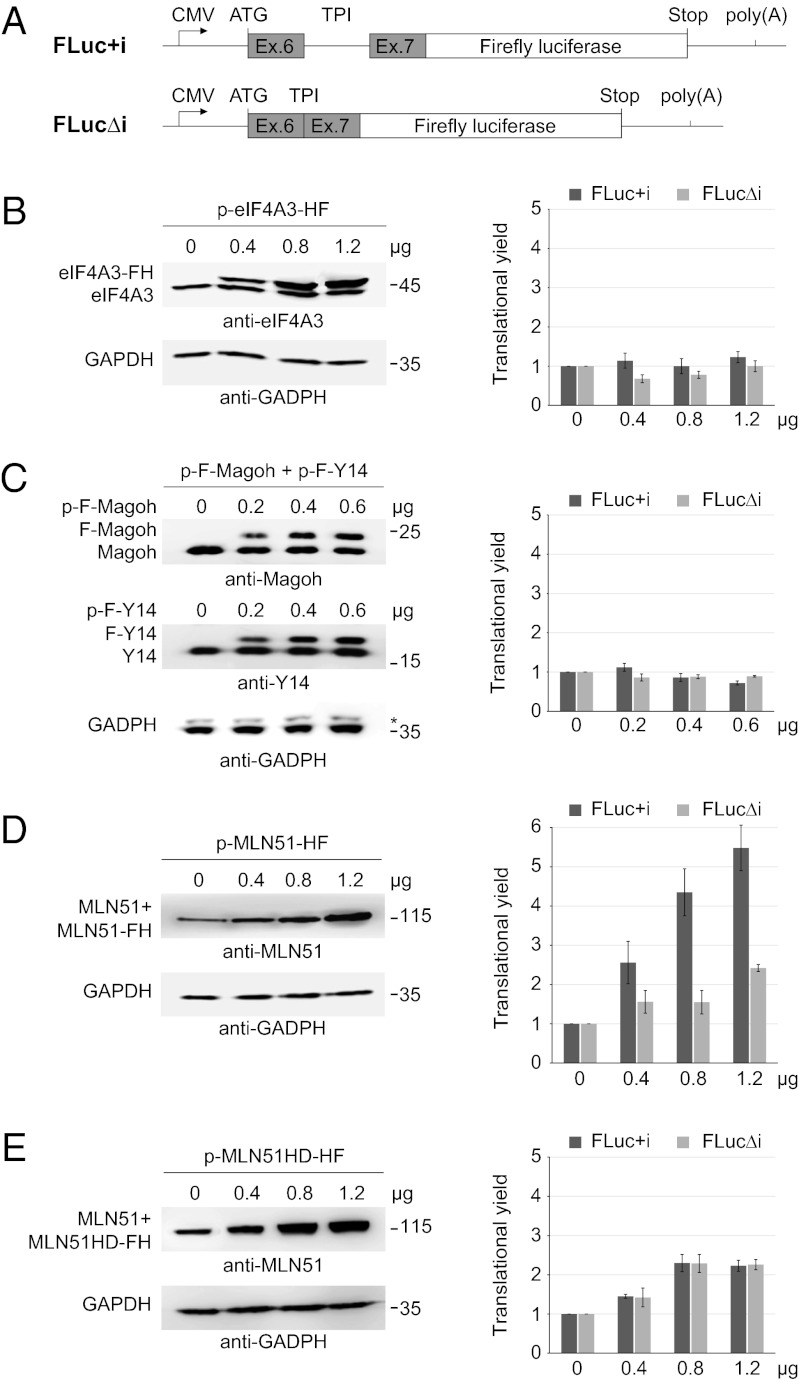
MLN51 Overexpression Enhances Translation in Human Cells.
So far, the role of EJC core components in translation has been assayed only by tethering experiments (17, 19). To investigate this EJC function in a different way, we measured the translational efficiency of reporter mRNAs in human HEK293 cells in which each core component was overexpressed. To do so, we designed two related reporters to monitor splicing effects. One contains intron 6 from human triose phosphate isomerase (TPI) flanked by its natural exons 6 and 7 and fused to the Firefly luciferase ORF (FLuc+i). The other is the corresponding intronless version (FLucΔi) (Fig. 1A). We also constructed vectors to overexpress eIF4A3 and MLN51 fused to a HA-Flag (HF) tag or to overexpress Magoh and Y14 fused to a Flag (F) tag. Because Magoh and Y14 form a stable heterodimer, the plasmids expressing each protein were cotransfected. Transfection of increasing amounts of these plasmids resulted in proportional increases in the level of each protein (Fig. 1 B–D). Endogenous and transiently expressed proteins were distinguished easily except in the case of MLN51, for which the two proteins comigrated because of the small size difference (Fig. 1D). Quantification confirmed the dose-dependent overexpression, which reached up to five times the level of the endogenous proteins (Fig. 1 B–D). To monitor the effect of overexpression on translation efficiency, we measured the luciferase activity of each reporter (FLuc+i and FLucΔi), which are normalized to the level of corresponding mRNAs measured by an RNA protection assay (RPA) (Fig. S1A). Overexpression of eIF4A3 or of Magoh and Y14 did not affect the reporters’ translation efficiency (Fig. 1 B and C). In contrast, overexpression of MLN51 enhanced the translation yield of spliced Firefly luciferase reporters in a dose-dependent manner (by more than fivefold) and, to a lesser extent, the translation yield of corresponding unspliced mRNA (by more than twofold) (Fig. 1D). The positive effect of MLN51 on translation also was observed for another intronless reporter coding Renilla luciferase (Fig. S1 B and C). Therefore, of the four EJC core proteins, only the overexpression of MLN51 positively affected mRNAs translation.
Fig. 1.
MLN51 overexpression preferentially enhances the translation of intron-containing reporters. (A) A schematic representation of luciferase constructs. TPI exons 6 and 7 (gray boxes) with or without intron 6 (thick line) were fused to the Firefly luciferase ORF (white box) to generate intron-containing (FLuc+i) and intronless (FLucΔi) FLuc reporters. (B–E) (Left) Western blots using indicated antibodies on extracts of HEK293 cells transfected with increasing amounts of the indicated plasmids. (Right) Bar charts representing the corresponding effects on translational yields of FLuc+i (dark blue bars) and FLucΔi (light green bars). The translational yield represents luciferase activities normalized by mRNA expression of each reporter measured by RPA. Data represent mean values ± SD measured in three independent experiments.
We next wondered whether the specific enhancement of spliced mRNA translation by MLN51 was linked to its presence within EJCs. To investigate this question, we overexpressed a mutant of MLN51 (H220A/D221A, MLN51HD) that previously had been shown to prevent MLN51 incorporation into EJCs both in vitro and in vivo (21, 22). MLN51HD was overexpressed to the same extent as the wild-type construct (Fig. 1E). Remarkably, like wild-type MLN51, this mutant increased the translation efficiency of the intronless reporter, but it failed to enhance specifically the translation of the spliced FLuc+i mRNAs (Fig. 1E). Thus, MLN51 overexpression strongly stimulated the translation of spliced mRNAs in an EJC-dependent manner and, to a lesser extent, the translation of intronless reporters.
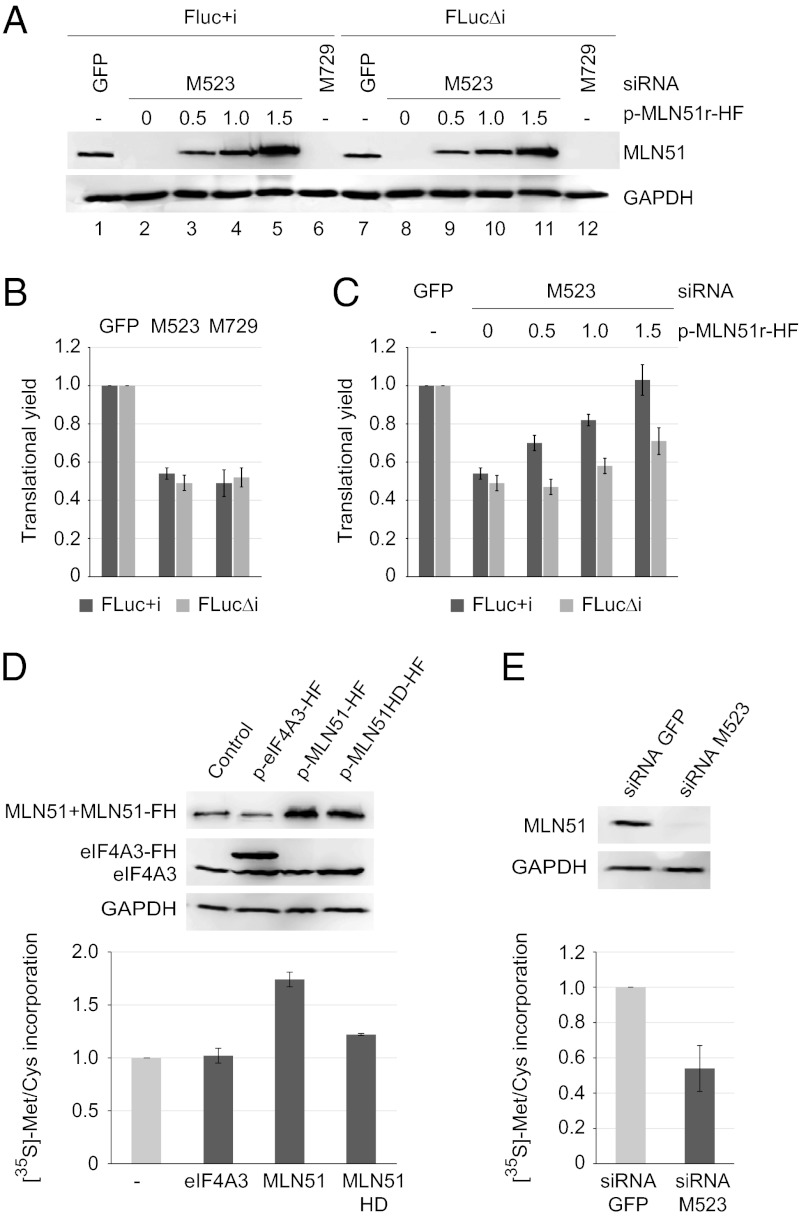
MLN51 Is a General Translation Activator.
To confirm the role of MLN51 in translation, we measured the translation efficiency of FLuc+i and FLucΔi reporter mRNAs in HEK293 cells in which MLN51 has been down-regulated with two different siRNAs (M523 and M729). In comparison with the effect of a control siRNA directed against GFP, M523 and M729 siRNAs reduced the amount of MLN51 to less than 10% (Fig. 2A, compare lanes 1 and 7 with lanes 2, 6, and 8, 12). MLN51 down-regulations were accompanied by an approximately twofold reduction in the translation efficiency of spliced and unspliced mRNAs (Fig. 2B). To substantiate the role of MLN51 in translation further, we gradually restored MLN51 expression in M523-RNAi knockdown cells by transfecting increasing amounts of an M523 siRNA-resistant plasmid (p-MLN51r-HF). Remarkably, this resistant protein fully restored the translation efficiency of FLuc+i mRNAs, confirming the role of MLN51 in translation (Fig. 2C). However, the translation efficiency of the intronless FLucΔi reporter was restored only partially, in agreement with the observation that MLN51 principally favors the translation of spliced mRNAs (Figs. 1D and 2C).
Fig. 2.
The MLN51 level affects the general translation efficiency. (A) Western blots of MLN51 in HEK293 cells treated with indicated siRNAs. Increasing amounts of p-MLN51r-HF plasmid were transfected in M523-treated cells to restore MLN51 expression (lanes 2–5 and 8–11). GADPH was used as loading control. (B) As in Fig. 1B, except that bar charts represent the translational yield of FLuc+i and FLucΔi reporters after siRNA silencing. (C) As in B in cells transfected with increasing amounts of p-MLN51r-HF. (D) (Upper) Western blots with antibodies specific to the indicated protein in HEK293 cellular extracts after overexpression of the indicated proteins. (Lower) Bar charts representing metabolic labeling after the overexpression of the indicated proteins measured by [35S]-Met/Cys incorporation after 1 h of incubation. Data represent mean values ± SD measured from three independent experiments. (E) As in D under the indicated siRNA knockdowns in HEK293 cells.
Finally, we monitored the effect of overexpression and down-regulation of MLN51 on general cellular translation by performing metabolic labeling. To this end, HEK293 cells overexpressing MLN51 or treated by siRNAs were pulsed in the presence of a radiolabeled methionine and cysteine mix before quantitation of total protein labeling. Interestingly, overexpression of MLN51 stimulated global cellular translation, but overexpression of eIF4A3 had no effect (Fig. 2D). In agreement with the results presented above, this general effect is largely EJC dependent; the overexpression of the MLN51HD mutant increased metabolic labeling only modestly, as is consistent with the effect observed with reporters (Fig. 1E). In contrast, down-regulation of MLN51 by siRNA reduced the overall cellular translation (Fig. 2E). Taken together, our results showed that MLN51 is a general enhancer of translation that operates mainly via the EJC.
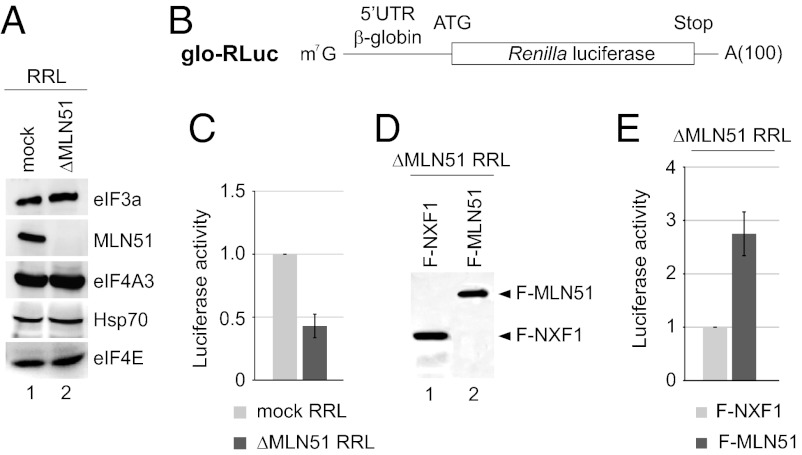
MLN51 Protein Level Modulates the Translation Efficiency in Vitro.
To investigate further the general role of MLN51 in translation, we pursued this study in a simplified in vitro translation system using nuclease-treated rabbit reticulocyte lysates (RRL), which contain a high amount of endogenous MLN51 protein (Fig. 3A, lane 1). RRL were immunodepleted with or without a specific anti-MLN51 antibody, resulting in ΔMLN51 and mock RRL (Fig. 3A). A nearly complete depletion of MLN51 (<5% remaining) was achieved, whereas other factors, including eIF4A3, heat shock protein 70, and the translation-initiating factors eIF3 and eIF4E, were not affected significantly (Fig. 3A, lanes 1 and 2). RRL then were used to translate an in vitro transcribed, capped, and poly(A)-tailed reporter mRNA encoding Renilla luciferase downstream of the globin 5′ UTR (glo-RLuc) (Fig. 3B). Consistently, a two- to threefold decrease in translation efficiency was observed upon MLN51 depletion as compared with the control (Fig. 3C). Similar results were obtained using untreated RRL that contains all endogenous mRNAs (Fig. S2 A and B).
Fig. 3.
MLN51 enhances translation in cell-free translational extracts. (A) Western blots with antibodies specific to the indicated protein in mock RRL (lane 1) and in MLN51-depleted (ΔMLN51) RRL (lane 2). (B) Schematic representation of the in vitro-transcribed glo-RLuc luciferase reporter. (C) Bar charts representing the luciferase activity of the glo-RLuc reporter in mock (gray bars) and ΔMLN51 (blue bars) RRL after 30 min of incubation. Data represent mean values ± SD measured from three independent experiments. (D) Western blots with anti-Flag. (E) As in C with ΔMLN51 RRL expressing F-NXF1 (gray bars) or F-MLN51 (blue bars).
We next restored the presence of MLN51 in ΔMLN51 RRL. To do so, in vitro-synthesized mRNAs encoding Flag-MLN51 (F-MLN51) or the unrelated RBP nuclear RNA export factor 1 (F-NXF1) as a control were incubated with ΔMLN51 RRL. Western blotting with anti-Flag antibody confirmed the similar expression of both proteins (Fig. 3D). Anti-MLN51 antibody revealed that the level of de novo-synthesized F-MLN51 protein was two to five times higher than the endogenous protein level in the control RRL (Fig. S2C). The expression of F-MLN51 in ΔMLN51 RRL increased the translation of glo-RLuc mRNA up to 2.5-fold compared with the RRL expressing F-NXF1 (Fig. 3E). Moreover, the presence of other EJC components or of preformed EJCs is not necessary for the translation-regulating function of MLN51 because Flag-MLN51 similarly enhanced translation in RRL that were codepleted for MLN51 and eIF4A3 (Fig. S2 D and E). Therefore, in a simplified cell-free system, MLN51 is able to enhance translation directly in an EJC-independent manner.
MLN51 Contacts the Translation-Initiating Machinery.
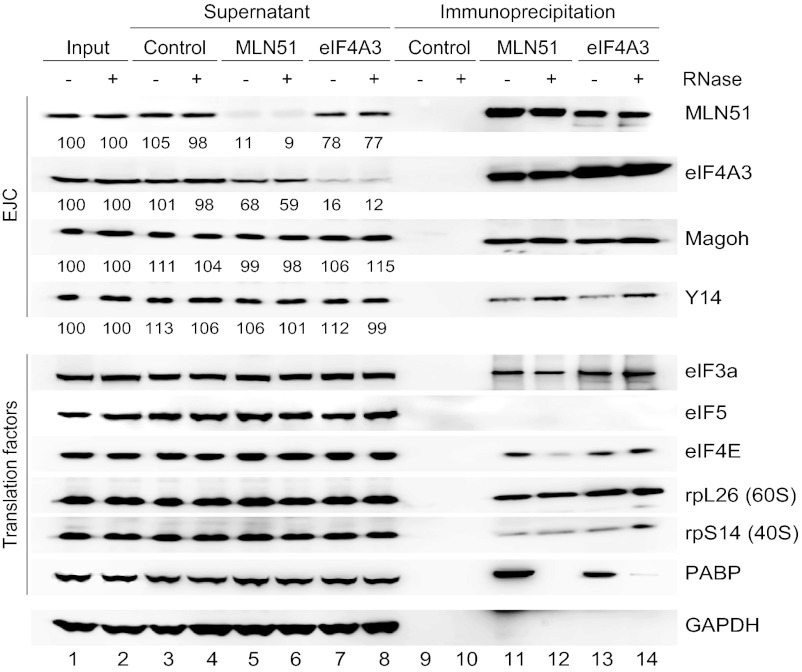
We next explored whether MLN51 and EJC core proteins are bound to translating mRNAs. The polysome profile of HeLa cell lysate analyzed by sucrose gradient centrifugation showed a typical fractionation of monosomes and polysomes (Fig. S3). Magoh and Y14 were found predominantly in the lighter mRNPs fractions, and a small proportion also was found within the monosome fractions. This profile is in agreement with the dissociation of EJC cores by translating ribosomes (5). In contrast, eIF4A3 and MLN51 cosedimented with the monosome fractions (40S, 60S, and 80S) but also were enriched in polysome fractions. This finding suggests that the proteins could be associated with mRNAs undergoing translation but also could be incorporated into other heavy complexes.
These data prompted us to analyze whether eIF4A3 and/or MLN51 interact with translation factors, notably those involved in initiation, in a more direct way. To do so, HEK293 proteins immunoprecipitated with anti-MLN51 and anti-eIF4A3 were blotted in parallel with a series of antibodies specific to EJC and eIFs factors (Fig. 4). Analysis of the supernatants showed that almost 90% of the endogenous proteins were precipitated. As expected, both anti-MLN51 and anti-eIF4A3 precipitated the other EJC core components independently of RNase treatment. The absence of association with the poly(A)-binding protein (PABP) demonstrated the efficiency of RNase treatment. In addition, MLN51 depletion was accompanied by a significant codepletion of eIF4A3, and vice versa; this observation was not as clear for Magoh and Y14. Thus, MLN51 and eIF4A3 are associated independently of the EJC, and only a small proportion of the four proteins is engaged in EJC cores at steady state. Of interest, both MLN51 and eIF4A3 are associated with several translation factors, including eIF3 (a subunit), eIF4A1, eIF4E, and components of the small (rpS14) and the large (rpL26) ribosomal subunits (Fig. 4 and Fig. S4A). In contrast, we were not able to detect the eIF2α subunit or eIF5 in the precipitates (Fig. 4 and Fig. S4A). A similar observation was made when immunoprecipitations were performed with HeLa cell extracts (Fig. S4B). Thus, MLN51 and eIF4A3, together and/or separately, exist in complexes containing the ribosomal subunits and several translation-initiating factors.
Fig. 4.
MLN51 associates with the translation machinery. Protein extracts from HEK293 cells treated with or without RNase were subjected to immunoprecipitation without antibody (Control) or with anti-MLN51 and anti-eIF4A3 antibodies. Inputs (lanes 1 and 2), supernatants (lanes 3–8) and immunoprecipitated fractions (lanes 9–14) were resolved by SDS/PAGE and immunoblotted with the indicated antibodies. Quantifications of EJC proteins are indicated.
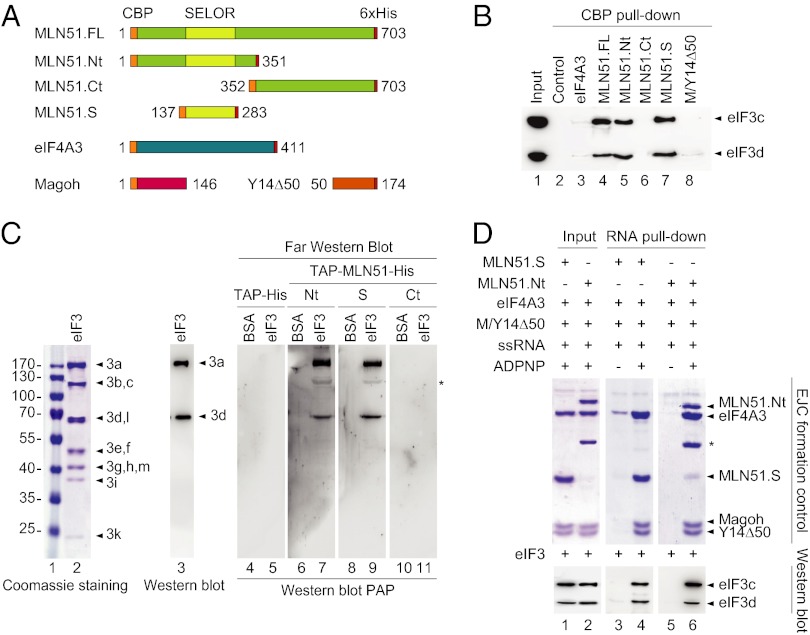
MLN51 Interacts Directly with eIF3 via Its SELOR Module.
To gain more insight into the links among MLN51, eIF4A3, and the translation-initiating machinery, we performed in vitro binding assays. Recombinant proteins corresponding to full-length MLN51 (MLN51.FL), eIF4A3, the heterodimer Magoh/Y14Δ50 (M/Y14Δ50) fused to an N-terminal calmodulin-binding protein (CBP) tag and eIF4E fused to a GST tag were purified from Escherichia coli (Fig. 5A and Fig. S5A). The 40S and 60S ribosomal subunits and eIF3 were purified from RRL or HeLa cells. We then performed binding assays using each purified CBP-EJC protein separately as bait and initiation factors as potential partners. Protein complexes were purified with calmodulin beads after extensive washing, and the eluted proteins were fractionated with SDS/PAGE and visualized either by direct Coomassie or silver staining or by Western blotting. No interaction was observed between any of the CBP-tagged EJC proteins and eIF4E or the 40S and the 60S ribosomal subunits under the test conditions (Fig. S5 B–D). In contrast, a clear interaction was detected between CBP-MLN51 and the purified eIF3 complex (Fig. 5B). This complex is formed by 13 subunits (eIF3a–eIF3m) and is organized around a core complex made of five subunits (eIF3a, b, c, g, and i), which can be probed with anti-eIF3c and anti-eIF3d antibodies (23). Remarkably, of the EJC proteins, only CBP-MLN51.FL coprecipitated eIF3 efficiently (Fig. 5B, lane 4 and Fig. S5E). As a positive control, we reproduced the previously described interaction between eIF3 and Upf1 (24) using CBP-Upf1 as bait (Fig. S5E, lane 3). To delineate the domain of MLN51 interacting with eIF3 more precisely, different deleted versions of MLN51 were purified (Fig. 5A and Fig. S5A): the N-terminal half (MLN51.Nt, amino acids 1–351), the SELOR domain (MLN51.S, amino acids 137–283) required for MLN51 incorporation into the EJC core (7, 22), and the unstructured C-terminal half (MLN51.Ct, amino acids 352–703) implicated in the assembly of stress granules (25). Remarkably, all the truncated proteins carrying the SELOR domain were able to precipitate eIF3 (Fig. 5B, lanes 4, 5, and 7). Therefore, MLN51 contacts eIF3 directly via its conserved SELOR domain.
Fig. 5.
MLN51 interacts directly with eIF3. (A) Schematic representation of recombinant EJC core proteins. (B) CBP pull-down assays. Immunoblotting with anti-eIF3c and anti-eIF3d antibodies monitoring eIF3 coprecipitation in the absence of CBP-tagged protein (lane 2) or in the presence of the indicated CBP-tagged EJC proteins. The input lane (lane 1) represents 30% of the total. (C) Far Western analyses. BSA and the eIF3 complex were resolved in 13.5% SDS/PAGE and were transferred to nitrocellulose. Membranes were incubated with the indicated TAP-tagged recombinant proteins before detection using PAP immunogene (lanes 4–11). In parallel, eIF3 subunits were revealed by Coomassie staining (lane 2) and by Western blotting (eIF3a and eIF3d, lane 3). The asterisk marks a nonspecific interaction revealed by PAP. (D) Protein coprecipitations with 3′-end–biotinylated ssRNA. The indicated EJC proteins were mixed with ssRNA with or without ADPNP before incubation with eIF3 and precipitation onto streptavidin beads (lanes 3–6). The input lanes (lanes 1 and 2) represent 10% of the total. EJC formation was controlled by Coomassie staining (Upper), and eIF3 coprecipitation by immunoblotting using anti-eIF3c and anti-eIF3d antibodies (Lower). The asterisk in the upper panel marks a degradation of MLN51.Nt recombinant protein.
MLN51 Interacts with eIF3a and eIF3d Subunits.
Next, we performed Far Western blotting experiments with the purified eIF3 complex to identify which subunit(s) interact with MLN51. Coomassie staining of the purified eIF3 complex revealed the presence of 11 or 12 detectable subunits after biochemical purification [Fig. 5C, lanes 1 and 2; our preparation probably lacks the labile eIF3j subunit (26)]. The eIF3 complex was transferred onto a nitrocellulose membrane before blotting with different tandem affinity-purified (TAP)-tagged versions of MLN51: TAP-MLN51.Nt, TAP-MLN51.Ct, or TAP-MLN51.S. TAP-tagged proteins bound to the immobilized eIF3 subunits were detected by Western blot using peroxidase anti-peroxidase (PAP). MLN51.Nt and MLN51.S proteins revealed eIF3 subunits migrating at 170 kDa and 65–70 kDa (Fig. 5C, lanes 7 and 9), but no signals were detected in control lanes containing BSA or TAP-His protein or with the TAP-MLN51.Ct protein (Fig. 5C, lanes 4–11). Although the largest protein is most likely eIF3a (170 kDa), the signal migrating around 70 kDa could correspond to either eIF3d or eIF3l. The use of a more resolving SDS/PAGE identified the smallest protein as eIF3d (Fig. S5F). Thus, MLN51, via its SELOR domain, interacts physically with two subunits of eIF3, eIF3a and eIF3d.
MLN51 Interaction with eIF3 Is Compatible with EJC Formation.
Given that the SELOR domain is necessary to assemble the EJC core stably onto mRNA (7, 22), we wondered whether its interactions with EJC core and eIF3 were mutually exclusive. We tested whether eIF3 still binds MLN51 within the EJC core reconstituted in vitro (22). The EJC core was formed with recombinant M/Y14Δ50, eIF4A3, and MLN51.S or MLN51.Nt and 3′-end–biotinylated ssRNA (Fig. 5D, lanes 1 and 2). Biotinylated ssRNAs were pulled down using streptavidin beads, and bound proteins were resolved on SDS/PAGE gels to verify EJC core formation (Fig. 5D, lanes 3–6). As expected, the EJC core was reconstituted only in the presence of Adenosine 5′-(β,γ-imido)triphosphate (ADPNP) but not in its absence (Fig. 5D, lanes 3–6), because ATP is required for EJC core assembly (22). The reconstituted EJC cores immobilized on beads were incubated with a stoichiometric amount of purified eIF3. After extensive washes, the elution contents were analyzed by Western blotting with anti-eIF3c and anti-eIF3d antibodies. We observed that eIF3 was stably bound to reconstituted EJC core but not to ssRNA alone (Fig. 5D, lanes 3–6). Moreover, eIF3 was equally retained by the EJC core containing MLN51.S or MLN51.Nt. Therefore, MLN51, alone or embedded within the EJC core, is capable of making direct and stable contact with the eIF3 complex, and this interaction takes place between MLN51 SELOR domain and the eIF3 subunits a and d.
Discussion
The EJC marks splicing events and communicates with downstream processes. However, how EJC proteins contribute to translation of newly synthesized mRNAs remained unclear. In this study we deciphered the role of individual core proteins in translation and discovered that MLN51 activates translation and contacts eIF3. Below, we discuss how our observations could lead to new models of translation activation by the EJC.
To study the role of EJC core components individually, we overexpressed each of them and monitored the translation efficiency of different reporters (Fig. 1). Overexpression of MLN51 increased the translation efficiency of all tested reporter mRNAs by at least twofold as well as overall cellular translation, whereas overexpression of other EJC core proteins had no effect. Knockdown of MLN51 and rescue of its expression supported its involvement in translation (Fig. 2). This result was confirmed with a cell-free system in which translation is independent of mRNA cellular history (Fig. 3). Here, MLN51 depletion reduced mRNA translation, whereas complementation of the depleted extracts by de novo-synthesized MLN51 restored translation efficiency, showing that MLN51 is a bona fide regulator of translation. The four EJC core proteins shuttle between the nucleus and the cytoplasm, but MLN51 is the only one whose localization is mainly cytoplasmic (21, 25). One could imagine that the role played by MLN51 in translation is independent of its role as part of the EJC core. Interestingly, although MLN51 is able to stimulate translation of mRNAs that have not experienced splicing, its effect is much more pronounced on the corresponding spliced mRNA (Fig. 1). This splicing-dependent function is relayed by MLN51 inside the EJC, because the overexpression of a specific mutant that prevents MLN51 incorporation into EJCs (21, 22) abolished the stimulation of translation (Figs. 1 and 2). Moreover, this mutant conserves its ability to stimulate (albeit more modestly) the translation of unspliced mRNAs, showing that MLN51 can act as a translation activator both within and without the EJC. Therefore, the EJC offers MLN51 the opportunity to be attached stably to mRNAs and subsequently to enhance their translation. This observation brings to light an additional example of how the EJC communicates advantageously with downstream machineries.
We also performed experiments to obtain insight into the molecular mechanism underlying the MLN51-related activation of translation. Analyses of polysome gradients suggested that MLN51 and eIF4A3, in contrast to Magoh/Y14, are associated with translating ribosomes (Fig. S3A). Several lines of evidence suggest that a significant proportion of MLN51 and eIF4A3 are associated in the cytoplasm and may bind mRNA independently of the EJC. Structural studies showed that the open conformation adopted by eIF4A3 in the absence of ATP and RNA is not detrimental to its interaction with MLN51 (6). In addition, immunodepletion of endogenous MLN51 from HEK293 cell lysate is accompanied by a significant codepletion of eIF4A3 (Fig. 4). Therefore, we performed immunoprecipitations of MLN51 and eIF4A3 and screened for potential association with translation factors. A subset of initiation factors, as well as the two ribosomal subunits, is associated with each protein in an RNase-insensitive manner (Fig. 4), but only MLN51 contacts the initiation factor eIF3 via its SELOR module (Fig. 5). This interaction combined with the reconstitution of the EJC core showed that MLN51, as part of the splicing mark, also could bind eIF3 (Fig. 5). Therefore, eIF3 is a potential additional member of the long list of EJC peripheral factors (6). We can speculate that eIF3 joins the EJC rapidly after mRNAs enter the cytoplasm. Our in vitro approach suggests that eIF3 binding to the EJC relies simply on MLN51; however, we cannot exclude the possibility that in vivo it is stabilized by other peripheral factors, such as SKAR. When isolated, the different parts of MLN51, including the SELOR module, can bind RNA with a low affinity (7), explaining how MLN51 could link eIF3 with mRNAs outside the EJC. However, this binding cannot be compared with the stable attachment of the tetrameric core onto mRNAs (22). Therefore, we propose that MLN51 overexpression favors the translation of spliced mRNAs rather than unspliced mRNAs because of its tighter association with RNA in the context of the EJC.
Appropriate initiation of translation is ensured by an intricate network of interactions between initiation factors, ribosome subunits, and mRNAs (27). It begins with the formation of the preinitiation 43S complex (PIC) composed of the 40S, the initiator transfer RNA (tRNA) associated with the eIF2 in a ternary complex with GTP, and several initiation factors, including eIF3. Then, mRNAs associated with eIF4F recruit the PIC at the 5′ extremity of the mRNA. This complex then scans the mRNA to reach the start codon where the 60S subunit joins the 40S subunit to constitute a competent 80S ribosome (27, 28). During this initiation phase, eIF3 plays multiple roles including (i) stabilization of the 40S interaction with the eIF2 ternary complex, (ii) recruitment of 43S through interaction with eIF4F, and (iii) assistance in the scanning step and in the fidelity of AUG start codon recognition (23). So, through its interaction with eIF3, MLN51 may stabilize the initiating mRNP and thus may promote the initiation of translation by constituting an additional link between the mRNA and translation factors. It is possible that after EJC disassembly during the first round of translation, MLN51 remains associated with mRNA and hence pursues its enhancer role during subsequent rounds of translation. Moreover, eIF3 also is important for the termination of translation, during which it helps release tRNA and mRNA from the 40S subunit and prevents its reassociation with the 60S subunit before the recognition of a new start codon (29). Therefore, we can imagine that the presence of EJCs near the stop codon may influence the termination of translation and ribosome recycling positively by recruiting eIF3. Finally, almost 50% of human mRNAs contain a short ORF upstream (uORF) of the main protein coding ORF (30) that finely modulates the expression of essential proteins, such as growth and transcription factors (31). Reinitiation downstream of a uORF depends mainly on the presence of remaining eIFs that maintain 40S subunits competent for further scanning and initiation. In several organisms, eIF3 crucially influences the efficiency of reinitiation (32–34). In this context, the presence of eIF3 bound to EJCs downstream of the uORF could help the 40S subunit resume scanning and reinitiate translation.
Nuclear capping and polyadenylation are essential for protecting mRNAs from degradation and also for cytoplasmic translation. Nuclear splicing also may contribute to the assembly of the translation apparatus via the recruitment of eIF3 onto EJCs. We and others recently have discovered that EJCs are not equally distributed onto spliced mRNAs (35, 36). Moreover, no one knows whether the composition of EJCs is identical at every junction onto every mRNA. MLN51 most likely is exchangeable without disrupting the complex (37). Therefore, additional experiments will be necessary to determine whether the variation in EJCs and notably the variability in MLN51 and eIF3 provide a way to fine-tune protein synthesis. Finally, MLN51 originally was isolated because of its overexpression in several types of breast cancers (7, 38, 39). It is tempting to speculate that the differences in MLN51 expression levels may enhance the expression of multiple mRNAs and hence may contribute to cancer progression. In this case, MLN51 could join the extended list of RBPs involved in tumorigenesis (40).
Materials and Methods
Cloning involved standard techniques. All antibodies are described in Supporting Information. All methods are detailed in SI Material and Methods.
Supplementary Material
Acknowledgments
We thank E. Izaurralde (Max Planck Institute, Tübingen, Germany) for providing anti-MAGOH antibodies; the Institut de Génétique et de Biologie Moléculaire et Cellulaire facilities for peptide synthesis and antibody production; C. Fraser for technical advice on eIF3 purification from HeLa cells; and the members of the H.L.H. laboratory for technical assistance and helpful advice and discussions. This work was supported by Centre National de la Recherche Scientifique Grants ATIP 2008 (to H.L.H) and ATIP 2007 (to B.S.); by the Institut National de la Santé et de la Recherche Médicale; by the University of Strasbourg (C.T.); by Agence Nationale de la Recherche Grants 2008-BLAN-0323 (to H.L.H. and C.T.) and 2011-BLAN-01801 (to H.L.H.); by the Ligue Nationale contre le Cancer (H.L.H., P-E.C., and C.T.); and by the Fondation pour la Recherche Médicale (H.L.H.). E.D. was a recipient of fellowships from the French Ministère de l'Enseignement Supérieur et de la Recherche and from the Fondation pour la Recherche Médicale.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218732110/-/DCSupplemental.
References
- 1.Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14(9):1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 4.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16(3):279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137(3):536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 6.Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18(1):112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Degot S, et al. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279(32):33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- 8.Ashton-Beaucage D, et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143(2):251–262. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Roignant JY, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143(2):238–250. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michelle L, et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol. 2012;32(5):954–967. doi: 10.1128/MCB.06130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kervestin S, Jacobson A. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Hir H, Séraphin B. EJCs at the heart of translational control. Cell. 2008;133(2):213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1(10):1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 15.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9(5):607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA. 1991;88(2):478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes Dev. 2004;18(2):210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA. 2003;100(20):11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11(12):1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133(2):303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 21.Daguenet E, et al. Perispeckles are major assembly sites for the exon junction core complex. Mol Biol Cell. 2012;23(9):1765–1782. doi: 10.1091/mbc.E12-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12(10):861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 23.Hinnebusch AG. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31(10):553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133(2):314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baguet A, et al. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120(Pt 16):2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- 26.Fraser CS, et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem. 2004;279(10):8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- 27.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19(6):568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 28.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jackson RJ, Hellen CU, Pestova TV (2012) Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86:45–93. [DOI] [PubMed]
- 30.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5’untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 31.Medenbach J, Seiler M, Hentze MW. Translational control via protein-regulated upstream open reading frames. Cell. 2011;145(6):902–913. doi: 10.1016/j.cell.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Szamecz B, et al. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22(17):2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy B, et al. The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA. 2010;16(4):748–761. doi: 10.1261/rna.2056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007;21(23):3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saulière J, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19(11):1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 36.Singh G, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151(4):750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7(5):e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degot S, et al. Metastatic Lymph Node 51, a novel nucleo-cytoplasmic protein overexpressed in breast cancer. Oncogene. 2002;21(28):4422–4434. doi: 10.1038/sj.onc.1205611. [DOI] [PubMed] [Google Scholar]
- 39.Tomasetto C, et al. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics. 1995;28(3):367–376. doi: 10.1006/geno.1995.1163. [DOI] [PubMed] [Google Scholar]
- 40.Wurth L. Versatility of RNA-Binding Proteins in Cancer. Comp Funct Genomics. 2012;2012:178525. doi: 10.1155/2012/178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.