Abstract
Cryptochromes are flavoproteins, structurally and evolutionarily related to photolyases, that are involved in the development, magnetoreception, and temporal organization of a variety of organisms. Drosophila CRYPTOCHROME (dCRY) is involved in light synchronization of the master circadian clock, and its C terminus plays an important role in modulating light sensitivity and activity of the protein. The activation of dCRY by light requires a conformational change, but it has been suggested that activation could be mediated also by specific “regulators” that bind the C terminus of the protein. This C-terminal region harbors several protein–protein interaction motifs, likely relevant for signal transduction regulation. Here, we show that some functional linear motifs are evolutionarily conserved in the C terminus of cryptochromes and that class III PDZ-binding sites are selectively maintained in animals. A coimmunoprecipitation assay followed by mass spectrometry analysis revealed that dCRY interacts with Retinal Degeneration A (RDGA) and with Neither Inactivation Nor Afterpotential C (NINAC) proteins. Both proteins belong to a multiprotein complex (the Signalplex) that includes visual-signaling molecules. Using bioinformatic and molecular approaches, dCRY was found to interact with Neither Inactivation Nor Afterpotential C through Inactivation No Afterpotential D (INAD) in a light-dependent manner and that the CRY–Inactivation No Afterpotential D interaction is mediated by specific domains of the two proteins and involves the CRY C terminus. Moreover, an impairment of the visual behavior was observed in fly mutants for dCRY, indicative of a role, direct or indirect, for this photoreceptor in fly vision.
Circadian clocks synchronize physiology and behavior of living organisms with 24-h environmental cycles. In Drosophila, the resetting of the clock depends mostly on light-mediated degradation of the clock protein TIMELESS (dTIM), which, in turn, affects the stability of its partner PERIOD (dPER). Light signals are received through the blue-light photoreceptor CRYPTOCHROME (dCRY), the expression of which is under clock control. dCRY associates with dTIM in a light-dependent manner and promotes its proteasome-mediated degradation (1). Cryptochromes are flavoproteins highly similar to photolyases, from which they have probably evolved, but across evolution they have lost or reduced the photolyase activity and gained roles in signaling (2). Cryptochromes consist of two protein domains: an N-terminal domain homologous to photolyases (Photolyase Related, or PHR), and a very divergent C-terminal tail (3). A class of cryptochromes, CRY-DASH (drosophila, arabidopsis, synechocystis, homo), with single-stranded DNA repair activity and without the C terminus tail, has been described in bacteria, plants, and animals (2). The role of cryptochromes in the circadian clock differs among the different species. Cryptochromes have merely a blue-light photoreceptor activity in plants whereas in mammals they are part of the central clock mechanism, and this function is not light dependent (4). In Drosophila, the unique CRY acts as a circadian photoreceptor in the master clock (5) whereas, in other insects, only the vertebrate-like CRYs play a role as transcriptional repressor (6). Moreover, dCRY has been shown to play a fundamental role in the fly’s magnetosensitivity, i.e., the use of the Earth’s magnetic field for orientation and navigation (7). dCRY is rhythmically expressed. Protein levels oscillate only under light–dark cycling conditions, with a peak in the late night; in constant darkness, they increase, reaching a plateau (8). dCRY resets the clock by interacting with dTIM in the presence of light: subsequent to this interaction, dTIM is phosphorylated and targeted for degradation through a ubiquitin-proteasome mechanism that involves JETLAG, an E3-ubiquitin ligase complex component (9). Upon light activation, dCRY also interacts with JETLAG and is degraded via proteasome (9). dCRY interacts also with the kinase shaggy/GSK3 (SGG), and the cryptochrome’s stability in light is considerably increased by this interaction whereas the inactivation of the kinase leads to the degradation of dCRY in darkness (10). The molecular mechanism by which dCRY is activated by light is still not fully understood, but a regulatory role for the C terminus of the protein has been demonstrated by several studies (3, 5, 11–13). The activation of dCRY by light requires a conformational change (13), but the release of a putative repressor cannot be excluded (11). In fact, it has been hypothesized that the activation of dCRY by light is mediated also by specific “regulators” that bind its C terminus, known to regulate the light dependence of dCRY activity (13). This hypothesis was supported by the observation that the C terminus of dCRY is a hotspot for molecular interactions: by in silico analysis and experimental validation, we could identify several protein–protein interaction motifs in this small region and, among them, two class III PDZ-binding motifs (3). PDZ (postsynaptic density protein 95, Drosophila disk large tumor suppressor, and zonula occludens-1 protein) domains are modular domains that play a crucial role in the assembly of large protein complexes involved in signaling processes. These domains have a conserved fold consisting of five or six β-strands and two to three α-helices forming a β-stranded sandwich. PDZ domains typically recognize the extreme C terminus of target proteins (14). Distinct PDZ domains bind to optimal sequences, and the structural analysis of known binding sites of PDZ domains and their ligands has provided insight into the specificity of PDZ protein–protein interactions (15). The preference of each residue of a binding peptide is related to the physical-chemical characteristics of different relevant residues on specific secondary structural elements forming the PDZ-binding pocket (16). Three major classes of PDZ-binding motifs have been established (17).
Here, we show that some functional linear motifs are evolutionarily conserved in the C terminus of cryptochromes, with class III PDZ-binding sites selectively maintained in animals. We detected the presence of dCRY in a multiprotein complex (the Signalplex) involved in the visual-signaling pathway (18), and we found that the interaction with this comlex is mediated by Inactivation No Afterpotential D (INAD), a scaffold protein with five structural PDZ domains. Moreover, we detected a role for dCRY in fly vision.
Results
Functional Motifs Are Conserved in CRY Across Species.
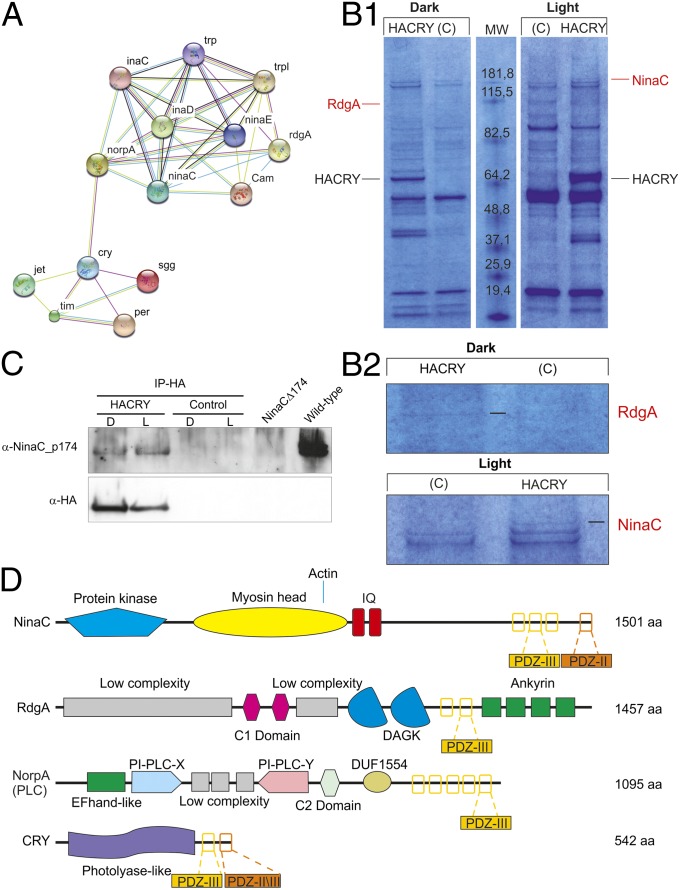
We searched for the evolutionary conservation of linear motifs in the C terminus of CRY throughout a broad range of organisms. Linear motifs are short sequences that mediate molecular interactions and very often reside in disordered or nonglobular regions of proteins. Unraveling the evolution of linear motifs is problematic, as these sites tend to be unstable over long evolutionary distances or to jump between different sequence positions inside nonglobular regions. dCRY is an excellent test case for this assumption, as it bears a highly variable C-terminal region that has undergone rapid evolution while maintaining overall similar roles in circadian rhythmicity. An unrooted neighbor-joining phylogenetic tree was constructed using amino acid sequences from various members of the CRY family from plants to humans (Fig. S1). Animal cryptochromes were clustered in four different groups: vertebrate, vertebrate-like (including invertebrate species), CRY4, and Drosophila-like. CRY sequences show many linear motifs that are not evenly distributed in all species investigated (Fig. S1). Among them, PDZ domains recognize short sequences at the C terminus of proteins and have an important role in mediating interactions for the assembly of large multiprotein complexes involved in signaling processes at specific subcellular locations. Interestingly, among the three major classes of PDZ-binding motifs, class III is evolutionarily conserved in the CRY C-terminal sequence across animal species (Fig. S1). We speculated that a protein partner of dCRY could be a PDZ domain-containing protein and searched the STRING database (19) for possible candidates. In this database, connections between proteins are based on several methods, including computational predictions. Fig. 1A shows the distribution of interactors for dCRY. The results showed a weak connection to No Receptor Potential A (NORPA), a protein belonging to the phototransduction complex (20).
Fig. 1.
Interaction of dCRY with the phototransduction complex. (A) Protein interaction network surrounding dCRY and INAD. The STRING interaction network is shown for dCRY, INAD, and their main interaction partners with edge colors representing different detection methods. Note that the edge between dCRY and NORPA is based on phenotypic enhancement assays and thus may not necessarily represent a true physical interaction. (B1) Coomassie blue-stained gel of heads of protein extracts coimmunoprecipitated with an anti-HA antibody. HACRY-overexpressing flies (HACRY, yw;tim-GAL4/+; UAS-HAcry/+) and relative controls (C, yw;tim-GAL4) were reared in 12:12 light:dark and collected in the dark (ZT24) and in the light (ZT24 + 15-min light pulse). Molecular masses of markers are indicated (BenchMark Pre-Stained Protein Ladder; Invitrogen). MW, molecular weight. Bands corresponding to HACRY are indicated in black, while stained proteins excised and characterized by mass spectrometry are indicated in red. (B2) Zoom of regions of the gel-bearing dRDGA and dNINAC bands. (C) Coimmunoprecipitation and Western blot confirming the interaction between HACRY and NINAC in HACRY-overexpressing flies (yw;tim-GAL4/+; UAS-HAcry/+). tim-GAL4 flies were used as control. Heads were collected as in B1. Membranes were probed with anti-NINACp174 and anti-HA antibodies. NinaCΔ174 and w1118 flies, collected at ZT1, were used as negative and positive control, respectively. (D) Schematic domain distribution for known and putative INAD interacting proteins. Each protein is drawn proportional to its size, with solid shapes representing different protein domains and their name from the Pfam database. Note that low-complexity regions, shown as light-gray rectangles, are not a proper domain. PDZ-binding motifs are shown as white rectangles with yellow (class III), orange (type II), or peach (overlapping classes II/III) borders.
dCRY Interacts with the Phototransduction Complex.
In an attempt to identify new partners of dCRY, a coimmunoprecipitation assay, followed by mass spectrometry analysis, was performed on transgenic flies overexpressing a hemagglutinin (HA)-tagged form of dCRY (HACRY; 13) raised in 12:12 light:dark cycles and collected at Zeitgeber Time 24 (ZT24), before lights on, and after a 15-min light pulse. An ∼115-kDa species was observed in the sample in the dark and an ∼180-kDa species after the light pulse, which were not present in the respective negative controls (Fig. 1B). These protein bands were digested in-gel, and the peptide mixtures were analyzed by liquid chromatography–mass spectrometry (LC-MS)/MS using an ESI-QTOF mass spectrometer (21). Analysis of the MS/MS data using the MASCOT software yielded the identification of two proteins involved in the fly visual-signaling pathway: Retinal DeGeneration A (RDGA) in the dark and Neither Inactivation Nor Afterpotential C (NINAC) after 15 min of light pulse (Fig. S2A) (18, 20). Although RDGA was identified on the basis of the MS/MS spectra of six different tryptic peptides, in the case of NINAC, the identification was based on the MS/MS spectrum of only one peptide displaying a significant score in MASCOT (Fig. S2B). The presence of NINACp174 in the complex with HA-tagged form of dCRY (HACRY) was also confirmed by Western blot with an antibody specifically raised against the p174 isoform of the protein that is localized in the rhabdomeres of photoreceptor cells in the fly’s eye (22). By this procedure, NINACp174 was also detected in the dark, albeit at lower levels than under light conditions (Fig. 1C). The difference between NINACp174/HACRY ratios under light and dark conditions was significant (P < 0.03, Mann–Whitney U test) (Fig. S2C).
dCRY Interacts with the Phototransduction Complex Through INAD.
Many of the elements of this visual cascade are assembled in a multiprotein-signaling complex (Signalplex) organized by INAD, a scaffold protein with five structural PDZ domains, each of which binds to a specific partner (20). A schematic representation of the functional domains of NINAC, RDGA, NORPA, and dCRY is given in Fig. 1D. To test whether dCRY interacts with the phototransduction complex through INAD, we searched for INAD in the immunocomplex formed by dCRY. Indeed, a Western blot with an anti-INAD antibody (23), performed on head protein extracts from HACRY-overexpressing flies immunoprecipitated with an anti-HA antibody, revealed that INAD interacts in vivo with dCRY (Fig. 2A). The interaction is quite strong in the light, but traces of INAD are visible also in the dark. The difference between INAD/HACRY ratios under light and dark conditions was significant (P < 0.02, Mann–Whitney U test) (Fig. S2D).
Fig. 2.
dCRY interacts with INAD. (A) Coimmunoprecipitation and Western blot confirming the interaction between HACRY and INAD in flies overexpressing HACRY (yw;tim-GAL4/+; UAS-HAcry/+). tim-GAL4 flies were used as control (“C”). Heads were collected as in Fig. 1B. Membranes were probed with anti-INAD and anti-HA antibodies. inaD1 and w1118 flies, collected at ZT1, were used as negative and positive controls of the antibody, respectively. (B) Identification of the interaction domains of dCRY and INAD using the yeast two-hybrid system. The five INAD PDZ domains are shown where modeled and assigned to putative PDZ subtypes depending on the residue types at the peptide-binding site. Relevant sequence motifs are shown as empty rectangles in the INAD and CRY sequence diagrams. Different domains of INAD were tested for interaction with the full-length dCRY in the presence of light, and different domains of dCRY were tested for interaction with the full-length INAD under both light and dark conditions (open and filled bars, respectively). Interacting fusions are shown in black, and relative β-galactosidase activity (Miller units) is reported for each fusion. Mean ± SEM of at least seven independent clones for each fusion, analyzed in triplicates, is shown. An extended version of the PDZ2–3 tandem, INAD (207–448), exhibits a significantly stronger affinity for dCRY compared with the whole protein (F14,87 = 67.81, P < 0.0001). The interaction between dCRY and INAD occurred in a light-dependent fashion with the C terminus of dCRY being crucial. On the other hand, these last 22 amino acids of the protein showed a light-independent affinity for INAD with a significantly stronger interaction in the light compared with the dark (t13 = 2.6, P = 0.02). (C) Yeast two- and three-hybrid assays highlighting that the interaction between dCRY and NINAC is mediated by INAD. The schematic shows the different proteins used as bait or prey fusion: C, dCRY; N, NINAC; I, INAD. Relative β-galactosidase activity (Miller units) is reported for each fusion. Mean ± SEM of at least six independent clones for each fusion, analyzed in triplicates, is shown. The expression of dCRY and NINAC alone does not result in the activation of the reporter gene. The expression of INAD in the yeast nucleus, to generate a three-hybrid system, shows that INAD acts as a structural bridge (BRIDGE) between the two proteins (F3,24 = 57.20, P < 0.0001). The interactions of dCRY–INAD and INAD–NINAC are also shown.
The physical interaction between dCRY and INAD was further analyzed using a yeast two-hybrid system (24), in which a full-length dCRY, directly fused to LexA (bait), was initially challenged with full-length INAD as prey (Fig. 2B and Table S1). A strictly light-dependent interaction between the two proteins was observed (Fig. 2B, dCRY), which is completely abolished when part of the dCRY C terminus (aa 521–540) is removed. As this region contains the binding motifs for PDZ domains, the 22 C-terminal amino acids of dCRY were tested for the ability to interact with INAD. A light-independent interaction between INAD and the extreme C-terminal tail of dCRY was observed (Fig. 2B, dCRY). To examine which domains of INAD are responsible for the interaction with dCRY, prey fusions expressing individual PDZs or different combinations of them were generated and tested for the interaction with full-length dCRY as bait (Fig. 2B, INAD). Single PDZ domains did not interact with dCRY, although all of the fusion proteins were correctly expressed in yeast cells. In fact, before the β-galactosidase assay, the expression of all fusion proteins was analyzed by Western blot on yeast lysate with an anti-HA antibody (Fig. S3 and Table S2). For PDZ1, PDZ3, and PDZ4, in addition to the expected signal, a band of molecular weight compatible with a dimeric organization was observed (Fig. S3). However, dimerization of PDZ domains seems not to influence binding to their partners, as the sites involved in the two events are different (25). Because some PDZ domains need other PDZ domains connected in tandem to fold properly and interact with their partners (25), the interaction between dCRY and INAD may also require tandem PDZ domains. However, prey fusions expressing tandems of PDZ linked by their native spacer sequences were still not able to interact with dCRY (Fig. 2B, INAD). An in silico analysis performed with CSpritz (26) revealed the presence of an α-helical motif upstream from the PDZ2 domain, specifically the motif MAKI (aa 235–238), which could form a unique extension of the PDZ domain and is also part of the known calmodulin-binding motif. An “extended” version of the PDZ2-PDZ3 tandem prey fusion was generated to include the predicted sites, ranging from residues 207 to 448, and this sequence showed high affinity for dCRY (Fig. 2B, INAD). These data suggest that the interaction between INAD and dCRY is mediated by the PDZ2-PDZ3 tandem, but that the PDZ2 domain needs to be extended upstream, with respect to the canonical PDZ domain boundary. Longer fusion sequences were prepared by adding a third PDZ domain; three different portions of INAD, including PDZ1–3 (aa 17–448), PDZ2–4 (aa 249–577), and PDZ3–5 (aa 364–664), respectively, were tested. Only the fusion expressing the N-terminal PDZ1–PDZ3 domains showed affinity for dCRY (Fig. 2B, INAD), suggesting that PDZ4 and PDZ5 are not involved in the interaction between dCRY and INAD. The higher binding affinity for the extended PDZ2-PDZ3 tandem compared with larger INAD fragments may be explained by the PDZ2 domain having a noncanonical structure, conferring a higher binding affinity for the dCRY motif. This affinity is likely reduced when PDZ1 is present due to entropy losses caused by increased structural rigidity. The expression levels of all fusions, analyzed by Western blot on yeast lysate with an anti-HA antibody, were comparable (Fig. S3).
The reported interaction between INAD and NINAC in the formation of the Signalplex (23), together with the interaction between INAD and dCRY that we observed, suggest that the interaction between dCRY and NINAC may be specifically mediated by INAD. To detect whether dCRY, INAD, and NINAC form a ternary protein complex, we devised a three-hybrid system, in which dCRY was used as bait and NINAC as prey and a FLAG-tagged form of INAD was selectively expressed in the yeast nucleus. The expression of all fusions was tested by Western blot on yeast lysate with anti-HA antibody for NINAC and anti-FLAG antibody for the nuclear INAD (Fig. S3). When we expressed dCRY as bait and NINAC as prey alone, no direct interaction between the two proteins was observed, whereas expression of INAD in the nucleus resulted in the activation of the reporter gene, indicating that the formation of a three-component complex is necessary to restore the activity of the transcription factor (Fig. 2C).
dCRY Is Involved in Visual Behavior.
The surprising presence of dCRY associated with the visual cascade complex could underline a role, direct or indirect, for this photoreceptor in fly vision, which has not been entertained as yet.
To investigate a possible involvement of dCRY in the fly eye-mediated light response, the electroretinogram (ERG) of flies in which dCRY was completely knocked out (cry01) (27) was analyzed. Moreover, we studied the optomotor and phototactic behavior of cry01 flies or flies in which dCRY lacked the C terminus tail (cryM) (5). Wild-type flies are known to show a diurnal rhythm in visual sensitivity determined by ERG recordings, with maximal sensitivity in the first half of the night (28). A comparable rhythm was found in control flies [Canton S (CS) × w1118] with a pronounced sensitivity and a maximum in the middle of the night (Fig. 3A). In contrast, the visual sensitivity of cry01 mutants was not dependent on the time of day albeit their ERG profiles were normal (Fig. S4D). The same was true for the optomotor turning response of the flies. Although the optomotor response of wild-type flies depended significantly on the time of the day (as already observed in ref. 29), it did not in cry01 mutants (Fig. 3B). cry01 mutants responded less to visual stimuli throughout the day than control flies, but this impairment was most evident during the first half of the night, around the wild-type flies’ maximum in optomotor turning response (Fig. 3B). The optomotor response was analyzed with two different setups (SI Materials and Methods) with similar results (Fig. 3 B and C and Fig. S4 A and B). Like cry01 mutants, cryM mutants also displayed a similar impairment in their optomotor turning response (Fig. S4 A and B). In a phototaxis assay using countercurrent distribution, in which wild-type flies orient and move toward a light source (30), cry01 and cryM mutants showed a reduced performance index of 0.41, compared with 0.63 of the progeny of the CS × w1118 cross used as control (Fig. S4C). To test whether the impaired optomotor response depends on CRY function in the compound eyes, we selectively rescued CRY in the eyes with the help of the upstream activating sequences (UAS)-GAL4 system, driving GAL4 under control of the eye-specific glass multiple reporter (gmrGAL4) (31). gmr-GAL4 is known to disturb the structure of the compound eyes in a dose- and temperature-dependent manner (32). As a consequence, gmrGAL4;cry01 control flies showed a lower optomotor response than the other cry01 mutants (Fig. 3C). Nevertheless, the expression of the HAcry7M construct (Fig. S5) in the compound eyes restored the optomotor response of cry01 mutants to almost wild-type levels.
Fig. 3.
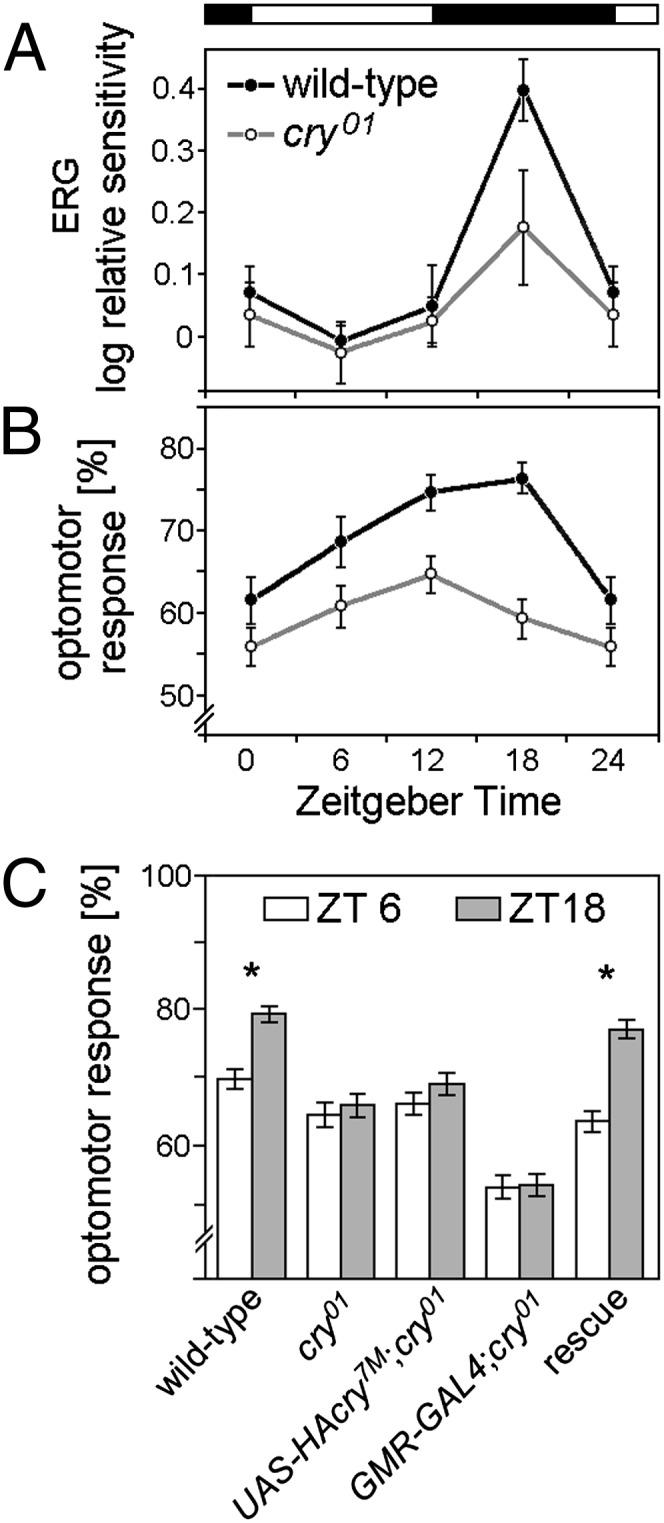
Visual behavior of wild-type flies, cry01 mutants, and cry01 mutants with CRY rescue in the eyes. (A) Visual sensitivity of cry01 and wild-type controls (CS × w1118) during the course of a day. Sensitivity was calculated as the reciprocal of the photon flux needed to evoke a criterion response of 6 mV in the ERG receptor potential. Within each genotype, sensitivity values were normalized to the average sensitivity at ZT6. Each point represents the average of values estimated for a minimum of 9 and a maximum of 13 flies. Mean values ± SEM are given. ANOVA revealed that sensitivity values were significantly dependent on the time of day for CS × w1118 (F3,38 = 15.649, P < 0.001) but not for cry01 (F3,39 = 1.775, P = 0.168). Note that the value at ZT4 is repeated at ZT0 to improve clarity. (B) Optomotor responses of cry01 and wild-type controls (CS × w1118) during the course of a day. Each point represents the average of 32 flies. The nonparametric Kruskal–Wallis test revealed that optomotor response values were significantly dependent on the time of day for CS × w1118 (P < 0.001) but not for cry01 (P = 0.181). Furthermore, two-way ANOVA showed that optomotor response was highly dependent on the genotype (F1,251 = 31.411, P < 0.001), meaning that wild-type flies generally showed a higher optomotor response than cry01 mutants. Note that the value at ZT24 is repeated at ZT0 to improve clarity. (C) Optomotor responses at ZT6 and ZT18 for wild-type flies, cry01 mutants, and flies with CRY rescued in the compound eyes (UAS-HAcry7M;cry01 × gmrGAL4;cry01). A total of 100 flies per genotype were analyzed in each experimental condition. Only wild-type and CRY-rescued flies showed a significant difference in optomotor response between the ZT6 and ZT18 (wild type: t198 = 5.23, P < 0.0001; rescued flies: t198 = 6.53, P < 0.0001).
Discussion
The analysis of the linear motifs present in the C terminus of CRYs showed that they were not evenly distributed in all species investigated. The class III PDZ motif is present in all animal phyla, suggesting a functional constraint on the evolving sequence, as the motif is maintained although it is not being conserved in the same sequence stretch. Our results clearly indicate that the circadian blue-light photoreceptor dCRY interacts with the visual transduction complex (Signalplex) through the scaffold protein INAD. The interaction between the two proteins is mediated by a specific region of INAD, which includes the PDZ2-PDZ3 tandem, but is extended upstream with respect to the canonical PDZ domain boundary to include a stretch of amino acids known to be part of a calmodulin-binding motif. Interactions modulated by multiple INAD PDZ domains have already been described (33). It has also been reported for other PDZ-containing proteins that two or three PDZ domains connected in tandem may exhibit different specificity in their target-binding properties compared with isolated domains (34). We also established that the 22-amino acid C-terminal sequence of dCRY is involved in binding to INAD, in accordance with the presence of either class III or class II/III PDZ-binding motifs predicted by the eukaryotic linear motif (ELM) program in the C terminus of the protein and also with the notion that PDZ domains preferentially interact with the absolute carboxyl-terminal ends of their target proteins (14). The interaction between dCRY and INAD is particularly effective in the light, and it is well recognized that the activity of both proteins is modulated by light. However, the light-independent interaction of the C-terminal fragment of dCRY with INAD suggests that the influence of light in the interaction of the full-length proteins is due to the PHR domain of dCRY. Supporting this hypothesis is the fact that the INAD PDZ4 and -5, known to be regulated by light-dependent conformational changes (33), are not involved in the interaction. The interaction between dCRY and NINAC observed in vivo represents quite an unexpected result. A connection between dCRY and a cardinal component of the fly visual cascade (23) was established, and the mediator role of INAD in the interaction was demonstrated. We also showed that this interaction has a functional importance for vision. In CRY-knockout flies, the diurnal cycling of photoreceptor sensitivity and motion vision typical of wild-type flies (28, 29) is abolished. Furthermore, the CRY-knockout flies are slightly but significantly impaired in motion vision. The diurnal rhythm in optomotor response was recovered when CRY was expressed in all photoreceptor cells of the compound eyes, showing that CRY in the photoreceptor cells is responsible for wild-type rhythms in motion vision. Motion detection depends mainly on intact vision in photoreceptors R1–6 with minor contribution from R7 and R8 (35, 36), whereas phototaxis is mediated by all eight photoreceptors in the compound eyes (37). dCRY is expressed in the entire cytoplasm of the photoreceptor cells and seems to have the highest density close to the rhabdomers, the place of the visual cascade (38). Therefore, dCRY may easily interact with INAD and eventually modulate the transient receptor potential (TRP) and TRP-like (TRPL) channel opening in interplay with the other PDZ proteins of the Signalplex. Interestingly, small amounts of CRY seem to be sufficient for this interaction as the optomotor response was highest at the end of the day until the middle of the night (ZT12–18) when CRY levels are low (9). Recently, dCRY was shown to be also involved in the membrane excitability (K+ channel conductance) of the large ventral Lateral clock Neurons (l-LNv) (39). These neurons fire action potential upon illumination with blue light, and this firing is dependent on dCRY. Although the way in which dCRY regulates the l-LNv firing rate in relation to K+ channel conductance remains unclear, our results further support an involvement of dCRY in membrane potential modulation. Here, we show that dCRY may be the link that couples the clock with the PDZ proteins of the Signalplex, in this way modulating vision in a circadian fashion. A functional circadian clock in the photoreceptor cells is obviously important to control visual coding efficiency in Drosophila and to optimize vision under different light intensity regimes (29). In fact, wild-type flies show circadian changes in the size of certain brain regions (e.g., optic lobes) and in photoreceptor cell terminals that control the sensitivity of photoreceptors to circadian variations in light levels (29). This structural plasticity is still maintained in period (per)01 flies, which lack a key component of the circadian machinery, but it is exclusively light-driven as there is no longer “anticipation” of the light/dark transitions (29). In most invertebrates, the components of visual signaling are localized on the rhabdomeres (40), whereas a ciliary vision (rods and cones) is predominant in the vertebrate retina (41). An important difference between the two kinds of photoreceptors is the biochemical cascade used to transduce photic signals in electric signals. In fact, rods and cones use a cascade involving cyclic guanyl monophosphate as a second messenger whereas rhabdomeric photoreceptors use a phosphoinositide-signaling cascade involving the enzyme phospholipase C (PLC) (41). Retinal photoreception in mammals includes a subset of retinal ganglion cells that are able to respond to light even in the absence of synaptic inputs (42). These cells, called “intrinsically photosensitive retinal ganglion cells” (ipRGCs), use melanopsin as photopigment and send their axons directly to the suprachiasmatic nucleus, the site of the primary circadian pacemaker in mammals (18, 41). ipGRCs have been shown to use a rhabdomeric-like phosphoinositide cascade involving the effector enzyme PLC (18, 41). Very recently, it has been observed that these melanopsin-expressing ganglion cells extend their projections toward the thalamo-cortical neurons implicated in pattern vision, establishing melanopsin-based photoreception as a significant source of visual information to the thalamo-cortical pathway, independent of rods or cones (43). The ipGRCs and the fly phototransduction mechanisms also share other similarities: both require a member of the Gq/11 family of G proteins as a mediator of the phototransduction cascade, and, in both cases, the phototransduction cascade is tightly coupled to the plasma membrane and involves light-sensitive channels belonging to the TRP family (44). The similarity of the photoreception cascade between Drosophila and the mammalian ipRGCs, and also the expression of CRY in both photoreceptor cells (45), raises the question of whether mammalian CRYs could contribute to the circadian functions of ipRGCs by specifically binding to the phototransduction complex. Although a homologous complex of the fly Signalplex has not been described in ipRGCs, several components of this multiprotein complex seem to be conserved (18). Specifically, a protein homolog of dINAD, INAD-like (INADL), bearing seven PDZ domains, has been identified in humans (46). A search for a functional protein interaction network, performed with the STRING database (Fig. S6), showed that INADL can be a functional partner of Crumbs homolog 1 precursor, a factor involved in retinal photoreceptor organization (47). This renders INADL a good candidate for a scaffold protein that organizes and maintains the phototransduction complex in ipRGCs. Our results extend the role of dCRY to fly visual biology and provide a tantalizing glimpse of a phylogenetically conserved possible role for CRY that may have circadian implications in mammalian vision also.
Materials and Methods
Bioinformatic Analyses.
The computational search for dCRY protein–protein interactions combined the results from the STRING database (19) of protein–protein interactions with the domain organization of proteins from Pfam (48). Relevant proteins were analyzed with CSpritz (26), which predicts intrinsic disorder in the sequence as well as linear motifs coding for common protein–peptide interactions taken from ELM (49). The X-ray structures of INAD PDZ domains were retrieved from the Protein Data Bank for domains 1 and 5 [Protein Data Bank (PDB) codes 1IHJ and 2QKT]. The three remaining domains were identified (50) and modeled based on PDB codes 2FNE (chain C) and 1Z87 (chain A) as templates for PDZ2-PDZ4 and PDZ3, respectively (Fig. S7).
Coimmunoprecipitation and Mass Spectrometry.
Head extracts from HACRY-overexpressing flies were subjected to coimmunoprecipitation as previously described (3). After the separation of proteins by SDS/PAGE, Coomassie-stained protein bands were excised, in-gel digested (21), and analyzed by LC-MS/MS on a Micromass CapLC unit (Waters) interfaced to a Micromass Q-Tof Micro mass spectrometer (Waters). MS/MS data were analyzed by MASCOT software (Matrix Science; www.matrixscience.com/) against the Drosophila sequences of the Swiss-Prot database (release 2011_03).
Western Blots.
Immunocomplexes were analyzed by Western blotting using the following antibodies: rabbit polyclonal anti-INAD (1:500) (25), rabbit polyclonal anti-NINACp174 (1:500) (22), and mouse anti-HA (Sigma; 1:5,000).
Yeast Two- and Three-Hybrid Tests.
dCRY, either full-length or fragments, was fused to the LexA moiety in the bait vector (pEG202), and INAD (full length or fragments) was fused to the “acid-blob” portion of the prey vector (pJG4-5) (24). In the yeast three-hybrid assay, dCRY was used as bait and NINAC as prey, and a FLAG-tagged full-length INAD was expressed in the nucleus. Quantification of β-galactosidase activity was performed in liquid culture as in Ausbel et al. (51).
Visual Sensitivity Determined by ERG Recordings.
Visual sensitivity was obtained from the irradiance response curves (IRC) recorded at four different ZTs. The ERG responses to light stimuli of different intensities were used to determine the IRCs. ERGs were recorded as in ref. 28.
Analysis of Optomotor Activity.
The walking optomotor test was performed as in ref. 52 (setup 1 in SI Materials and Methods). Details of setups 1 and 2 are given in SI Materials and Methods.
Phototaxis.
The experiments for phototaxis were performed as described in ref. 30. See details in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Michele Vidotto (University of Padova) for help with the initial linear motif analysis; Paola Cisotto (University of Padova) for technical support; Matteo Simonetti (University of Padova) for graphical support; Alberto Biscontin (University of Padova) for help with quantitative RT-PCR; Craig Montell (The Johns Hopkins University School of Medicine) for anti-INAD and anti-NINACp174 antibodies; Dr. Susan Tsunoda (Colorado State University) for inaD1 flies; the Bloomington Stock Center for NinaCΔ174 flies; and Dr. Taishi Yoshii (Okayama University) for gmrGAL4;cry01 flies. We thank Reinhard Wolf (Virchow Center, University of Würzburg) for help with the optomotor response experiments and Reinhard Wolf and Bambos Kyriacou (University of Leicester) for helpful comments on the manuscript. This work was funded by grants from the European Community (Sixth Framework Project Entrainment of the Circadian Clock 018741) and Fondazione Cariparo (Progetti di Eccellenza 2011–2012) (R.C.); University of Padova Grant CPDA099390/09 (to G.M.); and University of Padova Grants CPDA098382 and CPDR097328 and Fondo Investimento Ricerca di Base (FIRB) Futuro in Ricerca Grant RBFR08ZSXY (to S.C.E.T.). R.C. was supported by the Italian Space Agency [Disturbo del Controllo Motorio e Cardiorespiratorio (DCMC) grant] and the Ministero dell’Università e della Ricerca. C.H.-F. was supported by the Deutsche Forschungsgemeinschaft (Fo207/10-3), R.G. by the Graduate School of Life Sciences (University of Würzburg), and M.S. by the Hanns Seidel Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212317110/-/DCSupplemental.
References
- 1.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaves I, et al. The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 3.Hemsley MJ, et al. Linear motifs in the C-terminus of D. melanogaster cryptochrome. Biochem Biophys Res Commun. 2007;355(2):531–537. doi: 10.1016/j.bbrc.2007.01.189. [DOI] [PubMed] [Google Scholar]
- 4.Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6(5):220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304(5676):1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 6.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24(4):948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 7.Gegear RJ, Foley LE, Casselman A, Reppert SM. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature. 2010;463(7282):804–807. doi: 10.1038/nature08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95(5):669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 9.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19(3):241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 10.Stoleru D, et al. The Drosophila circadian network is a seasonal timer. Cell. 2007;129(1):207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 11.Rosato E, et al. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr Biol. 2001;11(12):909–917. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- 12.Dissel S, et al. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7(8):834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- 13.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci USA. 2011;108(2):516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saras J, Heldin CH. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21(12):455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 15.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275(5296):73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 16.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain-peptide interactions from primary sequences. Nat Biotechnol. 2008;26(9):1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonikian R, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6(9):e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montell C. Drosophila visual transduction. Trends Neurosci. 2012;35(6):356–363. doi: 10.1016/j.tins.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454(5):821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 21.Wilm M, et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 22.Porter JA, Hicks JL, Williams DS, Montell C. Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J Cell Biol. 1992;116(3):683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wes PD, et al. Termination of phototransduction requires binding of the NINAC myosin III and the PDZ protein INAD. Nat Neurosci. 1999;2(5):447–453. doi: 10.1038/8116. [DOI] [PubMed] [Google Scholar]
- 24.Golemis EA, Brent R. In: Searching for Interacting Proteins with the Two-Hybrid System III. In the Yeast Two-Hybrid System. Bartel PL, Field S, editors. New York: Oxford University Press; 1997. pp. 43–72. [Google Scholar]
- 25.Feng W, Shi Y, Li M, Zhang M. Tandem PDZ repeats in glutamate receptor-interacting proteins have a novel mode of PDZ domain-mediated target binding. Nat Struct Biol. 2003;10(11):972–978. doi: 10.1038/nsb992. [DOI] [PubMed] [Google Scholar]
- 26.Walsh I, et al. CSpritz: Accurate prediction of protein disorder segments with annotation for homology, secondary structure and linear motifs. Nucleic Acids Res. 2011;39(Web Server issue):W190–W196. doi: 10.1093/nar/gkr411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177(1):329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen DM, Christianson JS, Sapp RJ, Stark WS. Visual receptor cycle in normal and period mutant Drosophila: Microspectrophotometry, electrophysiology, and ultrastructural morphometry. Vis Neurosci. 1992;9(2):125–135. doi: 10.1017/s0952523800009585. [DOI] [PubMed] [Google Scholar]
- 29.Barth M, Schultze M, Schuster CM, Strauss R. Circadian plasticity in photoreceptor cells controls visual coding efficiency in Drosophila melanogaster. PLoS ONE. 2010;5(2):e9217. doi: 10.1371/journal.pone.0009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benzer S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc Natl Acad Sci USA. 1967;58(3):1112–1119. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87(4):651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 32.Kramer JM, Staveley BE. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet Mol Res. 2003;2(1):43–47. [PubMed] [Google Scholar]
- 33.Liu W, et al. The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye. Cell. 2011;145(7):1088–1101. doi: 10.1016/j.cell.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Long JF, et al. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J Mol Biol. 2003;327(1):203–214. doi: 10.1016/s0022-2836(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi S, Wolf R, Desplan C, Heisenberg M. Motion vision is independent of color in Drosophila. Proc Natl Acad Sci USA. 2008;105(12):4910–4915. doi: 10.1073/pnas.0711484105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wardill TJ, et al. Multiple spectral inputs improve motion discrimination in the Drosophila visual system. Science. 2012;336(6083):925–931. doi: 10.1126/science.1215317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi S, Heisenberg M. Photoreceptors and neural circuitry underlying phototaxis in insects. Fly (Austin) 2011;5(4):333–336. doi: 10.4161/fly.5.4.16419. [DOI] [PubMed] [Google Scholar]
- 38.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508(6):952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 39.Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331(6023):1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shieh BH, Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron. 1995;14(1):201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- 41.Graham D. Melanopsin ganglion cells: A bit of fly in the mammalian eye. In: Kolb H, Fernandez E, Nelson R, editors. Webvision: The Organization of the Retina and Visual System. Salt Lake City: University of Utah, Health Sciences Center; 2008. Available at http://webvision.med.utah.edu/book/part-ii-anatomy-and-physiology-of-the-retina/elanopsin-ganglion-cells-a-bit-of-fly-in-the-mammalian-eye. [PubMed] [Google Scholar]
- 42.Bailes HJ, Lucas RJ. Melanopsin and inner retinal photoreception. Cell Mol Life Sci. 2010;67(1):99–111. doi: 10.1007/s00018-009-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown TM, et al. Melanopsin contributions to irradiance coding in the thalamo-cortical visual system. PLoS Biol. 2010;8(12):e1000558. doi: 10.1371/journal.pbio.1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham DM, et al. Melanopsin ganglion cells use a membrane-associated rhabdomeric phototransduction cascade. J Neurophysiol. 2008;99(5):2522–2532. doi: 10.1152/jn.01066.2007. [DOI] [PubMed] [Google Scholar]
- 45.Thompson CL, et al. Expression of the blue-light receptor cryptochrome in the human retina. Invest Ophthalmol Vis Sci. 2003;44(10):4515–4521. doi: 10.1167/iovs.03-0303. [DOI] [PubMed] [Google Scholar]
- 46.Vaccaro P, et al. Distinct binding specificity of the multiple PDZ domains of INADL, a human protein with homology to INAD from Drosophila melanogaster. J Biol Chem. 2001;276(45):42122–42130. doi: 10.1074/jbc.M104208200. [DOI] [PubMed] [Google Scholar]
- 47.den Hollander AI, et al. Isolation of Crb1, a mouse homologue of Drosophila crumbs, and analysis of its expression pattern in eye and brain. Mech Dev. 2002;110(1–2):203–207. doi: 10.1016/s0925-4773(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 48.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gould CM, et al. 2010. ELM: The status of the eukaryotic linear motif resource. Nucleic Acids Res 38(Database issue):D167–D180.
- 50.Bindewald E, Cestaro A, Hesser J, Heiler M, Tosatto SC. MANIFOLD: Protein fold recognition based on secondary structure, sequence similarity and enzyme classification. Protein Eng. 2003;16(11):785–789. doi: 10.1093/protein/gzg106. [DOI] [PubMed] [Google Scholar]
- 51.Ausbel FM, et al. Current Protocols in Molecular Biology. New York: Green Publishing Associated; 1989. [Google Scholar]
- 52.Zordan MA, et al. Post-transcriptional silencing and functional characterization of the Drosophila melanogaster homolog of human Surf1. Genetics. 2006;172(1):229–241. doi: 10.1534/genetics.105.049072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.