Abstract
Efficient total synthesis of insulin is important to enable the application of medicinal chemistry to the optimization of the properties of this important protein molecule. Recently we described ‘ester insulin’ – a novel form of insulin in which the function of the 35 residue C-peptide of proinsulin is replaced by a single covalent bond – as a key intermediate for the efficient total synthesis of insulin. Here we describe a fully convergent synthetic route to the ester insulin molecule from three unprotected peptide segments of approximately equal size. The synthetic ester insulin polypeptide chain folded much more rapidly than proinsulin, and at physiological pH. Both the D-protein and L-protein enantiomers of monomeric DKP ester insulin (i.e. [AspB10, LysB28, ProB29]ester insulin) were prepared by total chemical synthesis. The atomic structure of the synthetic ester insulin molecule was determined by racemic protein X-ray crystallography to a resolution of 1.6 Å. Diffraction quality crystals were readily obtained from the racemic mixture of {D-DKP ester insulin + L-DKP ester insulin}, whereas crystals were not obtained from the L-ester insulin alone even after extensive trials. Both the D-protein and L-protein enantiomers of monomeric DKP ester insulin were assayed for receptor binding and in diabetic rats, before and after conversion by saponification to the corresponding DKP insulin enantiomers. L-DKP ester insulin bound weakly to the insulin receptor, while synthetic L-DKP insulin derived from the L-DKP ester insulin intermediate was fully active in binding to the insulin receptor. The D- and L-DKP ester insulins and D-DKP insulin were inactive in lowering blood glucose in diabetic rats, while synthetic L-DKP insulin was fully active in this biological assay. The structural basis of the lack of biological activity of ester insulin is discussed.
Introduction
Human insulin was the first protein to have its complete covalent structure elucidated, by Fred Sanger who received the Nobel Prize in 1958 for this achievement.1-2 The insulin molecule is made up of two polypeptide chains containing a total of 51 amino acids. There are two inter-chain disulfide bonds and one intra-chain disulfide bond. Immediately after the report of its covalent structure, insulin became the target of total chemical synthesis. Starting in the 1960s, a number of successful chemical syntheses of insulin were reported.3-9 All of these syntheses relied on the preparation of the 21 residue insulin A chain and the 30 residue B chain as separate polypeptides, and subsequent recombination of the A and B chains to give the folded insulin protein molecule containing the three native disulfide bonds. Several of these synthetic routes gave crystalline insulin, but the ‘Achilles heel’ for total chemical synthesis of this protein molecule has remained the poor yields of native insulin obtained by recombination of the individual A and B chains. It has been reported that even under highly optimized conditions, an overall atom efficiency10 of just 7% is obtained.11
How can this low recombination yield be improved? In 1967 Steiner showed that in nature insulin is produced as ‘proinsulin’ - a single polypeptide chain of 86 amino acids in which the C-terminal of the B-chain is connected to the N-terminal of the A-chain by a 35 residue C-peptide.12 In vivo, the proinsulin polypeptide folds efficiently in the endoplasmic reticulum (ER) of the pancreatic ß-cells, and is converted to the mature two-chain insulin molecule by proteolytic processing that removes the C-peptide.13 The recombinant production of human insulins for therapeutic use relies on the folding and enzymatic processing of recombinant proinsulin.14 In vitro, the overall yield for folding proinsulin and its enzymatic processing to insulin is as high as 70%.15
Even prior to the discovery of proinsulin, it had been shown that chemical tethering of the insulin A and B chains gave high yields of folded insulin containing native disulfides.16-21 However, the resulting cross-linked insulin molecules were inactive and for that reason the chemical tethering concept was not useful for the total synthesis of insulin, despite studies describing a chemically removable tether.21 Similarly, mini-proinsulin molecules in which the C-peptide connecting the B and A chains was shortened or eliminated in most cases gave folded insulin analogues devoid of biological activity.22 A single chain insulin construct with LysB29 connected directly to residue GlyA1 folded efficiently and was enzymatically converted to fully active desThrB30 insulin.23 Nonetheless, until recently the efficient total chemical synthesis of native human insulin has remained an unsolved challenge.
1. Ester Insulin
Recently we reported the successful prototyping of a novel concept for a simpler and more practical total chemical synthesis of human insulin.24 Our synthetic design involved a key intermediate in which the insulin A and B chains were linked by an ester bond formed between the γ-carboxyl group of GluA4 and the β-hydroxyl group of ThrB30. In order to explore its folding properties the 51 residue ester-linked polypeptide was prepared by native chemical ligation of two peptide segments, one of 18 residues and the other of 33 residues. The resulting ester-linked polypeptide folded efficiently with concomitant formation of three disulfide bonds, and could be converted to fully active human insulin by saponification at reduced temperature. 24
A more optimized total chemical synthesis of human insulin would employ a convergent condensation strategy from readily prepared synthetic peptide building blocks to give the key ester-linked polypeptide described above, followed by folding and chemical conversion to the native insulin molecule. Such convergent synthesis enables the most efficient use of peptide segment starting materials.25-26 Such a synthetic route would provide a practical alternative to recombinant methods for the production of human insulins for therapeutic use and could provide an alternative route to the manufacture of generic forms of off-patent insulins. Optimization of the ester insulin synthetic route will also enhance the versatility of this synthetic approach for the preparation of novel analogues, and will enable us to systematically tune the properties of the insulin molecule. This promises the expansion of chemical space to exploit modern medicinal chemistry for the development of insulin therapeutics.
Here, we report an efficient modular total synthesis of ester insulin, based on the fully convergent chemical ligation of three synthetic peptide building blocks. The 51 residue ester-linked polypeptide chain folded much more rapidly than proinsulin, and at near physiological pH. Both the L-protein and D-protein enantiomers of ester insulin were prepared using this synthetic route and were used for the determination of the high resolution X-ray structure of ester insulin by racemic protein crystallography.27 Synthetic ester insulin was converted to human insulin by simple saponfication. We report the biological activities of all these forms of insulin.
2. Design of the Synthesis
Convergent synthesis
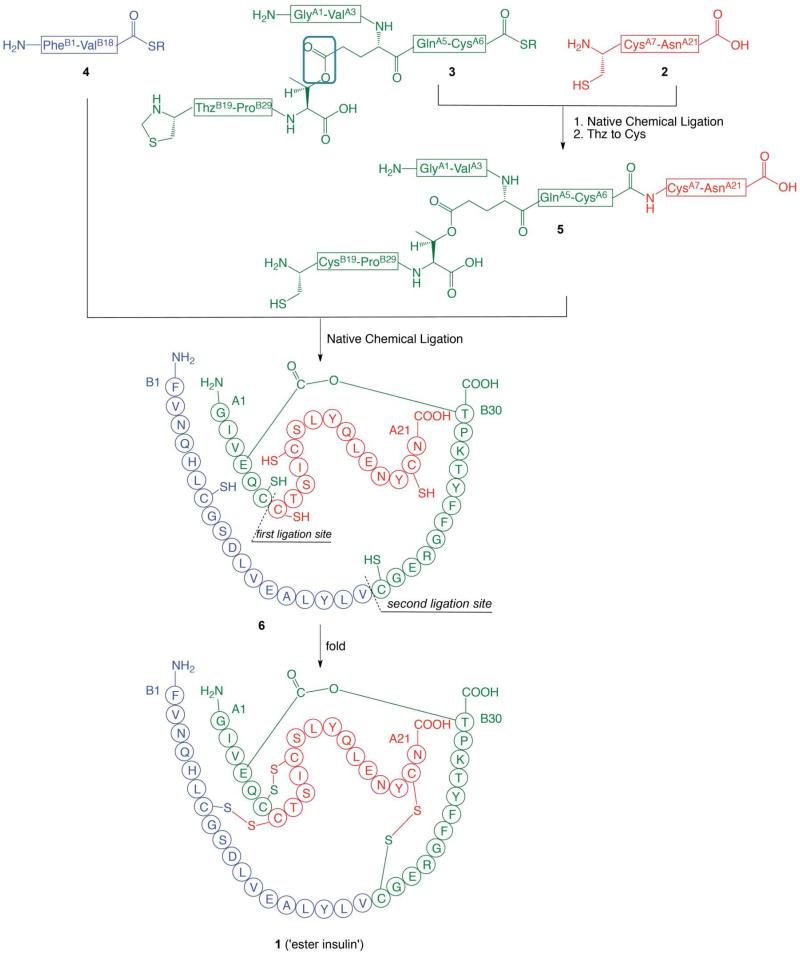
The ester insulin polypeptide (compound 6, Scheme 1) consists of 51 amino acids and contains six Xaa-Cys sites, each of which represents a potential site for native chemical ligation.28,29 In our original proof-of-concept preparation of the 51 residue ester insulin polypeptide just two peptide segments were used as building blocks, and one of the segments was relatively long.24 In the work reported here, a fully convergent strategy was designed to make use of three peptide segments of approximately equal size (Scheme 1). The three peptide building blocks used here are: GlyA1-GluA4[OβThrB30-ThzB19]-CysA6-thioester (3), PheB1-ValB18-thioester (4); and CysA7-AsnA21 (2) (Scheme 1). These peptides have 18 amino acid residues, 18 residues, and 15 residues, respectively.
Scheme 1.
Convergent synthetic strategy from three peptide segments of approximately equal size used for the total chemical synthesis of DKP ester insulin by ‘one pot’ native chemical ligations (R = –CH2CH2CO–Ala–COOH). The amino acid sequences are given in the single letter code. Color-coding corresponds to the peptide segments used in the synthesis. The key ester moiety is boxed in the top panel. The illustrations for ester insulin follow the convention used in Sohma et al. Angew Chem 2010.24
Protecting group tactics
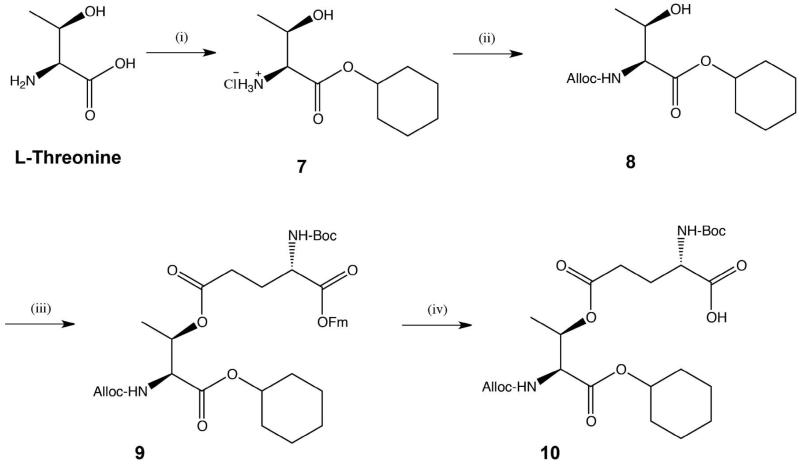
Synthesis of the ester containing peptide GlyA1-GluA4[OβThrB30-ThzB19]-CysA6-thioester (3) requires the use of several different types of protecting groups, so that each can be stable or removed at a given step, as needed. The key building block is a suitably protected Glu(OβThr) ester-linked dipeptide with three protecting groups removable under distinct sets of conditions. Since segment 3 is a thioester peptide, the Fmoc protecting group cannot be employed because the piperidine used for Fmoc deprotection will attack the thioester moiety. Therefore, we designed the key ester-linked Boc-Glu[Oβ(Alloc-Thr-α-O-cHex)]-OH dipeptide (10) (Schemes 2 & 3). The Boc and Alloc protecting groups can each be removed in the presence of the other, and they can each be removed without affecting the thioester moiety of 3. The cyclohexyl protecting group was employed to protect the C-terminal carboxylic functionality of ThrB30 during the solid phase peptide synthesis and is removed by anhydrous HF in the cleavage/deprotection step. Synthesis of the Boc-Glu[Oβ(Alloc-Thr-α-O-cHex)]-OH (10) is shown in Scheme 2. The chemical tactics used for the solid phase synthesis of the ester-linked branched peptide building block 3 are shown in Scheme 3. In addition to the differential protection of the amino groups in the ester-linked dipeptide, it was necessary to ‘permanently’ protect the N-terminal amino functionality of Gly1 in the A-chain while the sequence ThzB19-ProB29 was assembled by Boc chemistry solid phase peptide synthesis (SPPS) on the ester-linked ThrB30 (Scheme 3). For this purpose, chlorobenzyloxycarbonyl (Cl-Z) was employed.
Scheme 2. Synthesis of ester-containing dipeptide Boc-L-Glu[Oβ(Alloc-L-Thr-α-O-cHex)]-OH (10).
(i) cyclohexanol, SOCl2, 100 °C, 2.5 h; (ii) allyl chloroformate, Et3N, THF, rt, overnight (94% for both steps); (iii) Boc-Glu-OFm, EDC·HCl, DMAP, DCM, rt, overnight, (78%); (iv) 20 % piperidine in DCM, rt, 3 h, (91%).
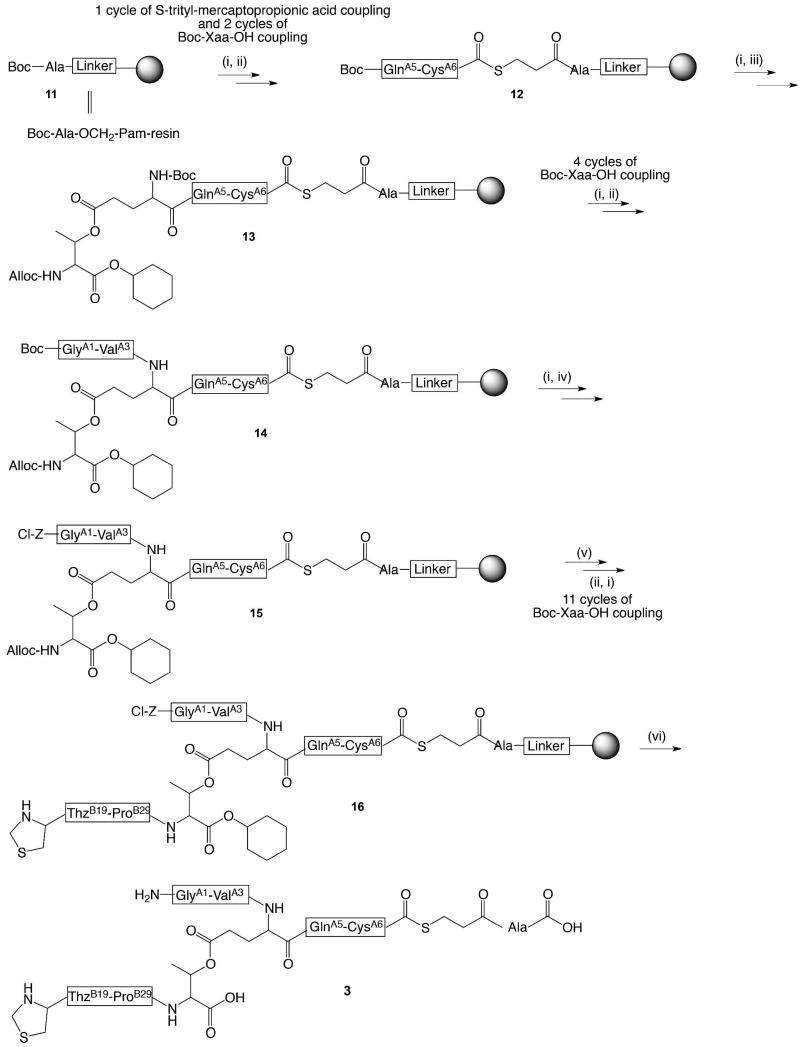
Scheme 3. Synthesis of ThzB19-[A1-GluA4(Oβ ThrB30)-A6-αCOSR.
(i) TFA for Boc deprotection. Trityl moiety was removed with a cocktail of 95:2.5:2.5 TFA:H2O:triisopropylsilane; (ii) Boc-Xaa-OH, HBTU, DIEA, DMF; (iii) Boc-Glu(Oβ(Alloc-Thr-α-O-cHex))-OH (10), HBTU, DIEA, DMF; (iv) DIEA, DMF, Z(2-Cl)-OSu; (v) Pd(PPh3)4, PhSiH3, DCM; (vi) HF, p-cresol.
Chemical ligations
The fully convergent synthesis of ester insulin from the three peptide segments was accomplished by two native chemical ligation steps, starting from the CysA7-AsnA2 segment 2 (Scheme 1). A ‘one pot’ synthesis approach was employed in order to minimize losses from multiple intermediate handling and purification steps.30 The N-terminal Cys of the thioester peptide 3 was introduced as the 1,3-thiazolidine-4-carboxo (Thz) group in order to prevent intramolecular reaction of the N-terminal Cys with the thioester moiety of the same peptide.30 After the first chemical ligation, the Thz-moiety was simply converted to the Cys-peptide product by treatment with methoxylamine·HCl at reduced pH. Without purification, the product solution was then readjusted to pH 7 and the second native chemical ligation was carried out. The full-length polypeptide was recovered by precipitation, and was folded with the formation of the three native disulfide bonds and then purified to give the desired ester insulin product (1).
3. Results and Discussion
We chose to make DKP insulin (i.e. [AspB10, LysB28, ProB29]-insulin). Relative to wild-type insulin this analog exhibits enhanced receptor binding and enhanced stability, and can be stably formulated as a zinc-free monomer.31 The amino acid sequence and covalent structure of the ester insulin target molecule is shown in Scheme 1.
Synthesis of ester–linked dipeptide (10)
The synthesis of the ester linked dipeptide Boc-Glu[Oβ(Alloc-Thr-α-O-cHex)]-OH (10) is shown in Scheme 2. Esterification of L-threonine with cyclohexanol generated compound 7 which was easily recovered by evaporation of the cyclohexanol and used without further purification. Compound 7 was treated with allyl chloroformate to afford carbamate 8 in high yields. Condensation with Boc-Glu-OFm generated ester 9, which was then treated with piperidine to remove the OFm moiety and to thus yield ester-linked dipeptide 10. This dipeptide was then incorporated in the synthesis of the branched ester-containing peptide (3) (Scheme 3). This four-step synthesis of dipeptide 10 was efficient and simple. More importantly it resulted in recovered yields of approximately 70% over the four steps.
Synthesis of peptide segments
The two peptide-thioesters PheB1-ValB18-αCOSR (4) (R=–CH2CH2CO–Ala–COOH) and GlyA1-GluA4[OβThrB30-ThzB19]-CysA6-αCOSR (3), and the Cys-peptide CysA7-AsnA21 (2) building blocks were prepared by manual stepwise Boc chemistry “in situ neutralization” solid phase peptide synthesis.32 Synthetic peptides were purified by preparative reverse phase HPLC and were characterized by analytical LC-MS. Full details are given in the Supporting Information.
Synthesis of the ester-containing peptide GlyA1-GluA4[OβThrB30-ThzB19]-CysA6-αCOSR (3) is shown in Scheme 3. This synthesis was performed on Boc-Ala-OCH2-Pam-resin. After one cycle of S-trityl mercaptopropionic acid coupling and two cycles of Boc-Xaa-OH coupling up to residue GlnA5, the dipeptide Boc-Glu[Oβ(Alloc-Thr-α-O-cHex)]-OH (10) containing Boc and Alloc protecting groups on GluA4(αNH2) and ThrB30(αNH2) respectively was coupled to GlnA5. The rest of chain A was constructed up to GlyA1; then the GlyA1 N-terminus was protected by chlorobenzyloxycarbonyl (Cl-Z), which is stable during Boc chemistry SPPS and can be removed in the final HF deprotection/cleavage step. The Alloc moiety was removed to reveal the αNH2- of ThrB30, and residues ProB29 to ThzB19 were assembled, followed by global deprotection and cleavage of the peptide from the resin by treatment with HF.
Native Chemical Ligation
Analytical data for a representative synthesis of L-ester insulin are given below. The synthesis of the enantiomer D-ester insulin was performed in the same manner. Analytical data for the syntheses of L- and D-peptide segments and D-ester insulin are shown in Figures S1-S5 (Supporting Information).
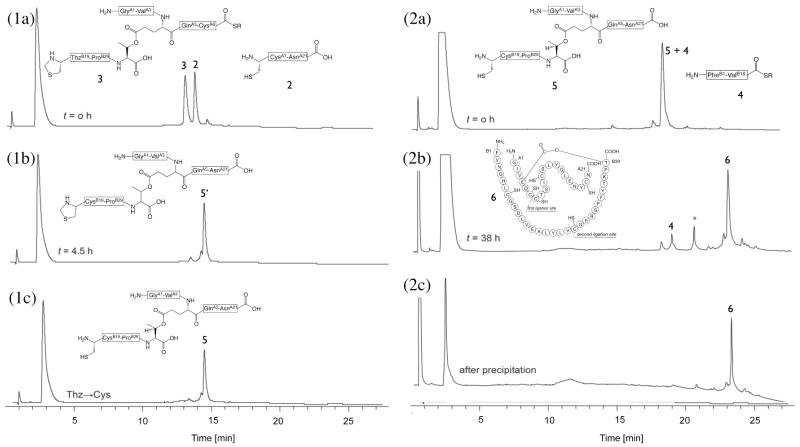
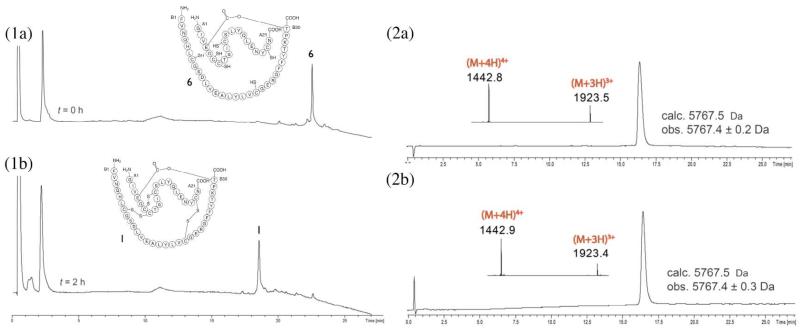
Based on the synthetic strategy shown in Scheme 1, the first native chemical ligation was performed between the C-terminal peptide segment CysA7-AsnA21 (2) and the ester-containing peptide GlyA1-GluA4[OβThrB30-ThzB19]-CysA6-thioester (3) in a buffer solution of pH 6.9 to give the ligation product GlyA1-GluA4[OβThrB30-ThzB19]-AsnA21 (5′) within 4.5h (Figure 1, panel 1a-b). A slight excess (1.07 eq.) of the Cys-peptide segment was used to insure complete reaction of the more ‘expensive’ ester-linked peptide-thioester segment. Without purification, the N-terminal ThzB19 was converted to CysB19 by treatment with methoxylamine·HCl at pH 4 to give GlyA1-GluA4[OβThrB30-CysB19])-AsnA21 (5) (Figure 1, panel 1c). After readjusting the product-containing solution to pH 6.9, the second native chemical ligation was performed in the same reaction mixture with peptide thioester segment PheB1-ValB18-thioester (4) (Figure 1, panel 2a-b). Because this ligation was at a hindered Val-Cys site, a high concentration of the aryl thiol catalyst mercaptophenylacetic acid (MPAA) was used.33 Within 38 h the reaction was complete and the full length polypeptide GlyA1-GluA4[OβThrB30-PheB1]-AsnA21 (6) was recovered by precipitation from water (Figure 1, panel 2c) and lyophilized. This ‘one pot’ synthesis proved to be an efficient way of carrying out sequential native chemical ligations; avoiding purification steps during the polypeptide assembly shortened the overall synthesis time since it excluded purification and lyophilization times which can take several days. In addition, this method minimized handling losses and therefore improved the overall yields.
Figure 1. One pot reactions for the total chemical synthesis of the ester insulin polypeptde 6.
(1) Native chemical ligation for the synthesis of GlyA1-GluA4[OβThrB30-CysB19]-AsnA21 (5). (1a-b) Reaction of peptide segments GlyA1-GluA4[OβThrB30-ThzB19]-CysA6-αCOSR (3) (R = –CH2CH2CO–Ala–COOH) and CysA7-AsnA21 (2). (1c) Conversion of Thz- to Cys-. The chromatographic separations were performed on a C18 column using a linear gradient (9%–53%) of buffer B in buffer A over 22 min (buffer A = 0.1% TFA in water, buffer B = 0.08% TFA in acetonitrile). (2) Native chemical ligation in the synthesis of GlyA1-GluA4[OβThrB30-PheB1]-AsnA21 (6). (2a-b) Reaction of GlyA1-GluA4[OβThrB30-CysB19])-AsnA21 (5) and PheB1-ValB18-αCOSR (4). Both peptide segments have the same retention time, as confirmed by LC-MS. Upon completion, excess PheB1-ValB18-αCOSR (4) remained. *: PheB1-ValB18-αCO MPAA thioester. (2c) Crude polypeptide (6) after precipitation from water. The chromatographic separations were performed on a C18 column using a linear gradient (4%–48%) of buffer B in buffer A over 22 min. All analyses were by LC-MS; only the HPLC data are shown.
Folding
The crude lyophilized full-length ester insulin polypeptide was folded at pH 7.6 in 1.5M guanidine hydrochloride aqueous buffer containing a cysteine/cystine redox couple. Folding with concomitant disulfide bond formation was rapid and nearly complete after 30 minutes. Folding was essentially complete after 2 h, as evidenced by the disappearance of linear polypeptide and the formation in ~85% HPLC yield of a sharp peak with an earlier elution time than the unfolded polypeptide in reverse phase HPLC, and a mass decrease of 6 Da corresponding to the formation of three disulfides (Figure 2, panel 1a-b). Folded globular proteins typically have earlier elution times in reverse phase HPLC compared to the corresponding unfolded polypeptides because the hydrophobic residues become buried in the core of the folded protein molecule.
Figure 2. Folding of ester insulin.
Folding was carried at room temperature in a pH 7.6 buffer solution containing 1.5 M GnHCl, 20 mM Tris, 8 mM cysteine, 1 mM cystine·HCl, at a starting polypeptide concentration of 0.3 mg/mL. (1a) Crude linear polypeptide ester insulin (6) at t = 0 h. (1b) Folding reaction after 2 h. The chromatographic separations were performed on a C18 column using a linear gradient (4%–48%) of buffer B in buffer A over 22 min (buffer A = 0.1% TFA in water, buffer B = 0.08% TFA in acetonitrile). (2a) Folded L-ester insulin after prep HPLC purification(Inset: on-line ESI-MS spectra of the main peak). (2b) Folded D-ester insulin after prep HPLC purification (Inset: on-line ESI-MS spectra of the main peak). The chromatographic separations were performed on a C18 column using a linear gradient (9%–53%) of buffer B in buffer A over 22 min.
It is important to note that the ester-linked polypeptide 6 folded in very high yield at near physiological pH 7.6, in a buffer in which the cysteine reducing component of the redox couple is in considerable excess. Proinsulin itself gives very poor (<10%) folding yields under similar conditions.34 Efficient (~60-70%) in vitro folding of proinsulin34 or DKP proinsulin35 occurs under kinetically controlled conditions and requires a much more basic pH 10.5 and a substantial excess of the oxidized component of the redox couple. The inability to fold proinsulin in vitro at physiological pH has been attributed to aggregation of partially-folded or misfolded polypeptide chains; at pH 10.5 correct folding of proinsulin is faster than aggregation and so folding proceeds successfully.34 In our case, the ester insulin polypeptide folds rapidly and in high yield at near-physiological pH under redox conditions that favor the reduction and reshuffling of misfolded products without aggregation problems. A possible molecular explanation for the lack of aggregation in folding ester insulin is as follows. The structure of hydrogen-bonded aggregates calls for the A and B chains to be parallel in two-layered beta sheets in the unfolded or partially-folded molecule. As is the case for single chain insulins,36 because of the constraints imposed by the additional ester bond, the ester insulin molecule cannot adopt this unfolded conformation that is required for aggregation. This efficient folding at physiological pH is a substantial advantage of ester insulin over proinsulin for insulin production.
The folded product solution was acidified to pH ~3 and purified by preparative HPLC followed by lyophilization to afford ester insulin 1. Essentially identical synthetic results were obtained for both L-ester insulin and D-ester insulin. The overall yield starting from the first ligation was ~15%, based on limiting peptide 3. The products were characterized by LC-MS (ESI); observed mass: L-ester insulin, 5767.4 ± 0.2 Da; D-ester insulin, 5767.4 ± 0.3 Da; calculated mass, 5767.5 Da (average isotopes) (Figure 2, panel 2a-b). From a total of 193 mg of peptide starting materials, the amount of the isolated folded product obtained was 23 mg, which is a yield of 12% in terms of ‘atom economy’.10 Based on our observed HPLC yield data we estimate an overall atom economy of at least 35% could be obtained with further optimization of the reaction sequence at increased scale.
Racemic Crystallization
We set out to obtain a high resolution X-ray structure of the ester insulin molecule. Initially, crystallization trials were carried out with L-ester insulin alone using the commercially available 96-Index screen conditions obtained from Hampton Research. However, these trials were not successful and no crystals were obtained even after several months. Recently we have had considerable success in crystallizing recalcitrant proteins from racemic mixtures.37 In order to implement racemic crystallization in the present case we used the efficient convergent synthesis described above to prepare both the L-protein and D-protein enantiomers of ester insulin, and then used in the same Hampton Index screen a racemic mixture where both L- and D- ester insulin were present in equal amounts; within a week micro-crystals were obtained from several conditions. More conditions were screened with protein concentrations of 2.5 and 5 mg/mL (i.e., 1.25 mg/mL of D enantiomer + 1.25 mg/mL of L enantiomer; and, 2.5 mg/mL of D enantiomer + 2.5 mg/mL of L enantiomer, respectively) using Hampton Crystal Screen, Crystal Screen 2, and Crystal Screen Lite. We obtained micro-crystals from several conditions, all of which contained either PEG (= polyethylene glycol), MPD (= (+/−)-2-methyl-2,4-pentanediol) or Jeffamine. We focused on these conditions and optimized them using PEGRx 1 kit and Grid Screen MPD obtained from Hampton Research, as well as other optimization methods including changing the precipitant and buffer concentrations. Finally, several conditions containing PEG afforded X-ray diffraction quality crystals.
X-ray structure of Ester Insulin
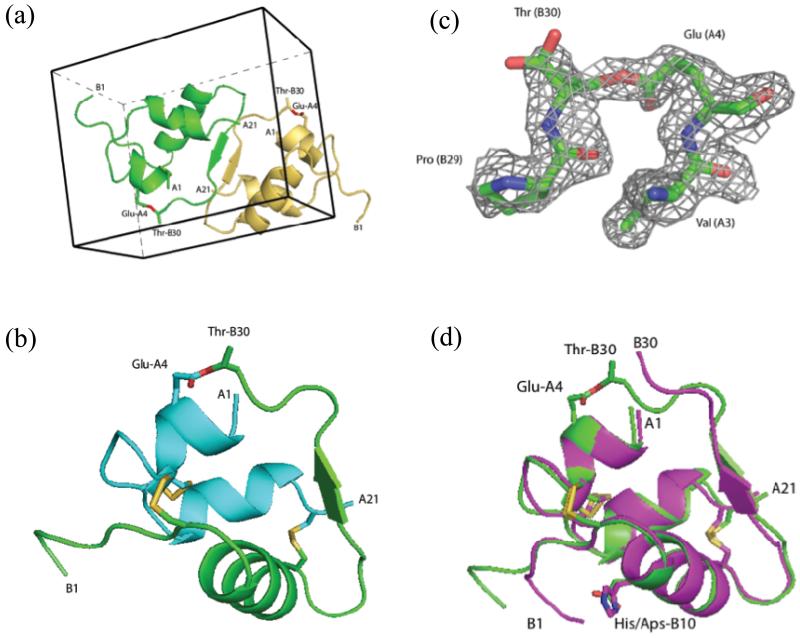
Diffraction data from four crystals were collected at the Advanced Photon Source. For structure determination, we used data acquired from a racemic crystal that diffracted to a resolution of 1.6 Å (see Experimental Section). Diffraction intensity statistics revealed that the protein racemate had crystallized in the centrosymmetric space group and had unit cell parameters of a = 20.5 Å, b = 27.8 Å, c = 34.8 Å, α = 96.4°, ß = 98.6°, γ = 104.5°. There was one enantiomer in the asymmetric unit and two molecules (one L-ester insulin, and one D-ester insulin) in the unit cell (Figure 3a). The solvent content for this crystal was relatively low at 25%. This compares with solvent contents of 30-40% reported for 3E7Y, 1XDA, 1B9E, 1QIY insulins in the PDB. The structure was determined by molecular replacement using the program MOLREP38 with a human insulin molecule from PDB code 3E7Y as a starting model. After protein model building and placement of solvent molecules, the final model was refined to crystallographic R-factor and R-free values of 22.1% and 25.0% respectively by REFMAC539 (an R-factor of 22.1% for a centrosymmetric structure corresponds to about 14% for an acentric one40).
Figure 3. X-ray structure of ester insulin (1.6 Å), obtained from racemic crystallization of D-DKP ester insulin and L-DKP ester insulin.
(a) Racemic crystal packing in centrosymmetric unit cell, showing the L-DKP ester insulin molecule (green) and the D-DKP ester insulin molecule (gold). The ester linkage is shown in sticks (O, red). (b) Cartoon representation of the A chain (cyan) and B-chain (green) of L-DKP ester insulin. The three disulfide bonds of the folded protein and the ester linkage are shown as sticks (O, red; S, gold). (c) SigmaA-weighted 2Fo-Fc electron density map, contoured at 1.5σ, showing the ester linkage between GluA4-ThrB30 (C, green; O, red; N, blue). (d) Superposition of X-ray crystal structures the T-form of KP insulin (PDB ID: 1LPH) (magenta) and DKP ester insulin (green). The three disulfide bonds and the ester linkage are shown as sticks (O, red; N, blue S, gold).
The packing of the D- and L-enantiomers of ester insulin in the unit cell is shown in Figure 3a. There are intermolecular hydrogen bonding interactions between the antiparallel ß-strands at positions PheB24-TyrB26 of the two enantiomeric molecules. As shown in Figure 3a, interactions in this region contribute to the centrosymmetric arrangement of the D- and L-enantiomers. Interestingly, this interface is an achiral antiparallel ß-sheet, in contrast to the native antiparallel ß-sheets found in L-proteins which are chiral. Features similar to this special ß-strand-type intermolecular interaction at the centrosymmetric interface between L- and D-enantiomers have been seen previously by us in a protein racemate crystal.41,42 Figure 3b and 3c show a cartoon representation of the observed X-ray structure of ester insulin monomer and the 2Fo - Fc electron density map of the ester link portion. In addition to the ß-sheet mentioned above, the structure of ester insulin features three α -helices: two relatively short helices in the A chain (IleA2-CysA7, LeuA13-CysA19), and a longer one in the B chain (SerB9-CysB19). Residues GlyB20-GlyB23 form a ß-turn. The disulfide bonds of ester insulin involve the same pairs of Cys residues as in native insulin. There are three disulfide bonds; two intermolecular disulfide bonds CysA7-CysB7, CysA20-CysB19, and one intramolecular disulfide bond CysA6-CysA11 with right-, left-, and left-hand chiral conformations, respectively.
In order to learn more about the effect of the ester bond on the structure of the insulin molecule, we compared the X-ray structure of our DKP ester insulin to the known X-ray structure of KP insulin (PDB code 1LPH).43 In both these insulin molecules the native ProB28-LysB29 sequence is inverted i.e. LysB28-ProB29; in addition, in DKP insulin the HisB10 that is involved in coordinating zinc ions found in hexamers of native insulin is replaced by AspB10. KP human insulin crystallizes as a hexamer, made up of three insulin dimers, that contains two zinc ions and three phenol molecules. In that crystalline form, the two insulin monomers in each dimer are different from each other in the N-terminus of the B-chain: in one insulin monomer (designated the ‘T-monomer’), residues SerB9-CysB19 are α-helical and residues PheB1-GlyB8 are in extended conformation, while in the other insulin monomer the α-helix is longer and includes residues GlnB4- CysB19 (designated the ‘Rf - monomer’). In contrast to KP-insulin, because DKP ester insulin does not have the His residue at the B10 position, it is a monomer and not a hexamer,44 and in any case no zinc ion (or any other divalent ion) has been used in the crystallization of DKP ester insulin. Being a monomer, DKP ester insulin has only the one conformation at the N-terminus of the B chain, in which residues SerB9-CysB19 are α-helical and residues PheB1-GlyB8 are in extended conformation. This is consistent with previous observations of the process of the native insulin hexamer’s dissociation to the biologically active monomer, where a dynamic structural change occurs at the N-terminus of the B chain giving the conformation observed in DKP ester insulin.44
Figure 3d shows the superposition of the DKP ester insulin and the T monomer of KP insulin (PDB ID: 1LPH). Superposition of these two molecules gave root mean square (r.m.s) Cα deviations of 0.8 Å for chain A and 2.6 Å for chain B. The distances between pairs of Cα atoms has been plotted as a function of residue number for DKP ester insulin versus KP insulin and can be found in Supporting Information Graph S1. The two X-ray structures are generally similar although backbone deviations were found at the unconstrained N-terminus of the B chain, and at the C-terminus of the B chain caused by the ester link. The ester bond connects the side chain of ThrB30 at the C-terminus of the B chain with the side chain of GluA4 near the N-terminus of the A chain and thus causes the C-terminal backbone of the B chain to move towards the N-terminus of the A chain. In addition, the disulfide bond between CysA6-CysA11 is left-handed in ester insulin and right-handed in KP-insulin.
Chemical Conversion of Ester Insulin into DKP Insulin
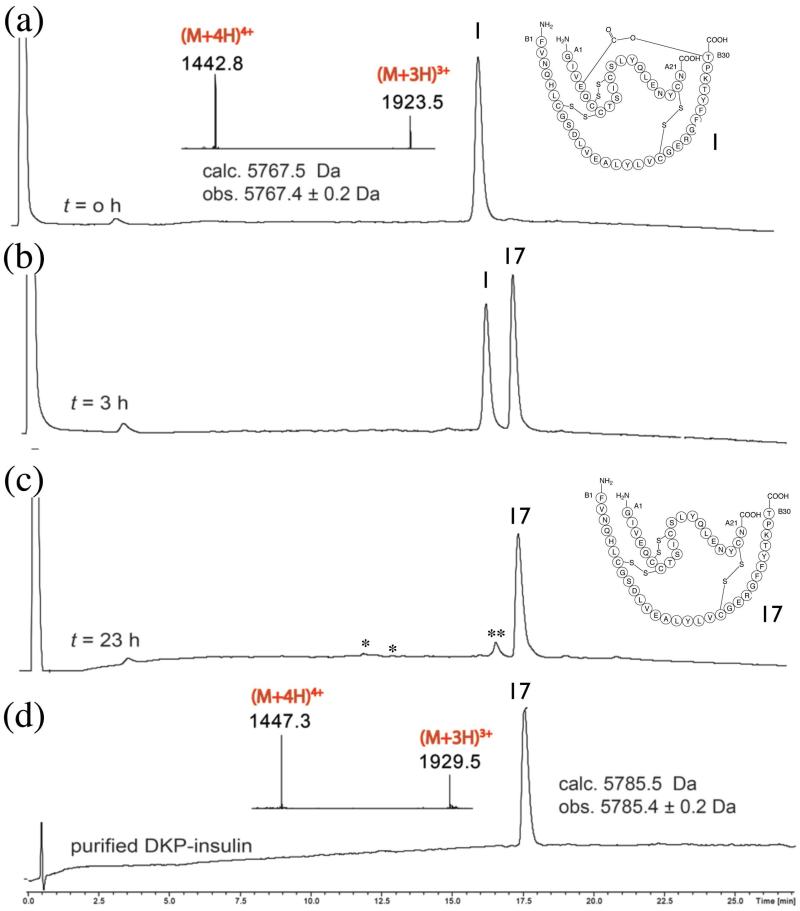
In order to confirm that the convergent synthesis of ester insulin will enable us to obtain the native biologically active insulin, we hydrolyzed the ester link between the A and B chains. Saponification of ester insulin 1 was performed under the following conditions:24 0.12 mg/mL of ester insulin 1, 25 mM of sodium hydroxide, 25 % acetonitrile (in water), at 4 °C. As shown in Figure 4, after 23 h the DKP insulin was obtained in approximately 92% yield as determined by HPLC analysis. As side products, derivatives of the A chain and the oxidized form of the B chain were observed in amounts of a few percent, probably because of disruption of disulfide bonds under the basic saponification conditions. The product DKP insulin 17 had an 18 Da higher mass than the DKP ester insulin, which confirmed the addition of the elements of water during the saponification.
Figure 4. Conversion of DKP ester insulin to native DKP insulin.
Saponification of ester insulin 1 to give DKP insulin 17: (a) Ester insulin 1 at t = o h (Inset: on line ESI-MS spectra of the main peak). (b,c) Reaction mixture at (b) t = 3 h and (c) t = 23 h. *: derived from A chain. **: oxidized B-chain. (d) Purified DKP insulin (Inset: on-line ESI-MS spectra of the main peak). The observed increase of 18 Da for the hydrolyzed DKP insulin confirms the conversion of ester insulin into DKP insulin by addition of the elements of water. The chromatographic separations were performed on a C18 column using a linear gradient (9%–53%) of buffer B in buffer A over 22 min.
Biological Activity
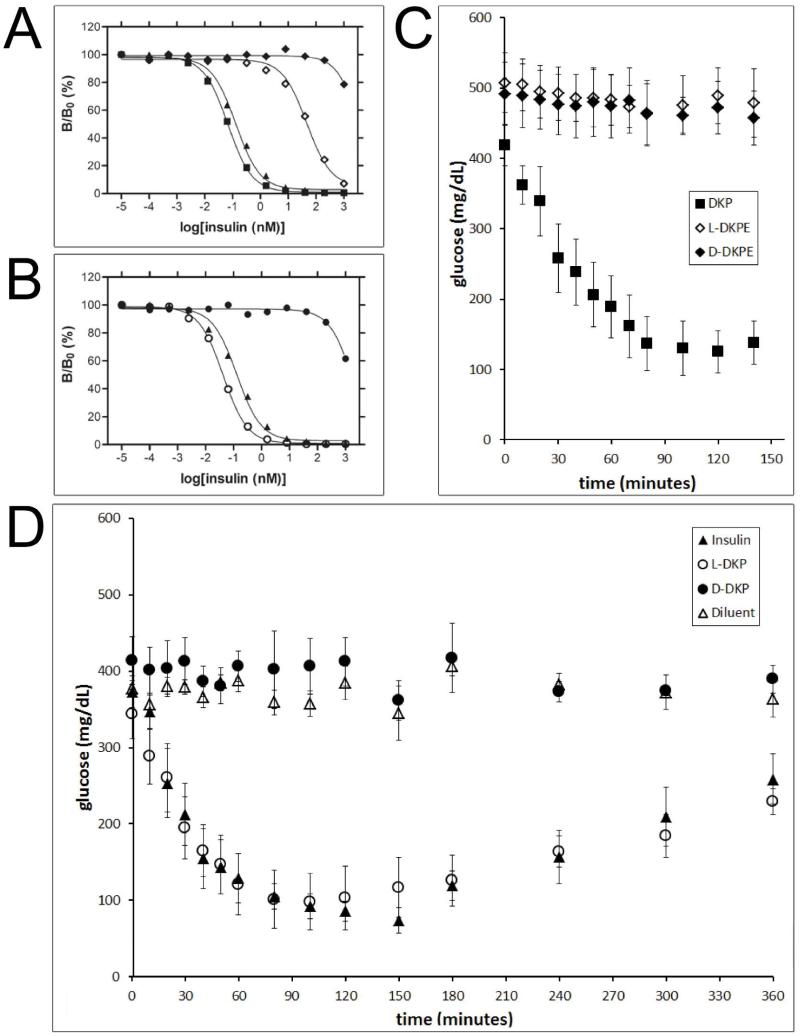
Synthetic D- and L-DKP ester insulins were characterized in receptor binding and rat blood sugar potency assays (Figure 5, panels A and C). As a control, the synthetic D- and L-DKP insulins generated by hydrolysis of the corresponding ester insulins were characterized in the same assays (Figure 5, panels B and D). In all the assays, the activities of the insulin analogues were compared with wild type (wt) biosynthetic insulin or biosynthetic DKP insulin.
Figure 5. Receptor-binding and diabetic rat assays.
(A) receptor-binding assay: biosynthetic DKP insulin (■), biosynthetic wild-type (wt) insulin (▲), synthetic L-DKP ester insulin (L-DKPE) (◇), synthetic D-DKP ester insulin (D-DKPE) (◆). (B) Receptor-binding assay: biosynthetic wt insulin (▲), synthetic L-DKP insulin (○), synthetic D-DKP insulin (●). (C) Diabetic rat assay: biosynthetic DKP insulin (■), synthetic L-DKPE (◇), synthetic D-DKPE (◆). The experiment presented in panel C was abbreviated since both ester insulins were not active. (D) Diabetic rat assay: biosynthetic wt insulin (▲), synthetic L-DKP insulin (○), synthetic D-DKP insulin (●).“Diluent” (△) in panel D means a buffer control (i.e., no protein). The same symbols have the same meanings throughout.
As shown in panel A of Figure 5, biosynthetic wt insulin and biosynthetic DKP insulin were highly active in binding to the insulin receptor. Synthetic L-DKP ester insulin had low activity comparing to synthetic L-DKP insulin (~103 less than DKP insulin), which is consistent with what was previously published for a related single-chain insulin analog.36,45 Synthetic D-DKP ester insulin had only background-level activity (~105 less than DKP insulin). As shown in panel B of Figure 5, synthetic L-DKP insulin had insulin receptor-binding activity comparable to that of biosynthetic DKP insulin, while synthetic D-DKP insulin was at least ~105 less active than L-DKP insulin in this assay. Note that in both panels A and B, L-DKP insulins (biosynthetic and synthetic) are somewhat more active than the wt insulin used as a positive control.
The insulin receptor-binding assays results are summarized in Table 1.
Table 1.
| WT insulin (biosynthetic) |
DKP insulin (biosynthetic) |
L-DKP ester insulin |
D-DKP ester insulin |
L-DKP insulin |
D-DKP insulin |
|
|---|---|---|---|---|---|---|
| Kd (nM) |
0.060 ± 0.009 | 0.029 ± 0.005 | 30.0 ± 0.3 | > ~2000 | 0.016 ± 0.003 |
>~1200 |
Assays for the reduction of blood glucose levels in diabetic rats gave results that were in agreement with the pattern of results obtained in the insulin receptor-binding assays. Active insulin causes the blood glucose concentration to fall for a few hours, then the effect wears off and the blood glucose concentration increases again to the baseline concentration (the baseline is high because the rats used are diabetic). Biosynthetic wt insulin (Figure 5, panel D), and biosynthetic DKP insulin (Figure 5, panel C) are both fully active in this assay. Synthetic DKP insulin was also fully active, being equipotent with biosynthetic wt insulin (Figure 5, panel D). Neither D-DKP ester insulin nor L-DKP ester insulin were active (Figure 5, panel C), and synthetic D-DKP insulin (Figure 5, panel D) was also inactive, in agreement with the low activities found in the receptor-binding assays (Table 1).
Structural Basis of the Lack of Biological Activity of Ester Insulin
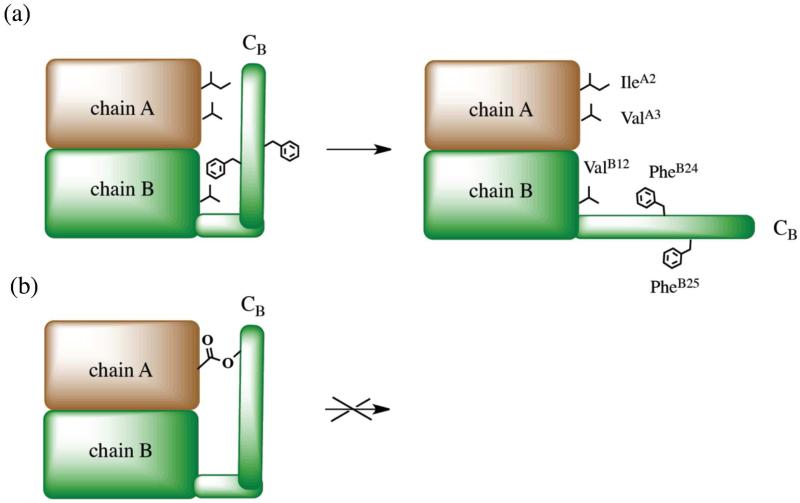
Although the X ray structure of DKP ester insulin is very similar to the structure of KP insulin, ester insulin shows greatly reduced activity in receptor-binding (Table 1) and is not biologically active (Figure 5C). This is consistent with previous studies suggesting that the C-terminus of the B-chain is flexible, enabling a conformational change in which the C-terminus of the B-chain moves away from the core of the insulin molecule exposing the residues responsible for the binding of the insulin to its receptor.15,46-48 There are several important sites participating in high affinity receptor activation, among them: residues PheB24 and PheB25, which associate with the amino- and carboxy-terminal domains of the α-subunit in the insulin receptor;49 the α-helix at GlyA1–ThrA8, which interacts with the C-terminal domain of the α-subunit of the insulin receptor;50 and residues ValB12 and TyrB16 which associate with the N-terminal region of the α-subunit of the insulin receptor.51-54 It was proposed that C-terminal B-chain movement during the receptor-binding process leads to a C-terminal ß-strand (PheB24-TyrB26) separation from the α-helix of chain B and from the N-terminus of chain A, which enables the interaction of the A chain N-terminal α-helix and residues ValB12, PheB24 and PheB25 with the receptor.15 These studies and the suggested conformational change receptor-binding mechanism are consistent with the inactivity of the ester insulin protein molecule: the ester linkage between the side chains of ThrB30 and GluA4 prevents the C-terminal region of the B chain from moving away from the insulin core to expose the high affinity binding elements within the insulin molecule (Figure 6).49
Figure 6. Conformation affects insulin binding to its receptor.
(a) Schematic illustration of conformational change of insulin upon its binding to the receptor, after Weiss et al.49 Chains A and B are schematically shown as brown and green regions, respectively. Residues IleA2, ValA3, ValB12, PheB24, and PheB25 implicated in binding to the insulin receptor are schematically shown. (b) Schematic model of ester insulin ‘locked’ in an unproductive conformation by the ester tether.
4. Summary and Conclusions
We have designed and implemented a modular, convergent synthesis of ester-insulin from three peptide segments of approximately equal size. Racemic crystallization was used for the facile production of diffraction quality crystals of ester insulin, a protein that was reluctant to crystallize as the L-protein form alone. The experimentally determined X-ray structure of DKP ester insulin is very similar to the T-form conformation in the crystal structure of KP insulin, and showed that the ester insulin contained three native disulfide bonds. Ester insulin was devoid of biological activity in a diabetic rat assay. This finding is consistent with previous studies suggesting that the binding of the insulin molecule to the insulin receptor requires a conformational change in insulin at the C-terminus of the B-chain, which is not possible for the ‘locked’ conformation of the ester insulin. Because the inactive ester insulin can be readily converted to the fully active insulin molecule by simple saponification, the convergent synthetic route to ester insulin reported here will enable the efficient generation of insulin analogues, and also promises to enable cost-effective industrial scale chemical manufacture of improved human insulins.
6. Experimental Section
Materials
Boc-amino acids (Peptide Institute, Osaka) and 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were obtained from Peptides International, Inc. Side-chain protecting groups used were Arg(Tos), Asn(Xan), Asp(OcHex), Cys(4-CH3Bzl), Glu(OcHex), His(DNP), Lys(2Cl-Z), Ser(Bzl), Thr(Bzl), Tyr(2Br-Z). Aminomethyl-resin was prepared from Biobeads S-X1 (BioRad, California).55 Boc-L-Asn-OCH2-phenylacetic acid and Boc-L-Ala-OCH2-phenylacetic acid were purchased from NeoMPS, Strasbourg France. Boc-D-Asn-OCH2-phenylacetamidomethyl resin, Boc-Glu-OFm and N-2-Chlorobenzyloxycarbonyloxysuccinimide (Z(2-Cl)-OSu) were obtained from Chem-Impex International Inc. (Wood Dale, IL). Trifluoroacetic acid (TFA) was purchased from Halocarbon Products (New Jersey). N,N-Dimethylformamide (DMF) was purchased from VWR international. HF was purchased from Matheson. All other chemical reagents were purchased from Sigma-Aldrich or Fisher Scientific.
Peptide synthesis
The L-amino acid and D-amino acid Cys-peptide segments CysA7-AsnA21 were synthesized on Boc-Asn-OCH2-Pam-resin of the appropriate chirality, using manual “in situ neutralization” Boc chemistry protocols for stepwise SPPS.32 The PheB1-ValB18 D- and L-peptide-thioester segments were synthesized on trityl-SCH2CH2CO-Ala-OCH2-Pam-resin. After removal of the Nα-Boc group, peptides were cleaved from the resin and deprotected by treatment with anhydrous HF/p-cresol at 0°C for 1 h.
LC-MS analysis and preparative HPLC
Peptide compositions were evaluated by analytical reverse phase LC-MS using a gradient of 0.08% TFA in acetonitrile (buffer B) versus 0.1% TFA in water (buffer A). Analytical HPLC was carried out as follows: self-packed C8 or C18 (3 μm, 300 Å) 2.1 × 50 mm column at 40 °C with a flow rate of 0.5 mL/min. The eluent was monitored at 210 nm, with on-line electrospray ionization mass spectrometry (ESI-MS) using an Agilent 1100 LC-ion trap. Peptides were purified on C18 (10 μm, 300 Å) silica with columns of dimension 10 × 250 mm. The silica used was TP Vydac. Crude peptides (~100 mg) were dissolved in 5% B or 6M GnHCl (at pH ~3), loaded onto the prep column and eluted at a flow rate of 5 mL per minute with a shallow gradient (e.g. 15-35 %B over 100 minutes) of increasing concentrations of solvent B in solvent A. Fractions containing the purified target peptide were identified by LC, and aliqouts of the selected fractions were combined and checked by LC-MS. Selected fractions were combined and lyophilized.
Synthesis of HCl·H-Thr-OcHex (7)
Cyclohexanol (120 mL) was cooled on ice and SOCl2 (4.9 mL, 67 mmol) was added dropwise under argon atmosphere. After the addition was complete, H-Thr-OH (4.0 g, 33.5 mmol) was added as a single portion and the reaction mixture was stirred for 2.5 h at 100°C and the reaction monitored by TLC. Upon completion, the reaction mixture was concentrated in vacuo to yield a yellow oil. This crude product was carried on to the next step without further purification. 1H NMR (500MHz, CDCl3): δ = 8.37 (br. s, 3H), 4.93-4.89 (m, 1H), 4.32-4.30 (m, 1H), 4.01-4.06 (m, 1H), 1.91-1.89 (m, 2H), 1.75-1.73 (m, 2H), 1.57-1.48 (m, 4H), 1.40-1.23 (m, 5H); 13C NMR (125 MHz, CDCl3): δ = 167.6, 75.6, 66.1, 59.5, 31.3, 31.2, 25.1, 23.5, 20.4; APCI-MS (m/z): [M(C10H20NO3)]+ calculated: 202.1; found: 202.1.
Synthesis of Alloc-Thr-OcHex (8)
To a solution of compound 7 in THF (60 mL), Et3N (12 mL) was added at 0°C. The reaction mixture was stirred for 5 min, followed by dropwise addition of a solution of Alloc-Cl (3.2 mL, 30.1 mmol) in THF (8 mL). The ice bath was removed and the reaction mixture was stirred at room temperature overnight under argon atmosphere. The reaction was monitored by TLC. After evaporation of the solvent, the crude product was dissolved in EtOAc, and the organic phase was washed sequentially with 1 N HCl (2x) and brine, dried over anhydrous MgSO4 and evaporated to give a brownish oil, which after purification by silica gel column chromatography (EtOAc, Hex) afforded 8 as a yellow oil (9.0 g, 31.5 mmol, 94% yield for both steps). 1H NMR (500MHz, CDCl3): δ = 5.98-5.91 (m, 1H), 5.66 (d, J = 8.5, 1H), 5.35 (d, J = 17.5 Hz, 1H), 5.24 (d, J = 10.5 Hz, 1H), 4.90-4.85 (m, 1H), 4.62-4.61 (m, 2H), 4.33-4.30 (m, 2H), 2.34 (d, J = 5 Hz, 1H), 1.92-1.85 (m, 2H), 1.74-1.73 (m, 2H), 1.56-1.46 (m, 3H), 1.43-1.31 (m, 3H), 1.28 (d, J = 6.5 Hz, 3H); 13C NMR (125 MHz, CDCl3): δ = 170.5, 156.6, 132.6, 117.8, 74.2, 68.2, 65.9, 59.2, 31.4, 31.3, 25.2, 23.5, 19.9 167.6, 75.6, 66.1, 59.5, 31.3, 31.2, 25.1, 23.5, 20.4; APCI-MS (m/z): [M(C14H23NO5)]+ calculated: 286.2; found: 286.1.
Synthesis of Boc-Glu(Oβ(Alloc-Thr-α-OcHex))-OFm (9)
EDC·HCl (4.5 g, 23.5 mmol) was added to a stirring solution of Alloc-Thr-OcHex (8) (3.5 g, 12.2 mmol), Boc-Glu-OFm (10 g, 23.5 mmol) and DMAP (0.17 g, 1.4 mmol) in DCM (55 mL) at 0°C. The reaction mixture was stirred at room temperature overnight under an argon atmosphere. The reaction was monitored by TLC. Upon completion, the solvent was evaporated; the crude product was dissolved in AcOEt, and washed successively with saturated NH4Cl solution (2x), saturated NaHCO3 solution (2x) and brine. The organic layer was dried over MgSO4 and the solvent was removed in vacuo. The residue was purified by silica gel column chromatography (AcOEt, Hex) to yield Boc-Glu(Oβ(Alloc-Thr-α-O-cHex))-OFm (9) as a white foamy solid (6.8 g, 9.5 mmol, 78%). 1H NMR (500MHz, CDCl3): δ = 7.77 (dd, Jmeta = 7.7 Hz, Jortho = 3 Hz, 2H), 7.62-7.59 (m, 2H), 7.43-7.39 (m, 2H), 7.35-7.31 (m, 2H), 5.96-5.89 (m, 1H), 5.75 (d, J = 9.5 Hz, 1H), 5.47-5.46 (m, 1H), 5.33 (dd, Jtrans = 17 Hz, Jgem = 1.5 Hz, 1H), 5.22 (dd, Jcis = 10.5 Hz, Jgem = 1.5 Hz, 1H), 5.06 (d, J = 8.5 Hz, 1H), 4.82-4.77 (m, 1H), 4.61-4.60 (m, 2H), 4.49-4.45 (m, 3H), 4.42-4.37 (m, 1H), 4.23 (t, J = 7 Hz, 1H), 2.35-2.22 (m, 2H), 2.11-2.05 (m, 1H), 1.90-1.73 (m, 3H), 1.72-1.65 (m, 2H), 1.51-1.40 (m, 3H), 1.46 (s, 9H), 1.39-1.25 (m, 3H), 1.33 (d, J = 6.5 Hz, 3H); 13C NMR (125 MHz, CDCl3): δ = 172.1, 171.4, 169.2, 156.6, 155.4, 143.5, 143.3, 141.3, 141.2, 132.6, 127.9, 127.2, 125.0, 120.0, 117.8, 80.0, 74.3, 70.8, 67.2, 66.0, 57.7, 52.7, 46.7, 31.5, 31.4, 30.1, 28.4, 27.4, 25.2, 23.5, 23.4, 17.0; LC-MS (ESI): [M(C38H48N2O10)+Na]+ calculated: 715.3 Da; found: 715.4 Da.
Synthesis of Boc-Glu(Oβ(Alloc-Thr-α-O-cHex))-OH (10)
To 5.3 g of Boc-Glu(Oβ(Alloc-Thr-α-OcHex))-OFm (9) (7.6 mmol), 40 mL of 20% piperidine in DCM was added and the reaction was maintained at room temperature for 3 h. The reaction was monitored by TLC. The reaction was evaporated to dryness, dissolved in EtOAc, and extracted with 1 N HCl (2x). The organic layer was dried over anhydrous MgSO4 and the solvent was removed under vacuum. The crude product was then purified by silica gel column chromatography (DCM, MeOH, AcOH) to yield Boc-Glu(Oβ(Alloc-Thr-α-OcHex))-OH (10) as a white powder (3.6 g, 6.9 mmol, 91%). 1H NMR (500MHz, CDCl3): δ = 5.99-5.89 (m, 2H), 5.45-5.44 (m, 1H), 5.33 (dd, Jtrans = 17.5 Hz, Jgem = 1 Hz, 1H), 5.22 (dd, Jcis = 10.5 Hz, Jgem = 1 Hz, 1H), 4.86-4.77 (m, 1H), 4.61-4.60 (m, 2H), 4.46 (dd, J1 = 9.5 Hz, J2 = 2 Hz, 1H), 4.33-4.29 (m, 1H), 2.45-2.36 (m, 2H), 2.17-2.10 (m, 1H), 2.00-1.94 (m, 1H), 1.86-1.77 (m, 2H), 1.72-1.66 (m, 2H), 1.54-1.41 (m, 2H), 1.45 (s, 9H), 1.39-1.26 (m, 4H), 1.31 (d, J = 6.5 Hz, 3H); 13C NMR (125 MHz, CDCl3): δ = 176.1, 171.5, 169.5, 156.7, 155.6, 132.5, 117.9, 80.4, 74.6, 71.0, 66.1, 57.7, 52.6, 31.4, 31.2, 30.3, 28.3, 27.6, 25.2, 23.5, 23.4, 17.1; APCI-MS (m/z): [M(C24H38N2O10)-H] calculated: 513.2; found: 513.0.
Synthesis of peptide (GlyA1-GluA4[OβThrB30-ThzB19]-CysA6)-thioester (3) (Scheme 3)
Protected Boc-GlnA5-CysA6-SCH2CH2CO-Ala-OCH2-Pam-resin (12) was prepared on a 0.5 mmol scale on Boc-Ala-OCH2-Pam-resin (11) via HSCH2CH2CO-Ala-OCH2-Pam-resin56 by manual Boc chemistry stepwise solid-phase peptide synthesis (SPPS) using “in situ neutralization” protocols.32 All amino acids were coupled for the standard coupling time (12 min) unless otherwise indicated. The side chain ester-linked dipeptide Boc-Glu(Oβ(Alloc-Thr-α-O-cHex))-OH 10 and Boc-ValA3-OH were sequentially coupled using this SPPS protocol but with longer coupling duration (1 h). Three additional cycles of standard SPPS yield compound 14. The Nα-Boc group was removed by treatment with TFA, and after washing with DMF and neutralization with 25% DIEA in DMF for 10 seconds, the revealed α-amino group was re-protected by reaction with N-2-chlorobenzyloxycarbonyl succinimide (Z(2-Cl)-OSu) (12 min) to give compound 15. The Nα–Alloc protecting group of ThrB30 was selectively removed using Pd(PPh3)4 (62 μmol) as catalyst in the presence of excess phenylsilane (7.3 mmol) in degassed DCM (4 mL) for 20 min. Boc-ProB29-OH was coupled to the revealed αNH2- of ThrB30 for 30 min, and the remainder of the peptide was constructed by standard stepwise SPPS and continued through ThzB19 to yield resin-bound compound 16. The Nα-deprotected peptide-resin was thoroughly washed with DMF and DCM and dried under vacuum. The peptide was then cleaved from the resin support, and side-chain protecting groups were simultaneously removed by treatment for 1 h at 0 °C with anhydrous HF containing p-cresol (90:10 v/v). After complete evaporation of the HF under reduced pressure, crude peptide products were precipitated and triturated with chilled diethyl ether, and the peptide products were dissolved in 50% aqueous acetonitrile containing 0.1% TFA and lyophilized. The crude product was purified by preparative HPLC and lyophilized to give 291 mg (0.132 mmole, 26% yield based on 0.5 mmol of Boc-Ala-OCH2-Pam-resin (11)) of peptide 3. The product was characterized by LC-MS (ESI); observed: L-(GlyA1-GluA4[OβThrB30-ThzB19]-CysA6)-thioester, 2206.8 ± 0.3 Da; D-(GlyA1-GluA4[OβThrB30-ThzB19]-CysA6)-thioester, 2206.6 ± 0.3 Da; calculated, 2206.5 Da (Figure S1).
Synthesis of the ester insulin polypeptide chain GlyA1-GluA4[OβThrB30-PheB1]-AsnA21) (6)
Using the synthetic strategy shown in Scheme 1 the C-terminal peptide segment CysA7-AsnA21 (2, 50.8 mg, 28.9 μmol, 3.6 mM) was reacted with ThzB19-[A1-GluA4(OβThrB30)-A6]-thioester (3, 59.8 mg, 27.1 μmol, 3.9 mM) under the following conditions: aqueous 6 M GnHCl, 100 mM sodium phosphate at pH 6.9, 100 mM MPAA and 20 mM TCEP·HCl for 4.5h, to give the ligation product GlyA1-GluA4[OβThrB30-ThzB19])-AsnA21 (5′) (Figure 1, panel 1). Without purification of the crude product, the N-terminal ThzB19 was converted to CysB19 by addition of methoxylamine·HCl to a concentration of 0.15 M and overnight reaction at pH 4.0 to give GlyA1-GluA4[OβThrB30-CysB19])-AsnA21 (5). Then, PheB1-ValB18-thioester (4, 82.7 mg, 37.9 μmol, 5.4 mM), MPAA (200 mM) and TCEP·HCl (50 mM) were added to the same reaction mixture, and the pH was readjusted to 6.9 (Figure 1, panel 2). Another portion of TCEP·HCl was added after 20 h and the pH was readjusted to 6.9. Within 38 h the reaction was complete and the full-length polypeptide GlyA1-GluA4[OβThrB30-PheB1]-AsnA21 (6) was recovered by precipitation from water (Figure 1, panel 2c). The resulting white precipitate was washed with water to afford crude linear polypeptide polypeptide GlyA1-GluA4[OβThrB30-PheB1]-AsnA21 (6), which was then redissolved in 50% B and lyophilized.
Folding/disulfide formation
The crude ester-linked polypeptide 6 was then folded at room temperature at a concentration of 0.3 mg/mL in a buffer solution containing 1.5 M GnHCl, 20 mM Tris, 8 mM cysteine, 1 mM cystine·HCl, pH 7.6. Completion of the folding was revealed by the near quantitative formation (after 2 h) of the earlier eluting folded product (Figure 2, panel 1). The folding buffer was then acidified with 30% (v/v) HCl (this caused the pH of the solution to drop to ~3) and the folded product was purified by preparative HPLC followed by lyophilization of selected fractions to afford 23 mg (3.99 μmol) of ester insulin 1. The yield starting from the first ligation was 15% (based on limiting peptide 3). The product was characterized by LC-MS (ESI); observed: L-ester insulin, 5767.4 ± 0.2 Da; D- ester insulin, 5767.4 ± 0.3 Da; calculated, 5767.5 Da (Figure 2, panel 2).
Protein Crystallization
Racemic crystallization of ester insulin was performed by mixing equal amounts (by weight) of purified lyophilized L-ester insulin and D-ester insulin in water at the following concentrations: 5 mg/mL (2.5 mg/mL of L- and 2.5 mg/mL D-) and 2.5 mg/mL (1.25 mg/mL of L- and 1.25 mg/mL D-). Each solution was centrifuged to remove any particulate matter present and then used directly for crystallizations. Crystallization screening was conducted at 19 °C using Index Screen (cat. No. HR2-144), Crystal Screen (cat. No. HR2-110), Crystal Screen 2 (cat. No. HR2-112), Crystal Screen Lite (cat. No. HR2-128), PEGRx 1 (cat. No. HR2-082) and Grid Screen MPD (cat. No. HR2-215) obtained from Hampton Research. Crystallization screens were performed by the hanging drop vapor diffusion method. The drops were generated by mixing 1μL of protein solution with 1μL of reservoir solution and 1μL of water and placed against 0.5 mL of reservoir solution. Crystals appeared within 10 days. X-ray diffraction data were collected from crystals grown at 2.5 mg/mL protein concentration from 0.2M sodium citrate tribasic dihydrate, 0.1M HEPES sodium, 13% (v/v) MPD, pH 7.5, or crystals grown at 5 mg/mL protein concentration from 0.05M citric acid, 38% (v/v) PEG-200, pH 3.6. Diffraction data obtained from a racemic crystal grown in the latter condition was used for structure solution.
Data Collection
For data collection, selected crystals were briefly transferred to the cryoprotectant (reservoir solution plus 20% (v/v) glycerol) and flash-frozen in liquid nitrogen. The X-ray diffraction data were collected at 100 K using 0.97921 A° wavelength at the Argonne National Laboratory (Advanced Photon Source, beamline 24IDE). Crystal diffraction images were integrated, scaled, and merged with HKL-2000.57 Data were collected to a resolution of 1.6 Å.
X-ray Structure Determination
The structure of racemic ester insulin in the space group was solved by molecular replacement using the MOLREP program38 and the model of the human insulin molecule from PDB code 3E7Y. Electron density and model examinations were done using COOT.58 The restrained positional and anisotropic B-factor refinement was performed in REFMAC5.39 The main chain torsion angles for all residues are in the allowed and additional allowed regions of the Ramachandran plot. The data collection and refinement statistics are summarized in Table S1. The coordinates and structure factors have been deposited in the Protein Data Bank with PDB ID code 4IUZ.
Receptor-Binding Assays and Rat Bioassays
Procedures for these assays are described in the Supporting Information (S4-S6).
Supplementary Material
Acknowledgements
We thank J. Whittaker and L. Whittaker for insulin receptor-binding assays; N. Strokes and F. Ismail-Beigi for rat assays; and N. Wickramasinghe and Z.-L. Wan for discussions regarding the crystal structure. This work was supported in part by grants from the National Institutes of Health (DK04049 to MAW and DK089934 to SBK) and by a pilot grant from the American Diabetes Association (NBP).Use of NE-CAT beamline 24-ID at the Advanced Photon Source is supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Supporting Information Available. Amino acid sequence of DKP insulin; synthesis of peptide segments; chemical synthesis of D-DKP ester insulin; chemical conversion of D-DKP ester insulin into D-DKP insulin; receptor-binding assay; rodent potency assay; X-ray data collection and refinement statistics for racemic DKP ester insulin PDB ID code 4IUZ; distances between Cα atoms of the (a) A chain and (b) B-chain of the DKP ester insulin and KP insulin (PDB ID: 1LPH) crystal structures. This information is available free of charge via the internet at http://pubs.acs.org
References
- 1.Sanger F. Science. 1959;129:1340. doi: 10.1126/science.129.3359.1340. [DOI] [PubMed] [Google Scholar]
- 2.Brown H, Sanger F, Kitai R. Biochem. J. 1955;60:556. doi: 10.1042/bj0600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meienhofer J, Schnabel E, Bremer H, Brinkhoff O, Zabel R, Sroka W, Klostermeyer H, Brandenburg D, Okuda T, Zahn H. Z Naturforsch. 1963;18b:1120. [PubMed] [Google Scholar]
- 4.Katsoyannis PG. Science. 1966;154:1509. doi: 10.1126/science.154.3756.1509. [DOI] [PubMed] [Google Scholar]
- 5.Kong YT, Du YC, Huang WT, et al. Sci Sin. 1966;15:544. as cited in: Zhang, Y.S. Science China 2010, 53, 16. [PubMed] [Google Scholar]
- 6.Marglin A, Merrifield RB. J. Am. Chem. Soc. 1966;88:5051. doi: 10.1021/ja00973a068. [DOI] [PubMed] [Google Scholar]
- 7.Katsoyannis PG, Tometsko AM, Zalut C. J. Am. Chem. Soc. 1967;89:4505. doi: 10.1021/ja00993a046. [DOI] [PubMed] [Google Scholar]
- 8.Sieber P, Kamber B, Hartmann A, Jöhl A, Riniker B, Rittel W. Helv. Chim. Acta. 1974;57:2617. doi: 10.1002/hlca.19740570839. [DOI] [PubMed] [Google Scholar]
- 9.Akaji K, Fujino K, Tatsumi T, Kiso Y. J. Am. Chem. Soc. 1993;115:11384. [Google Scholar]
- 10.Trost BM. Angew. Chem. Int. Ed. 1995;34:259. [Google Scholar]
- 11.Hua Q, Chu Y, Jia W, Phillips N, Wang R, Katsoyannis P, Weiss MA. J. Biol. Chem. 2002;277:43443. doi: 10.1074/jbc.M206107200. [DOI] [PubMed] [Google Scholar]
- 12.Steiner DF. Trans. N. Y. Acad. Sci. 1967;30:60. doi: 10.1111/j.2164-0947.1967.tb02452.x. [DOI] [PubMed] [Google Scholar]
- 13.Steiner DF. Curr. Opin. Chem. Biol. 1998;2:31. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 14.Cowley DJ, Mackin RB. FEBS Letters. 1997;402:124. doi: 10.1016/s0014-5793(96)01511-6. [DOI] [PubMed] [Google Scholar]
- 15.Mayer JP, Zhang F, DiMarchi RD. Biopolymers. 2007;88:687. doi: 10.1002/bip.20734. [DOI] [PubMed] [Google Scholar]
- 16.Brandenburg D, Wollmer A. Hoppe-Seyler’s Z. Physiol. Chem. 1973;354:613. doi: 10.1515/bchm2.1973.354.1.613. [DOI] [PubMed] [Google Scholar]
- 17.Wollmer A, Brandenburg D, Vogt HP, Schermutzki W. Hoppe-Seyler’s Z. Physiol. Chem. 1974;355:1471. [PubMed] [Google Scholar]
- 18.Brandenburg D, Schermutzki W, Zahn H. Hoppe-Seyler’s Z. Physiol. Chem. 1973;354:1521. [PubMed] [Google Scholar]
- 19.Geiger R, Obermeier R. Biochem. Biophys. Res. Commun. 1973;55:60. doi: 10.1016/s0006-291x(73)80059-2. [DOI] [PubMed] [Google Scholar]
- 20.Obermeier R, Geiger R. Hoppe-Seyler’s Z. Physiol. Chem. 1975;356:1631. [PubMed] [Google Scholar]
- 21.Busse WD, Carpenter FH. Biochemistry. 1976;15:1649. doi: 10.1021/bi00653a010. [DOI] [PubMed] [Google Scholar]
- 22.Rajpal G, Liu M, Zhang Y, Arvan P. Mol. Endocrinol. 2009;23:679. doi: 10.1210/me.2008-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tofteng AP, Jensen KJ, Schaffer L, Hoeg Jensen T. Chem. Bio. Chem. 2008;9:2989. doi: 10.1002/cbic.200800430. [DOI] [PubMed] [Google Scholar]
- 24.Sohma Y, Hua QX, Whittaker J, Weiss MA, Kent SBH. Angew. Chem. Int. Ed. 2010;49:5489. doi: 10.1002/anie.201001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrickson JB. J. Am. Chem. Soc. 1977;99:5439. [Google Scholar]
- 26.Bang D, Pentelute BL, Kent SBH. Angew. Chem. Int. Ed. 2006;45:3985. doi: 10.1002/anie.200600702. [DOI] [PubMed] [Google Scholar]
- 27.Pentelute BL, Gates ZP, Tereshko V, Dashnau JL, Vanderkooi JM, Kossiakoff AA, Kent SB. J. Am. Chem. Soc. 2008;130:9695. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson PE, Muir TW, Clark Lewis I, Kent SBH. Science. 1994;266:776. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 29.Kent SBH. Chem. Soc. Rev. 2009;38:338. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 30.Bang D, Kent SBH. Angew. Chem. Int. Ed. 2004;43:2534. doi: 10.1002/anie.200353540. [DOI] [PubMed] [Google Scholar]
- 31.Hua QX, Nakagawa S, Hu SQ, Jia W, Wang S, Weiss MA. J Biol Chem. 2006;281:24900. doi: 10.1074/jbc.M602691200. [DOI] [PubMed] [Google Scholar]
- 32.Schnolzer M, Alewood P, Jones A, Alewood D, Kent SBH. Int. J. Pept. Res. Ther. 1992;40:180. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 33.Luisier S, Avital-Shmilovici M, Weiss MA, Kent SBH. Chem. Commun. 2010;46:8177. doi: 10.1039/c0cc03141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter J, Lilie H, Rudolph R. Anal. Biochem. 2002;310:148. doi: 10.1016/s0003-2697(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 35.Mackin RB, Choquette MH. Protein Expression Purif. 2003;27:210. doi: 10.1016/s1046-5928(02)00643-5. [DOI] [PubMed] [Google Scholar]
- 36.Markussen J. Int. J. Pept. Protein Res. 1985;25:431. doi: 10.1111/j.1399-3011.1985.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 37.Yeates TO, Kent SBH. Annu. Rev. Biophys. 2012;41:41. doi: 10.1146/annurev-biophys-050511-102333. [DOI] [PubMed] [Google Scholar]
- 38.Vagin A, Teplyakov A. J. Appl. Crystallogr. 1997;30:1022. [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Luzzati PV. Acta. Cryst. 1952;5:802. [Google Scholar]
- 41.Mandal K, Pentelute BL, Tereshko V, Kossiakoff AA, Kent SBH. J. Am. Chem. Soc. 2009;131:1362. doi: 10.1021/ja8077973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandal K, Pentelute BL, Tereshko V, Thammavongsa V, Schneewind O, Kossiakoff AA, Kent SBH. Protein Sci. 2009;18:1146. doi: 10.1002/pro.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciszak E, Beals JM, Frank BH, Baker JC, Carter ND, Smith GD. Structure. 1995;3:615. doi: 10.1016/s0969-2126(01)00195-2. [DOI] [PubMed] [Google Scholar]
- 44.Weiss MA, Hua QX, Lynch CS, Frank BH, Shoelson SE. Biochemistry. 1991;30:7373. doi: 10.1021/bi00244a004. [DOI] [PubMed] [Google Scholar]
- 45.Hua QX, Hu SQ, Jia W, Chu YC, Burke GT, Wang SH, Wang RY, Katsoyannis PG, Weiss MA. J. Mol. Biol. 1998;277:103. doi: 10.1006/jmbi.1997.1574. [DOI] [PubMed] [Google Scholar]
- 46.Mirmira RG, Nakagawa SH, Tager HS. J. Biol. Chem. 1991;266:1428. [PubMed] [Google Scholar]
- 47.Dodson EJ, Dodson GG, Hubbard RE, Reynolds CD. Biopolymers. 1983;22:281. doi: 10.1002/bip.360220137. [DOI] [PubMed] [Google Scholar]
- 48.Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Bing X, Markussen J. J. Mol. Biol. 1991;220:425. doi: 10.1016/0022-2836(91)90022-x. [DOI] [PubMed] [Google Scholar]
- 49.Xu B, Hu SQ, Chu YC, Huang K, Nakagawa SH, Whittaker J, Katsoyannis PG, Weiss MA. Biochemistry. 2004;43:8356. doi: 10.1021/bi0497796. [DOI] [PubMed] [Google Scholar]
- 50.Wan Z, Xu B, Chu YC, Li B, Nakagawa SH, Qu Y, Hu SQ, Katsoyannis PG, Weiss MA. Biochemistry. 2004;43:16119. doi: 10.1021/bi048223f. [DOI] [PubMed] [Google Scholar]
- 51.Huang K, Xu B, Hu SQ, Chu YC, Hua QX, Qu Y, Li B, Wang S, Wang RY, Nakagawa SH, Theede AM, Whittaker J, De Meyts P, Katsoyannis PG, Weiss MA. J. Mol. Biol. 2004;341:529. doi: 10.1016/j.jmb.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz GP, Burke GT, Katsoyannis PG. Int. J. Pept. Protein Res. 1981;17:243. doi: 10.1111/j.1399-3011.1981.tb01989.x. [DOI] [PubMed] [Google Scholar]
- 53.Hu S, Burke GT, Schwartz GP, Ferderigos N, Ross JBA, Katsoyannis PG. Biochemistry. 1993;32:2631. doi: 10.1021/bi00061a022. [DOI] [PubMed] [Google Scholar]
- 54.Nakagawa SH, Tager HS, Steiner DF. Biochemistry. 2000;39:15826. doi: 10.1021/bi001802+. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell AR, Kent SBH, Engelhard M, Merrifield RB. J. Org. Chem. 1978;43:2845. [Google Scholar]
- 56.Hackeng TM, Griffin JH, Dawson PE. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10068. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otwinowski Z, Minor W. Macromol. Crystallogr., Pt A. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Acta Cryst. D. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.