Abstract
Molecular genetic diagnostic testing for mitochondrial disease has evolved continually since the first genetic basis for a clinical mitochondrial disease syndrome was identified in the late 1980s. Owing to global limitations in both knowledge and technology, few individuals, even among those with strong clinical or biochemical evidence of mitochondrial respiratory chain dysfunction, ever received a definitive molecular diagnosis prior to 2005. Clinically available genetic diagnostic testing options improved by 2006 to include sequencing and deletion analysis of an increasing number of individual nuclear genes linked to mitochondrial disease, genome-wide microarray analysis for chromosomal copy number abnormalities, and mitochondrial DNA whole genome sequence analysis. To assess the collective effect of these tests on the genetic diagnosis of suspected mitochondrial disease, we report here results from a retrospective review of the diagnostic yield in patients evaluated from 2008 to 2011 in the Mitochondrial-Genetics Diagnostic Clinic at The Children’s Hospital of Philadelphia. Among 152 patients aged 6 weeks to 81 years referred for clinical evaluation of multisystem presentations concerning for suspected mitochondrial disease, a genetic etiology was established that confirmed definite mitochondrial disease in 16.4 % and excluded primary mitochondrial disease in 9.2 %. Substantial diagnostic challenges remain owing to the clinical difficulty and frank low yield of a priori selecting individual nuclear genes to sequence based on particular symptomatic or biochemical manifestations of suspected mitochondrial disease. These findings highlight the particular utility of massively parallel nuclear exome sequencing technologies, whose benefits and limitations are explored relative to the clinical genetic diagnostic evaluation of mitochondrial disease.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-012-0174-1) contains supplementary material, which is available to authorized users.
Key Words: Next generation sequencing, massively parallel sequencing, whole exome sequencing, retrospective study, mitochondrial disease diagnosis
Introduction
Mitochondrial respiratory chain disease is an increasingly well-recognized, but notoriously heterogeneous, group of multisystemic energy deficiency disorders [1]. Its extensive heterogeneity has presented a substantial obstacle for establishing a definitive genetic diagnosis and clear pathogenic understanding in individual patients with suspected mitochondrial disease [2]. While known genetic causes of “classical” mitochondrial DNA (mtDNA)-based disease syndromes have been readily diagnosable, the overwhelming majority of patients with clinical and/or biochemical evidence of suspected mitochondrial disease have had no identifiable genetic etiology for their debilitating or lethal disease [3]. Here, we review the temporal evolution and context of molecular genetics diagnostic testing for individuals with suspected mitochondrial disease, from the recognition of the first mtDNA-based disorders in the late 1980s through to the recent advent of massively parallel sequencing technologies that can be used to diagnose essentially all known nuclear gene causes of mitochondrial disease. We also report the results of a retrospective study performed to evaluate the clinical diagnostic efficacy of traditional and emerging molecular genetic testing in the Mitochondrial-Genetics Diagnostic Clinic at The Children’s Hospital of Philadelphia over a 3-year period from 2008 to 2011, which immediately preceded the clinical advent of massively parallel, or next generation, sequencing. Indeed, these data clearly elucidate the specific diagnostic challenges in suspected mitochondrial disease that will, in many cases, be met through incorporation of whole exome, and possibly whole genome, sequencing in the clinical setting. Even at this relatively early stage in the history of these new sequencing technologies, it is evident that they will revolutionize the clinical diagnostic process for highly heterogeneous disorders, such as mitochondrial disease.
Traditional Molecular Diagnostic Approach (1988–2005)
Mitochondrial disease became an increasingly recognized clinical entity during this time period [4]. For example, in 1994 an average of 12 specialists were seen by a given patient in the University of California San Diego health system before a referral was made for their evaluation in a mitochondrial disease center, but, by 2004, this had improved to an average of 2–3 specialists before a mitochondrial disease clinic referral was considered (Robert K. Naviaux, personal communication). However, establishing a definitive diagnosis of mitochondrial disease in patients referred for this possibility proved highly complex [5]. To address this challenge, several iterations of consensus clinical diagnostic criteria were proposed by expert clinicians to categorize individuals with suspected mitochondrial disease as “Definite”, “Probable”, “Possible”, or “Unlikely”, thereby conveying diagnostic confidence levels that were heavily weighted on clinical and biochemical findings typical of known mitochondrial disease syndromes [5–7]. However, genetic diagnostic options to identify a precise etiology for a given individual who met clinical diagnostic criteria for even “Definite” mitochondrial disease were fairly limited [8].
Remarkably, the first mtDNA mutations causative of human mitochondrial disease were not identified until 1988 [9, 10], although these have since expanded rapidly to include hundreds of distinct pathogenic mutations involving all 37 mtDNA genes throughout the 1990s and 2000s [11]. Despite these gains in understanding the genetic basis of mitochondrial disease, clinical diagnostic testing options to evaluate an individual patient for all potentially pathogenic mtDNA gene mutations were limited throughout much of this period [12]. Indeed, genetic testing was limited in most clinical diagnostic cases to testing for a panel of approximately a dozen common mtDNA point mutations underlying well-recognized classic mitochondrial diseases, including mitochondrial encephalomyopathy lactic acidosis and stroke-like episodes (MELAS), myoclonic epilepsy and ragged red fibers (MERRF), and neurogenic ataxia and retinitis pigmentosa (NARP), as well as mtDNA deletions that result in Pearson syndrome, progressive external ophthalmoplegia (PEO), or Kearns–Sayre Syndrome [13]. Should such “common mtDNA mutation panel” testing be unrevealing in an individual patient with suspected mitochondrial disease, the clinically-based genetic diagnostic evaluation for a specific molecular etiology was often halted. Indeed, whole mtDNA genome analysis by polymerase chain reaction (PCR)-based Sanger sequencing was not widely available on a clinical diagnostic basis until the mid-2000s. Furthermore, even when such genetic diagnostic testing was pursued on a research basis, it was recognized that the capacity to sensitively detect low-level heteroplasmy for a given mtDNA mutation was technologically limited by the “gold standard” Sanger method, whose lower heteroplasmy detection sensitivity limit is in the range of 30–50 % mutant load [14]. Thus, it was not uncommon practice to repeat mtDNA point mutation analysis in a target tissue such as muscle should it be unrevealing in blood [2].
It has long been recognized that nuclear genes play a major role in causing mitochondrial disease, as they are estimated to cause approximately three-quarters of pediatric and at least one-third of adult mitochondrial disease [4]. However, should a nuclear gene disorder be suspected, clinically-based genetic diagnostic testing options were generally limited to tests that were rather generic in application and typically had low yield in cases of suspected mitochondrial disease. Nuclear chromosome analysis (blood karyotype) could be used to evaluate for large aneuploidies. Targeted fluorescence in situ hybridization assays could be used to investigate for a dozen identifiable microdeletion syndromes, such as velocardiofacial (DiGeorge) syndrome or, perhaps, Williams syndrome [15]. Very few individual nuclear genes were available to be sequenced in a clinical diagnostic laboratory, even should a given patient be suspected to have a nuclear gene disorder similar to what may have been reported previously in the literature in even one other case. Rather, targeted PCR amplification and Sanger sequencing-based mutation analysis only existed prior to 2005 in the clinical diagnostic setting for a handful of nuclear genes known to cause primary mitochondrial disease [4]. For example, discovery was only made in 2004 that the genetic cause of Alpers syndrome was POLG, which is now recognized to be among the most common nuclear gene causes of primary mitochondrial disease [16]. To complicate matters further, many clinical genetic diagnostic laboratories at that time only offered screening for known pathogenic mutations in specific nuclear genes, with a trend toward sequencing entire genes only emerging in the late 2000s. Research-based linkage analysis was an option that could be explored to identify a candidate gene region that might harbor a disease-causing nuclear gene mutation(s) if an individual was part of a large family in which multiple individuals appeared to have symptoms of mitochondrial disease, which, although uncommon, was the main method by which most nuclear genes causative of primary mitochondrial disease were successfully identified [3].
Current Molecular Diagnostic Approach (2006–2011)
A significant improvement in the ability to definitively diagnose maternally-inherited mitochondrial disease came with widespread clinical diagnostic availability of whole mtDNA genome sequencing [14]. Rather than limiting the evaluation to one of a dozen “common” mtDNA mutations, whole mtDNA genome sequencing permitted identification of all known and potentially novel disease-causing mutations in a single platform. Several methodologies were utilized for mtDNA genome analysis by different clinical diagnostic laboratories during this time, ranging from the gold-standard PCR/Sanger based method to surveyor-based heteroduplex analysis to chip-based array analysis [14]. These methodologies differed significantly in their ability to detect low-level heteroplasmy, and substantial clinician energy was required to assure the proper tissue was assayed and testing methodology was employed in a given case. However, should an individual with clinical manifestations concerning for mitochondrial disease and biochemical evidence of mitochondrial electron transport chain enzyme activity deficiency, oxidative phosphorylation impairment, and/or abnormal histologic findings in skeletal muscle be found to have normal mtDNA genome sequence and no evidence of mtDNA genome large deletions or duplications, then their clinician could conclude confidently they did not have a maternally-inherited mtDNA cause for their disorder [17]. Symptomatic tissues could also be studied routinely by mtDNA content analysis, where mtDNA proliferation was a nonspecific, although often seen, finding in individuals with mtDNA sequence mutations in tRNA genes. Identification of mtDNA depletion or multiple deletion(s), however, was suggestive of a primary nuclear genetic disorder due to mutations in any of a small handful of nuclear genes involved in mitochondrial nucleotide metabolism [14].
Throughout the 2000s, an increasing number of nuclear gene causes of primary mitochondrial disease were identified, tabulated at 59 genes in 2007 [2], and 79 genes [18] to more than 100 genes by 2010 [3], with some variability depending on the precise definition of primary mitochondrial disease. However, most single nuclear gene disorders were identified in only one or a few families. Thus, the prevalence and phenotypic range of any one of these individual gene disorders was not readily apparent from their initial reports. As sequencing of individual genes was introduced continually into the clinical diagnostic setting, clinicians commonly selected one or a few of these nuclear genes to test in either a collective or step-wise fashion based upon the symptomatic or biochemical specific findings in their patients, with several complex algorithms available to guide the order in which individual tests might most efficiently be performed [13]. However, this genetic testing approach was expensive, costing, on average, several thousands of dollars to sequence each gene. Gene analysis did not commonly include testing for possible gene or exon-level deletions, although this did become clinically available over time, as exemplified by a targeted “MitoMet Array” that could be used to identify a deletion in a specific gene, as when only a single pathogenic mutation was identified in a patient for a gene in which disease was known to result only in a recessive fashion [14].
Despite such challenges in the rapidly changing landscape of potential mitochondrial disease gene candidates, clinicians generally embraced clinically available nuclear gene diagnostic testing even if a given gene disorder seemed fairly uncommon, an imprecise phenotypic match, or testable only at a relatively high cost in an effort to identify the cause of very severe disorders. Yet, the ultimate failings of an individual gene by gene diagnostic approach applied across the general group of all individuals with suspected mitochondrial disease is evident upon examining the genetic prevalence of POLG. POLG mutations have been estimated to be among the leading genetic causes of mitochondrial disease, accounting, potentially, for up to 8 % of cases with a wide range of clinical presentations and age at onset [19, 20]. While clinical diagnostic testing for POLG mutations in all cases of suspected mitochondrial disease might be prudent, it is unlikely to provide the genetic diagnosis in more than 90 % of individuals with suspected mitochondrial disease, and a POLG diagnosis might very will be missed if not considered by a clinician in a given individual whose presentation is not “classic” for a known POLG disease phenotype. This problem is compounded when considering the more than 100 known nuclear gene causes of mitochondrial disease and highlights the low diagnostic yield expected when sequencing genes on an individual basis.
Retrospective Analysis of Genetic Diagnostic Yield in the Mitochondrial-Genetics Diagnostic Clinic
Study Overview and Methods
To investigate the genetic diagnostic yield from available genetic diagnostic analyses and an individualized genetic testing approach, we performed an institutional review board-approved (#11-8431) retrospective study of the diagnostic yield from all patients referred for outpatient-based evaluation of suspected mitochondrial disease from June 2008 to October 2011 in the Mitochondrial-Genetics Diagnostic Clinic at The Children’s Hospital of Philadelphia. All medical records were reviewed by a Clinical Genetic Counselor in advance of the clinic appointment, at which time medical and family histories were reviewed in detail with the family. Physical, neurologic, and dysmorphologic examinations were performed by a Clinical Geneticist (M.J.F.) on all patients. Neuroimaging studies (brain magnetic resonance imaging or spectroscopy, and/or cerebrospinal fluid studies, such as amino acids, glucose, protein, or neurotransmitter levels) were reviewed or obtained as appropriate, based on individual symptoms. Routine metabolic screening studies in blood and urine were obtained on most patients at the time of the clinic visit, including comprehensive chemistry panel, blood count, thyroid function screening, lipoprotein profile, creatinine kinase, uric acid, ammonia, plasma amino acids, blood lactate and pyruvate, plasma carnitine analysis, urine organic acids, urine amino acids, and urinalysis. Additional laboratory studies were obtained to further evaluate for specific metabolic disorders, if clinically indicated. Muscle and/or skin biopsy analyses were reviewed, when available, or obtained based on individual presentations, particularly in adult patients, for the purposes of obtaining muscle histology, immunohistochemistry, electron transport chain enzymology, mtDNA genome sequence and deletion analysis, mtDNA content analysis, and coenzyme Q10 content. Most clinical encounters ranged from 90–120 minutes in duration, including genetic counseling. Patients were re-evaluated on an annual basis if no diagnosis was evident following initial evaluation.
Clinically-based genetic diagnostic studies pursued were individualized to patient presentation. Whole mtDNA genome sequencing was obtained in muscle (if available) or, otherwise, in blood if indicated based upon individual patient presentation. Genome-wide single nucleotide polymorphism microarray was obtained to evaluate for chromosomal copy number alterations (deletions/duplications) in cases with congenital anomalies or developmental delay. Select nuclear genes were sequenced in a step-wise fashion in a Clinical Laboratory Improvement Amendments (CLIA)-approved clinical diagnostic laboratory in an individualized fashion, often over the course of multiple years. Any genetic mutations identified in research studies in which the patient might be enrolled were confirmed in a CLIA-approved clinical diagnostic laboratory.
Study Results
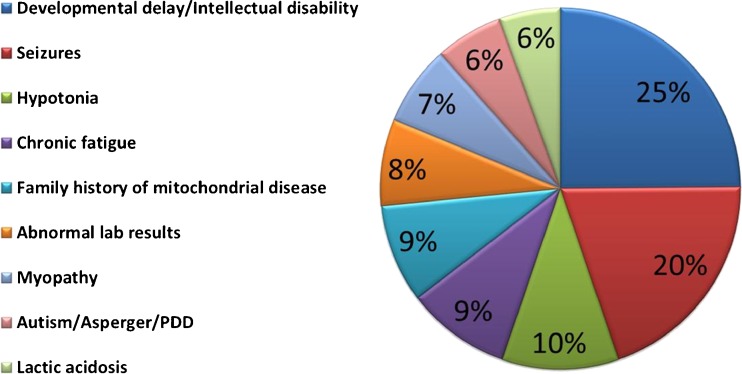
A total of 152 new patients were clinically evaluated during the 3.25-year time period studied. The leading referral indications included global developmental delay (25 %), seizures (20 %), and hypotonia (10 %), with most patients referred for multiple indications (Fig. 1), but with disease manifestations seen involving nearly every organ system. Diagnoses were grouped for this study into the traditional clinical diagnostic scheme of “Definite”, “Probable/Possible”, or “Unlikely” mitochondrial disease, or “Other non-mitochondrial primary genetic disease”, upon completion of all clinically-based diagnostic testing (Table 1). These groupings were influenced by common clinical and biochemical diagnostic criteria (i.e., modified Walker and Bernier criteria [6, 7]), but, for purposes of this study, a categorization of “Definite” mitochondrial disease was only made when a clear molecular etiology was identified that was consistent with their clinical presentation.
Fig. 1.
Leading indications for referral to Children’s Hospital of Philadelphia Mitochondrial-Genetics Diagnostic Clinic. One hundred and fifty-two participants were evaluated in the outpatient clinical setting between 2008 and 2011, with ages at presentation ranging from 6 weeks to 81 years. Most patients were referred for multiple indications, with the leading referral indications displayed that were present in > 5 % of patients
Table 1.
Current “pre-next generation sequencing” genetic diagnostic yield of individuals with suspected mitochondrial disease
| Definite primary mitochondrial disease | |
| mtDNA cytopathy | 13.2 % |
| Nuclear gene disorder | 3.2 % |
| Probable/possible mitochondrial disease | |
| mtDNA variant of unconfirmed pathologic significance | 5.3 % |
| Abnormal tissue biochemistry and no clear molecular etiology | 2 %* |
| Normal tissue biochemistry and no clear molecular etiology | 23 %* |
| Unlikely primary mitochondrial disease | 44.1 %* |
| Proven other genetic disorder | 9.2 % |
*Individuals in whom next generation sequencing-based analysis of large nuclear gene panels, the whole exome, or even the whole genome are likely to have the greatest effect in improving the diagnostic yield in the clinical evaluation of suspected mitochondrial disease
“Definite” mitochondrial disease categorization was made in 16.4 % (25/152) of patients who had clinical and/or biochemical manifestations consistent with mitochondrial disease, as well as confirmed pathogenic mutation(s) in a known disease gene, including 12.1 % (16/116) of children evaluated and 30.6 % (9/36) of adults evaluated. Among all patients evaluated, 13.2 % (20/152, including 11 children and 9 adults) had a genetically-confirmed primary mitochondrial disease due to a mtDNA cytopathy, including 19 individuals with clearly pathogenic mtDNA mutations detectable in blood (Table 2) and 1 woman with isolated chronic progressive external ophthalmoplegia who had a mtDNA deletion exclusively in muscle. In addition, nuclear gene causes of primary mitochondrial disease were confirmed in 3.2 % of patients (5/152) evaluated in the clinic, all of whom were children. The specific nuclear gene diagnoses made included RRM2B in 1 clinic patient (an infant and her deceased brother who had severe mtDNA depletion in skeletal muscle), POLG in 2 clinic patients (a boy with Leigh syndrome and epilepsy, and a presymptomatic infant whose older brother had died of intractable epilepsy and liver failure after valproate treatment), and AGC1 in 2 clinic patients (siblings with global developmental delay and epilepsy diagnosed following CLIA-certified clinical laboratory confirmation of mutations identified on a research basis by whole exome sequencing in collaboration with Dr Hakon Hakonarson in the Children’s Hospital of Philadelphia Center for Applied Genomics).
Table 2.
Mitochondrial diseases identified by mitochondrial DNA (mtDNA) whole genome sequencing. Pathogenic mtDNA mutations were identified by Sanger sequencing in blood in 18 patients from 8 kindreds. The m.12264C>T mutation in the nineteenth patient from the ninth kindred was not detectable by Sanger sequencing of the mtDNA genome in blood, but rather was detected initially in the proband’s muscle by Sanger sequencing as homoplasmic and confirmed subsequently by ARMS quantitative polymerase chain reaction to be present in 30 % mutant heteroplasmy load in the proband’s blood [34]. Bold indicates mutations that would be detected on the classical “common mtDNA mutation panel”, whereas others are only detectable by whole mtDNA genome sequencing
| mtDNA gene | mtDNA mutation | mtDNA mutation level | # Affected patients | Reference |
|---|---|---|---|---|
| tRNALEU | 3243A>G | Heteroplasmy | 3 | |
| tRNALEU | 3288A>G | Heteroplasmy | 6 | [33] |
| tRNALYS | 8344A>G | Heteroplasmy | 1 | |
| tRNASER(AGY) | 12264C>T | Heteroplasmy | 1 | [34] |
| tRNATRP | 5537_5538insT | Heteroplasmy | 2 | [35] |
| ND4 | 11778G>A / 14484T>C | Heteroplasmy/homoplasmy | 3 | [36] |
| ND6 | ||||
| ND4 | 11778G>A | Homoplasmy | 1 | |
| ND5 | 13513G>A | Heteroplasmy | 2 |
“Probable” or “Possible” primary mitochondrial disease categorization was made for 30.3 % of patients evaluated (46/152) (Table 1), as defined in this study as having clinical presentations that could be consistent with mitochondrial dysfunction, but where no definitive genetic etiology was clearly identified either because only limited genetic testing had been performed or a classical mitochondrial disease syndrome was not evident. Individuals were labeled as “Probable” mitochondrial disease if there was biochemical evidence of impaired respiratory chain oxidative phosphorylation or enzyme activity (<30 % mean), and as “Possible” mitochondrial disease if biochemical evidence of impaired respiratory chain oxidative phosphorylation or enzyme activity was either not performed or normal. However, as the goal of this study was to evaluate molecular genetics diagnostic yield, patients with “Probable” and “Possible” disease are reported here in a combined fashion. Patients in this category were subdivided based on having (1) mtDNA mutations of possible, but currently unconfirmed, pathogenesis in 5.3 % of patients (8/152) (where specific variants in question included ND2-4936C>T in 1, COXII-7962T>C in 1, ATP6-155A>T in 1, ATP8-8472C>T together with ND2-4960C>T in 2, tRNA-TYR-5836A>G in 2, and tRNA-GLN-4340A>G in 1); (2) abnormal respiratory chain polarographic and/or enzymatic studies below 20 % of the control mean owing to unclear genetic etiology in 2 % of patients (3/152); or (3) tissue biochemical studies either unrevealing or not performed, as in the case of family refusal of a muscle or liver biopsy owing to its invasive nature, in 23 % of patients (35/152). In regard to novel mtDNA mutations of possible pathogenic significance, demonstration of their pathogenicity often requires “cybrid” analysis, where a particular mtDNA variant of interest is placed in a common nuclear background to test the functional effects of that specific variant [21]. Unfortunately, cybrid-based functional analyses are not currently available on a clinical diagnostic basis in the USA, which results in potential pathogenic mtDNA variants often lingering as a possible, but unproven, diagnosis for patients. For example, the ATP6-A155T gene is predicted to have a possible pathogenic function, but respiratory chain enzyme activity analysis in the proband’s fibroblasts was unrevealing of a specific abnormality to confirm or refute its pathogenicity.
“Other non-mitochondrial primary genetic diseases” were definitively diagnosed in 9.2 % (14/152) of the overall patient cohort referred for suspected mitochondrial disease, including 10.3 % of children (12/116) and 5.6 % of adults (2/36). This category included a wide range of single gene disorders that cause other inborn errors of metabolism and/or neuromuscular diseases in 5.9 % of patients (9/152) (Table 3). In addition, a number of chromosomal abnormalities were diagnosed by high-resolution single nucleotide polymorphism microarray and/or karyotype in 3.3 % of patients (5/152), which ranged from complex chromosomal imbalances to deletion of submicroscopic chromosomal regions containing just one disease gene, as in the case of MEF2C [22].
Table 3.
Other primary genetic disorders confirmed in 14 patients who were clinically evaluated for suspected mitochondrial disease. Eight patients were found to have single gene disorders not related to primary mitochondrial disease, where the specific causative gene is shown in parentheses. Cantu syndrome was clinically diagnosed in 1 patient, with research-based testing in progress. Five patients were found to have chromosomal copy number alterations, thereby confirming nonmitochondrial primary genomic disorders as the cause for their presentation. The testing platform used for chromosomal analyses is shown in parentheses to include genome-wide single nucleotide polymorphism (SNP) microarray analysis and/or karyotype
| Single gene disorders not involving primary mitochondrial disease (9 patients) | Chromosomal copy number abnormalities (5 patients) |
|---|---|
| Molybdenum cofactor deficiency (MOCS2) | MEF2C deletion (SNP array) |
| Carnitine palmitoyl transferase 1 deficiency (CPT1) | IL1RAPL2 deletion (SNP array) |
| WFS1-related hearing loss (WFS1) | Chromosome 7q31.32q32.2 deletion of 7.91 Mb (SNP array) |
| Myotonia congenital (CLCN1) | Three-way unbalanced translocation (karyotype and SNP array) |
| Congenital myasthenic syndrome (CHRNE) | Isochromosome Xp (karyotype) |
| Rigid spine myopathy (SEPN1) | |
| Ullrich muscular dystrophy (COL6A1) | |
| Gitelman syndrome (SLC12A3) | |
| Cantu syndrome |
Perhaps the most intriguing category of patients evaluated, however, involves the patients (44.1 %; 67/152) in whom primary mitochondrial disease was thought to be “Unlikely”. This conclusion was often based on the presence of too few symptoms or severity to be suspicious of “classical” mitochondrial disease syndromes, and/or the presence of additional features such as dysmorphisms, congenital anomalies, or involvement of clinical symptoms (such as skeletal or rheumatologic problems, and/or prenatal-onset disorders) not typical of classically-defined cases of mitochondrial disease [1]. Ultimately, it is precisely this category of complex patients who might stand to benefit most from unbiased whole exome or genome sequencing approaches, where the molecular genetics findings might well inform their individual clinical disease understanding and management.
Study Conclusions
Overall, 25.6 % of patients (16.4 % “Definite” mitochondrial disease and 9.2 % “Other non-mitochondrial primary genetic disease”) who were referred for evaluation of suspected mitochondrial disease had the genetic basis for their disorder clearly identified by an individualized, albeit time-, labor-, and cost-intensive, genetic diagnostic approach. Diagnostic tests pursued were highly subjective based on individual presentations and family preferences, rather than being applied routinely in a standardized fashion to all cases with suspected mitochondrial disease. However, our clinic’s limited yield from genetic diagnostic testing in patients referred at any age and based upon any indication for evaluation of suspected mitochondrial disease are relatively consistent with reports in the literature among patients with more specific clinical or biochemical constellations of classic mitochondrial disease. In particular, we identified clearly pathogenic mtDNA mutations in 13.2 % of 152 patients, which is similar to the 11.5 % mtDNA mutation rate reported upon review of 113 pediatric patients with definite mitochondrial disease based on biochemical evidence by Scaglia et al. in 2004 [23]. Whereas we required identification of a specific molecular etiology before categorizing an individual as having “Definite” mitochondrial disease, results of prior analyses that reported higher genetic diagnostic yields may have been affected by studying populations that already met the consensus clinical diagnostic criteria for mitochondrial disease before genetic testing yield was evaluated. However, whether suspected mitochondrial disease patient cohorts have been grouped by common disease symptoms (such as Leigh syndrome or encephalopathy) [24] or common biochemical findings (such as complex I or IV deficiency) [8, 25], less than half of patients with suspected mitochondrial disease typically receive a confirmed molecular genetic diagnosis owing to the high genetic heterogeneity of mitochondrial disease, nonspecific correlation between genotype and phenotype, high cost and diagnostic odyssey of step-wise testing algorithms, and the lack of knowledge regarding all possible mitochondrial disease genes to test [3].
Emerging Molecular Diagnostic Approach (2012 and beyond…)
On 27 March 2012, the American College of Medical Genetics issued a policy statement on “Points to Consider in the Clinical Application of Genomic Sequencing” (http://www.acmg.net/StaticContent/PPG/Clinical_Application_of_Genomic_Sequencing.pdf). In this consensus statement, it was suggested that the utilization of:
… “whole genome sequencing (WGS) or whole exome sequencing (WES) should be considered in the clinical diagnostic assessment of a phenotypically affected individual when (a) The phenotype or family history data strongly implicate a genetic etiology, but the phenotype does not correspond with a specific disorder for which a genetic test targeting a specific gene is available on a clinical basis; (b) A patient presents with a defined genetic disorder that demonstrates a high degree of genetic heterogeneity, making WES or WGS analysis of multiple genes simultaneously a more practical approach; (c) A patient presents with a likely genetic disorder but specific genetic tests available for that phenotype have failed to arrive at a diagnosis; or (d) A fetus with a likely genetic disorder in which specific genetic tests, including targeted sequencing tests, available for that phenotype have failed to arrive at a diagnosis.”
The first 3 of these 4 general indications directly apply, at varying times in their diagnostic evaluation, to individuals with suspected, but unproven, nuclear gene causes of mitochondrial disease. The second criteria alone is met in nearly all cases of suspected mitochondrial disease just upon consideration of the already more than 100 known nuclear gene causes of mitochondrial disease [3], which are generally each relatively uncommon and caused by a plethora of many “private” mutations within each family, rather than a single or few common mutations within each gene (www.hgmd.org) [26]. In addition, mitochondrial diseases have been associated with all Mendelian inheritance patterns, most commonly involving autosomal recessive inheritance in the pediatric population and autosomal dominant inheritance in the adult population, but X-linked inheritance can also be seen. Furthermore, whole exome sequencing can identify not just known mitochondrial disease genes, but also mutations in a wide range of genetic disorders with overlapping clinical manifestations that may directly or indirectly cause secondary mitochondrial dysfunction. Most importantly, whole exome or genome sequencing offers a powerful opportunity to apply personalized medicine to mitochondrial disease, whereby a given individual or family can be diagnosed with the genetic basis of their specific complex presentation that may plausibly result from a mutation in either known disease genes or any of the thousands of novel genes not yet linked to human disease [27]. Many good biologic candidates exist, as up to 1500 nuclear genes are estimated to make proteins necessary for mitochondrial function, only 85 % of which have even been identified definitively as belonging to the “MitoCarta” gene set [28].
A major question currently facing the mitochondrial disease community relates to whether individuals with suspected mitochondrial disease should be tested for potential genetic causes in a simultaneous or sequential fashion. The latter might involve first sequencing the most likely individual genes that are well-known causes to explain a specific constellation of disease features, which, if unrevealing, would be followed by more expansive sequencing of large panels of “known” mitochondrial disease genes, and, ultimately, proceeding only in recalcitrant cases to whole exome (or genome) sequencing. One argument against general implementation of such a sequential approach to genetic diagnostic testing is the rapidly decreasing costs of massively parallel sequencing technologies. Indeed, the current costs are essentially equivalent when obtaining clinically-based Sanger sequencing for 2 or 3 individual genes as to obtaining simultaneous targeted enrichment with next generation sequencing (NGS) analysis of panels of 100 to 1000+ genes or even the entire 20,000+ genes that comprise the nuclear exome. Thus, cost consideration alone would support initial pursuit of the most comprehensive testing option possible.
However, another relevant factor to be considered by the clinician is test sensitivity. Targeted enrichment strategies that are used for whole exome sequencing currently do not, in actuality, generate coverage of 100 % of all known nucleotide positions in all coding exons in all known genes. Rather, depending on the specific testing platform, sequencing might best be expected to provide reliable coverage for perhaps 98 % of all coding exons in all nuclear genes. Thus, the ordering clinician needs to be cognizant that the relevant genes they may be particularly interested in having tested in a given patient may not be sequenced as comprehensively as if they were to order a more limited gene panel in which the full sequence coverage of particular genes of interest may have been optimized and guaranteed. While gene coverage is currently a relevant factor in the choice of which testing platform to pursue, this is likely to be less of a problem for whole exome sequencing based genetic diagnostic tests over time, as, while some systematic challenges—such as GC content of a given area—may remain, many areas can be optimized to improve capture of the most desired regions of relevant genes.
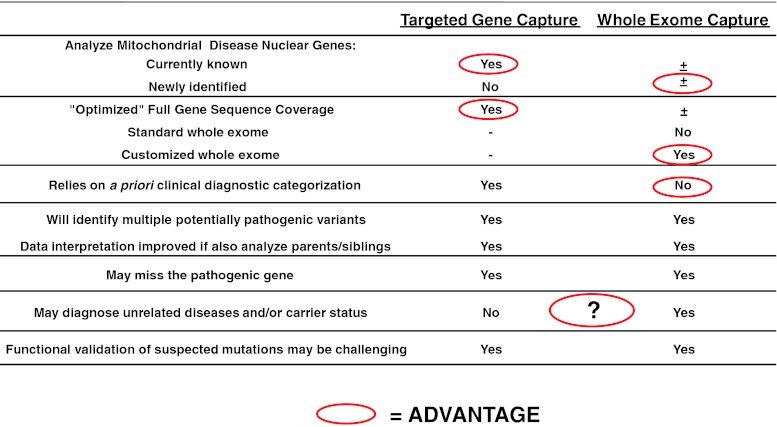
Time is yet another major consideration for clinicians to weigh in determining whether to pursue a simultaneous or sequential genetic testing approach in their patients with suspected mitochondrial disease. The diagnosis of mitochondrial disease has often entailed a prolonged “diagnostic odyssey” that massively parallel NGS-based genetic testing options stand to reduce substantially. Specifically, a comprehensive genetic diagnostic test that might reliably analyze all known mitochondrial disease genes in a simultaneous fashion in the course of weeks to months might be a reasonable first step that negates the need for invasive tissue biopsy in some cases. A visual overview of relative performance characteristics of targeted gene capture or whole exome sequencing that may aid the clinician when considering the optimal clinical diagnostic tool for their purposes is provided in Fig. 2.
Fig. 2.
Key considerations for pursuing targeted gene capture versus whole exome capture in the clinical diagnostic evaluation of suspected mitochondrial disease. Those factors that may generally be considered a particular advantage for a given approach are circled in red, where the question mark indicates this may be a relative consideration based on the context in which testing is pursued
Many questions facing the application of NGS technologies to mitochondrial disease relate to the general novelty of the technology and the new types of information that can now be learned from its use in a clinical setting. For example, while the general clinical genomics community currently remains undecided about a single optimal method for determining which genes should be analyzed and/or which mutations should be reported, the clinical diagnostic precedent already exists for reporting out suspected pathogenic mutations in genes not previously linked to human disease (https://www.bcm.edu/geneticlabs/test_detail.cfm?testcode=1500). Another possible diagnostic path that clinicians faced with the diagnostic evaluation of suspected mitochondrial disease may choose to pursue might involve a clinical diagnostic whole exome or genome-based method that initially captures all genes, but sets a bioinformatics “filter” on the sequence data to permit analysis of a prioritized subset of a few to thousands of specific genes that are postulated to be relevant to a given condition, and leaving the remaining sequence data for “second tier” analysis and/or for validation in a research setting. However, the boundaries between what is considered a clinical test versus a research study must be addressed specific to mitochondrial disease, such as whether there may be specific functional assays that will need to be commonly performed to determine whether a given disease gene and/or mutation satisfies criteria for being truly causative of a mitochondrial disease. In addition, it remains to be decided whether, and how, aggregation of the large volumes of sequence and/or variant data being generated by the NGS approaches might be shared with the broader clinical and/or research community, such that all patients might benefit from improved understanding of the frequency of individual mutations and/or disorders in individuals with suspected mitochondrial disease.
Yet another major issue facing the clinical implementation of NGS technologies is whether DNA samples should be tested only from the proband or also from their nuclear family. While it has long been clear that parental samples are needed for confirmation that identified mutations fit expected disease inheritance patterns to support their pathogenicity, the general clinical genomics community has not definitively answered whether the diagnostic processes that implement massively parallel sequencing of all, or many, nuclear genes might be made significantly more accurate and efficient by simultaneously analyzing the parental and/or sibling samples by the same NGS-based methodology as is applied to the affected individual’s sample, rather than simply using them for mutation validation purposes. This is an increasingly important consideration for whole exome sequencing technologies that may identify multiple potential disease-causing mutations whose significance would be clarified by knowing familial segregation patterns, a process that may be slow and labor intensive by classical Sanger sequencing methodologies and made much more efficient by generating all familial data up front. However, analyzing such large amounts of genetic information on the entire nuclear family may introduce new challenges of deciding the extent of genetic information to analyze and report on unaffected family members, as well as insurance-based issues of who the responsible party might be to cover family-based NGS costs. Clearly, the information explosion upon us presents previously unforeseen answers and questions alike.
The sensitivity of targeted capture and analysis of prioritized gene subsets has already begun to be analyzed in variably-defined mitochondrial disease populations. Select gene capture was investigated by PCR-amplification of 103 nuclear genes in a study of 103 patients with early-onset Leigh syndrome with biochemical evidence of complex I deficiency [24], demonstrating a diagnostic rate of 23 % (13/60) for patients where the molecular genetic etiology had not been known previously. Of the 103 nuclear genes selected for having presumed relevance to complex I deficiency, confirmed disease-causing mutations were identified in 11 known nuclear genes and 2 novel nuclear genes, NUBPL and FOXRED1. The novel genes were confirmed as disease-causing by cDNA complementation of the wild type versions of the respective disease genes in the patients’ cell lines. The same group subsequently reported the sensitivity of performing targeted capture of 1381 nuclear genes (including genes that comprise the “MitoCarta” panel of 1034 mitochondrial-localized genes and an additional 347 genes that cause inherited metabolic disease) and the mtDNA genome in 291 patients with demonstrated impairment of oxidative phosphorylation capacity [25]. The rationale for prioritizing analysis of the “MitoCarta” gene panel was based on the knowledge that 94 % of known nuclear disease genes encode proteins that are located in the mitochondria. However, this approach led to definite molecular genetic diagnosis for only 24 % (10/42) of unsolved cases, where mutations were overall identified in 77 known mitochondrial disease loci. The investigators extrapolated the potential diagnostic yield of NGS analysis of this large “MitoExome” gene set to conclude that should such analysis have been applied to all cases (both those that had a previously known and those with an unknown molecular genetic basis), a diagnosis could be established in 47 % of individuals with mitochondrial oxidative phosphorylation deficiencies. In addition, the investigators identified potential novel candidate gene mutations that would be consistent with autosomal recessive inheritance in another 20 % of cases that require research-based validation. These studies highlight the potential utility of massively parallel sequencing in the clinical diagnostic evaluation of variably defined mitochondrial disease cohorts both for nuclear genes known to cause disease, as well as for novel nuclear genes not previously linked to any known biologic function and/or human disease.
Regardless of the specific whole exome platform ultimately chosen, it is important to recognize that definitive nuclear gene mutations will not be evident in all individuals in whom mitochondrial disease will be suspected. Mutations that will be missed owing to technologic considerations include sequence variants in regions where the nuclear genome is not captured, as well as genes not prioritized either for capture and sequencing, or for bioinformatics analysis. This latter scenario occurs when certain base pairs or exons are not covered at sufficient sequencing depth to permit reliable interpretation of the nucleotide position at a given region, or when a specific laboratory test may only optimize their assay and guarantee reported sequence analysis of a specific subset of genes. A general lack of knowledge about whether specific variants truly are benign or pathogenic also contributes to difficulties with NGS data interpretation, such as whether a given mutation has a low enough population frequency that is consistent with its causing a rare disease. There are also inherent genetic characteristics that may lead to certain disease-causing mutations being systematically unrecognized by testing laboratories, which include the mutations being located in nonexonic gene regions, such as untranslated regions, intronic splice sites, or distant enhancers, for example, that may or may not be targeted by a given NGS testing platform. In addition, structural rearrangements, insertions or deletions, and trinucleotide repeat disorders are genomic alterations that are commonly missed by many enrichment technologies. Autosomal dominant disorders can be challenging to diagnose confidently if their full penetrance is not known and if parental samples are not analyzed simultaneously to permit determination of whether a given mutation is de novo or inherited. Complex inheritance of genetic disorders will also make ready diagnosis a challenge, as might be the case in the potential settings of synergistic heterozygosity in 2 or more different genes contributing to an individual patient’s disease, mutations in both nuclear and mitochondrial genomes working together to cause disease in a given patient, or epigenomic modifications underlying disease. Finally, it is important to recognize that mtDNA genome mutations will not be identified reliably unless the testing laboratory specifically includes capture and analysis of the mtDNA genome.
NGS technologies are improving the sensitivity of clinical diagnostic testing for mtDNA mutations. Indeed, NGS-based analyses are increasingly recognized to offer improved sensitivity for low-level heteroplasmy detection down to perhaps 1–10 %, depending on the specific mtDNA enrichment strategy and NGS platform used, thereby becoming the new “gold-standard”, even relative to Sanger-based sequencing methods [29–31]. Further, they provide a ready estimate of mutant heteroplasmy load at the same time as mtDNA mutation detection, where mutations detected by Sanger sequencing methods require a second molecular test, such as amplification refractory mutations system (ARMS) quantitative PCR [32], to be performed at great effort with test development for each individual mutation for purposes of determining mutation heteroplasmy load. In addition, recently developed mtDNA enrichment methods, such as long-range PCR-based amplification of mtDNA prior to NGS analysis, allows for sensitive and simultaneous low-level heteroplasmy and mtDNA deletions/duplication determination in a single assay [31]. While the possibility exists of a single clinical diagnostic NGS-based testing platform to simultaneously analyze both the nuclear exome and the mtDNA genome, this is not yet available clinically. Thus, clinicians need recognize that they must obtain appropriate mtDNA genome sequencing and/or deletion testing in addition to nuclear whole exome sequencing should mitochondrial disease be high in their differential diagnosis.
In summary, while the potential impact of NGS on the mitochondrial disease diagnostic approach cannot be overstated, significant questions regarding its optimal clinical implementation do remain. Ultimately, many of the deciding factors for these questions are likely to relate to standard issues of clinical value, such as testing cost, turn-around-time, avoidance of invasive testing, and the degree of diagnostic certainty gained through a given test’s sensitivity and specificity. Answers will surely evolve over time as technologies continue to mature and desired applications grow clearer. There are likely to be unique algorithms amenable to distinct clinical scenarios. Regardless of the way in which the diagnoses are reached, it is important to recognize that the emergence of NGS now permits a more sophisticated understanding of mitochondrial disease as a group of disorders (Fig. 3). Mitochondrial disease has long been a clinical entity raised in the setting of multi-organ dysfunction, as exemplified by the “common rule” teaching that if a patient’s unexplained disease involves 3 or more unrelated organs, one should consider mitochondrial disease [1]. However, the definition of primary mitochondrial disease has evolved to incorporate understanding that it directly involves an impaired ability to generate energy, such that tissue-based evaluation for oxidative phosphorylation capacity is the gold-standard diagnostic tool. Now, a widening ability to recognize the precise molecular genetic basis of disease permits individual mitochondrial diseases to be understood as resulting from mutations in genes in particular cellular pathways, which may or may not primarily impair the basic cellular ability to generate energy. Viewed in this light, molecular genetics knowledge must be integrated appropriately with clinical and biochemical findings to achieve a more accurate understanding of mitochondrial diseases. Thus, incorporation of NGS-based genetic testing will revolutionize not only the clinical diagnostic approach and diagnostic yield in suspected mitochondrial disease, but also the inherent ability of the mitochondrial disease community to define the spectrum of mitochondrial disease.
Fig. 3.

A more comprehensive understanding of mitochondrial diseases is obtainable by recognition of molecular genetics etiologies, as emerging next generation sequencing technologies will permit comprehensive genetic analyses to build upon and inform biochemical and clinical definitions of mitochondrial disease
Conclusion
The molecular diagnosis of suspected mitochondrial disease has evolved rapidly over the past two decades. A dedicated Mitochondrial-Genetics Diagnostic Clinic improves the diagnosis of primary mitochondrial diseases owing to a focus on recognizing “classic”, but complex, mitochondrial disease phenotypes, guiding optimal utilization and interpretation of metabolic screening laboratory studies, genetic diagnostic testing, and tissue biopsy studies, as well as identifying a wide range of phenotypically overlapping conditions. Since 2005, the molecular genetics diagnostic rate has improved for mitochondrial DNA cytopathies and for nuclear chromosomal copy number alterations detectable by genome-wide microarray technologies. While the molecular genetics diagnosis of individual nuclear gene disorders has improved modestly from the growing identification of mitochondrial disease causes, as well as small gene panel-based approaches, prioritizing individual genes for clinical diagnostic testing is excessively time- and labor-intensive for the clinician and family, as well as collectively cost- and labor-intensive at the diagnostic laboratory level—problems compounded by the shortage of specialists available to meet rising clinical demand. The emergence of massively parallel whole exome or genome sequencing technologies offers the ability to apply a more systematic and comprehensive approach to the identification of mutations in nuclear genes that affect mitochondrial function, either directly in the setting of primary mitochondrial disease or indirectly in the setting of other primary genetic disorders.
Electronic supplementary material
(PDF 907 kb)
Acknowledgments
We thank the many patients and families who contributed to this work, as well as the many clinicians who contributed to the multispecialty evaluations and care of these patients. This work was funded, in part, by a grant from the National Institutes of Health (R03-DK082446 to M.J.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The content of the article has not been influenced by the sponsors.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Elizabeth McCormick and Emily Place contributed equally to this work
References
- 1.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, et al. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 2.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, et al. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metaob. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucker EJ, Compton AG, Thorburn DR. Recent advances in the genetics of mitochondrial encephalopathies. Curr Neurol Neurosci Rep. 2010;10:277–285. doi: 10.1007/s11910-010-0112-8. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 5.Naviaux RK. Developing a systematic approach to the diagnosis and classification of mitochondrial disease. Mitochondrion. 2004;4:351–361. doi: 10.1016/j.mito.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–1411. doi: 10.1212/01.WNL.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- 7.Wolf NI, Smeitink JA. Mitochondrial disorders: a proposal for consensus diagnostic criteria in infants and children. Neurology. 2002;59:1402–1405. doi: 10.1212/01.WNL.0000031795.91814.D8. [DOI] [PubMed] [Google Scholar]
- 8.Tucker EJ, Compton AG, Calvo SE, Thorburn DR. The molecular basis of human complex I deficiency. IUBMB Life. 2011;63:669–677. doi: 10.1002/iub.495. [DOI] [PubMed] [Google Scholar]
- 9.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC, Zheng XX, Lott MT, Shoffner JM, Hodge JA, Kelley RI, et al. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 11.Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: a human mitochondrial genome database. Nucleic Acids Res. 1996;24:177–179. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiMauro S, Garone C. Historical perspective on mitochondrial medicine. Dev Disabil Res Rev. 2010;16:106–113. doi: 10.1002/ddrr.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong LJ, Scaglia F, Graham BH, Craigen WJ. Current molecular diagnostic algorithm for mitochondrial disorders. Molecular genetics and metabolism. 2010;100:111–117. doi: 10.1016/j.ymgme.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 14.Wong LJ. Molecular genetics of mitochondrial disorders. Dev Disabil Res Rev. 2010;16:154–162. doi: 10.1002/ddrr.104. [DOI] [PubMed] [Google Scholar]
- 15.Falk MJ, Robin NH. The primary care physician's approach to congenital anomalies. Primary care. 2004;31:605–619. doi: 10.1016/j.pop.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers' syndrome and mitochondrial DNA depletion. Ann Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 17.Falk MJ. MITO 101 – Genetic Counseling for Mitochondrial Disease: United Mitochondrial Disease Foundation; 2008. Available at: www.umdf.org. Accessed December 19, 2012.
- 18.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saneto RP, Naviaux RK. Polymerase gamma disease through the ages. Dev Disabil Res Rev. 2010;16:163–174. doi: 10.1002/ddrr.105. [DOI] [PubMed] [Google Scholar]
- 20.Falk MJ. Neurodevelopmental manifestations of mitochondrial disease. J Dev Behav Pediatr. 2010;31:610–621. doi: 10.1097/DBP.0b013e3181ef42c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trounce IA, Pinkert CA. Cybrid models of mtDNA disease and transmission, from cells to mice. Curr Top Dev Biol. 2007;77:157–183. doi: 10.1016/S0070-2153(06)77006-5. [DOI] [PubMed] [Google Scholar]
- 22.Zweier M, Rauch A. The MEF2C-Related and 5q14.3q15 Microdeletion Syndrome. Mol Syndromol. 2012;2:164–170. doi: 10.1159/000337496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scaglia F, Towbin JA, Craigen WJ, Belmont JW, Smith EO, Neish SR, et al. Clinical spectrum, morbidity, and mortality in 113 pediatric patients with mitochondrial disease. Pediatrics. 2004;114:925–931. doi: 10.1542/peds.2004-0718. [DOI] [PubMed] [Google Scholar]
- 24.Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet. 2010;42:851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012;Chapter 1:Unit1 13. doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Cui H, Wong LJ. Application of next generation sequencing to molecular diagnosis of inherited diseases. Top Curr Chem 2012 May 11 [Epub ahead of print]. [DOI] [PubMed]
- 28.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang S, Huang T. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. Biotechniques. 2010;48:287–296. doi: 10.2144/000113389. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Schonberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Cui H, Wong LJ. Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin Chem. 2012;58:1322–1331. doi: 10.1373/clinchem.2011.181438. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Venegas V, Li F, Wong LJ. Analysis of mitochondrial DNA point mutation heteroplasmy by ARMS quantitative PCR. Curr Protoc Hum Genet. 2011;Chapter 19:Unit 19 6. doi: 10.1002/0471142905.hg1906s68. [DOI] [PubMed] [Google Scholar]
- 33.Hadjigeorgiou GM, Kim SH, Fischbeck KH, Andreu AL, Berry GT, Bingham P, et al. A new mitochondrial DNA mutation (A3288G) in the tRNA(Leu(UUR)) gene associated with familial myopathy. J Neurol Sci. 1999;164:153–157. doi: 10.1016/S0022-510X(99)00062-3. [DOI] [PubMed] [Google Scholar]
- 34.Schrier SA, Wong LJ, Place E, Ji JQ, Pierce EA, Golden J, et al. Mitochondrial tRNA-serine (AGY) m.C12264T mutation causes severe multisystem disease with cataracts. Discov Med. 2012;13:143–150. [PMC free article] [PubMed] [Google Scholar]
- 35.Santorelli FM, Tanji K, Sano M, Shanske S, El-Shahawi M, Kranz-Eble P, et al. Maternally inherited encephalopathy associated with a single-base insertion in the mitochondrial tRNATrp gene. Ann Neurol. 1997;42:256–260. doi: 10.1002/ana.410420220. [DOI] [PubMed] [Google Scholar]
- 36.Brown MD, Allen JC, Van Stavern GP, Newman NJ, Wallace DC. Clinical, genetic, and biochemical characterization of a Leber hereditary optic neuropathy family containing both the 11778 and 14484 primary mutations. Am J Med Genet. 2001;104:331–338. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 907 kb)