Abstract
Osteoclasts are specialized bone resorbing cells that derive from monocyte precursors. We have identified three populations of cells with high osteoclastogenic potential in murine bone marrow, which expressed the phenotype: B220−CD3−CD11b−/low CD115+ and either CD117hi, CD117intermediate or CD117low. We have evaluated these populations for their ability to also generate macrophages and dendritic cells. At a single cell level, the population expressing higher CD117 levels was able to generate bone-resorbing osteoclasts, phagocytic macrophages and antigen-presenting dendritic cells in vitro with efficiencies of over 90 percent, indicating that there exists a common developmental pathway for these cell types.
Cells with osteoclastogenic potential also exist in blood and peripheral hematopoietic organs. Their functional meaning and/or their relationship with bone marrow progenitors is not well established. Hence, we characterized murine peripheral cell populations for their ability to form osteoclasts, macrophages and dendritic cells in vitro. The spleen and peripheral blood monocyte progenitors share phenotypic markers with bone marrow progenitors, but differ in their expression of CD11b, which was low in bone marrow but high in periphery.
We propose that circulating monocyte progenitors are derived from a common bone marrow osteoclasts/macrophage/dendritic cell progenitor (OcMDC), which we have now characterized at a clonal level. However, the lineage relationship between the bone marrow and peripheral monocyte progenitors has yet to be defined.
INTRODUCTION
Monocytes are a heterogeneous population of mononuclear cells that develop in the bone marrow from hematopoietic myeloid progenitors (1–3). Once developed, they can differentiate in the bone marrow and/or migrate via the bloodstream to peripheral organs where they further mature to maintain a pool of tissue-associated macrophages and dendritic cells (DCs), both of which play crucial roles in the maintenance of tissue homeostasis and immune functions (4–6). The concept of a common progenitor for macrophages and DCs was initially proposed after taking into consideration the shared functional characteristics between these cell types. Macrophages are tissue scavenging phagocytic cells that internalize cell debris and bacteria. They are also potent inducers of inflammatory mediators. DCs, although phenotypically and morphologically distinct, are also phagocytic cells with the ability to induce inflammatory responses and they are the quintessential professional antigen presenting cells that are crucial for the initiation of the adaptive immune responses.
Recently, several publications have identified and characterized a bone marrow common progenitor cell for these two functionally related cell types. Using flow cytometry combined with in vitro and in vivo differentiation assays, it was reported that cells with the phenotype Lin-CD115+ CD135+ C×3CR1+ correspond to an isolatable progenitor cell with the ability to generate macrophages and DCs (2, 5, 7).
Osteoclasts (OCs) are unique bone-resorbing multinuclear cells, which are critically involved in skeletal function such as bone remodeling, fracture repair and pathological bone resorption associated with inflammatory conditions (8). Osteoclasts are derived from myeloid progenitors through signals mediated by c-fms/CD115 and RANK (9, 10). The dependence of osteoclastogenesis on signaling through c-fms/CD115 indicates that they also belong to the monocyte lineage. In addition, because both cytokine pathways are important for DC development, a common origin of osteoclasts and DCs has been proposed (9). Miyamoto et al isolated osteoclast precursor cells bearing the phenotype CD117+ CD115+ RANK− from murine bone marrow and tested their ability to differentiate into osteoclasts and dendritic cells in vitro (11). They showed that DCs and osteoclasts could share a common progenitor. Given these findings, it is likely, that a common monocyte progenitor exists for macrophages, DCs, and osteoclasts.
Searching for osteoclast-committed progenitors in the bone marrow we identified a population of cells with high osteoclastogenic potential (12). This population is characterized by the phenotype: B220−CD3−CD11blow CD115+ CD117+, accounts for 0.1 - 0.3% of total nucleated cells, and has the ability to generate mature bone-resorbing osteoclasts when cultured in the presence of M-CSF and RANKL. In the present work, we evaluated this population for its ability to also generate macrophages and DCs, both at a population and at a clonal level. We report that at a clonal level, this population is able to generate mature bone-resorbing osteoclasts, phagocytic macrophages and antigen-presenting dendritic cells in vitro, indicating that there exists a common developmental pathway for all these cell types.
In addition to bone marrow, cells with osteoclastogenic potential exist in blood and peripheral hematopoietic organs (13). The functional meaning of these populations and/or their relationship with osteoclastogenic bone marrow progenitors is not well established. It has been proposed that these cells correspond to peripheral progenitors that are able to progress into multiple terminally differentiated monocyte cells, and that under pathological conditions, they could contribute to osteoclastogenesis with a direct impact on extra medullary osteolysis. Because several research groups have revealed a considerable heterogeneity within monocyte populations in the periphery and the association of cells within the various types of monocytic cells is not well defined (1), we thought it important to characterize any preferential populations with osteoclastogenic potential, and determine if these could also generate other types or terminally differentiated monocyte lineage cells. Using similar approaches to the ones utilized for the characterization and isolation of bone marrow osteoclast progenitors, we have identified populations with high osteoclastogenic potential in peripheral blood and spleen. As observed in the bone marrow populations, these peripheral populations contain common progenitors for osteoclast, macrophages and dendritic cells. These progenitors share phenotypic markers with the bone marrow progenitor, but differ in their expression of CD11b. Both spleen and peripheral blood monocyte progenitors share the phenotype: B220− CD3− NK1.1− CD11b+ Ly-6Chi CD115+ CCR2hi C×3CR1+.
MATERIALS AND METHODS
Mice
Eight weeks old C57BL/6 male mice were purchased from Charles River Laboratories (Wilmington, MA). All animals were housed in the Center for Comparative Medicine at the University of Connecticut Health Center and kept in sterile conventional or Thoren microisolators and given water and rodent chow ad libitum. All protocols involving animal usage were approved by the Animal Care Committee of the University of Connecticut Health Center.
Antibodies and Flow Cytometry
All of the antibodies used for the phenotypic analyses and sorting of mouse hematopoietic progenitors from bone marrow, spleen and peripheral blood are commercially available. These include: anti-B cell lineage antibody (mAb): anti-CD45R/B220 (RA3-6B2); anti-T cell lineage mAb: anti-CD3 (145-2C11); anti-NK cell mAb: NK1.1 (PK136); anti-monocyte/macrophage antibodies: anti-CD11b/Mac-1 (M1/70), anti-F4/80 (BM8); anti-dendritic cell mAb: anti-CD11c (N418); anti-granulocyte mAb: anti-Ly-6G/Gr-1 (RB6–8C5); anti-progenitor cell antibodies: anti-Ly-6A/E Sca-1 (E13.161.7), anti-ckit/CD117 (2B8), anti-c-fms/CD115 (AFS98), anti-CD135/Flt-3 (A2F10); anti-MHC and co-stimulatory molecules: anti-MHCI/H-2Kb (AF6–88.5), anti-MHCII/I-A/I-E (M5/114.15.2), anti-CD86/B7-2 (GL1), anti-CD80/B7-1 (16-10A1), anti-CD40 (1C10); anti-chemokine receptor antibodies: anti-CCR2 (475301), anti-CCR5/CD195 (C34–3448), anti-CCR7/CD197 (4B12), anti-adhesion molecules mAb: anti-CD62L/L-selectin (MEL-14), anti Ly6C (AL-21), anti Ly6G (1A8).
All these antibodies were obtained directly conjugated to fluorochromes or biotinylated from e-Biosciences (San Diego, CA, USA), BioLegend (San Diego, CA, USA), Pharmingen/BD Bioscience (San Diego, CA, USA) or R&D System (Minneapolis, MN, USA). Labeling of cells for flow cytometric analysis or cell sorting was performed by standard staining procedures wherein directly conjugated or biotinylated mAb plus second step reagents were sequentially added to the cell preparation of interest. All antibodies were titrated for optimal dilutions. All stainings were done on ice and dead cells were identified and excluded by their ability to incorporate propidium iodide (PI). Flow cytometry analysis and sorting was performed in a BD-FACS Aria (BD Biosciences. San Jose, CA, USA) equipped with five lasers and 18 fluorescence detectors. All the data was analyzed using FlowJo software from Tree Star Inc (Ashland, OR, USA).
Isolation of Hematopoietic Progenitors
Bone marrow cells were harvested for single cell suspensions by flushing femurs and tibias with 10ml of staining medium (1× Hank’s balanced salt solution (HBSS); 10 mM HEPES; 2% newborn calf serum) into 15mL tubes. Cells were spun down (1500rpm for 5 mins at 4°C), supernatant removed and the pellet containing leukocytes and red blood cells (RBC) was resuspended in 1 ml of red blood cell lysing buffer (Sigma, St Louis, MO), incubated for 5 minutes and washed with staining medium. The final pellet was resuspended in 10ml of staining medium and filtered through a 100 μm Nytex mesh. Live cells were counted in a haemocytometer using trypan blue exclusion.
Spleens were removed from mice and gently smashed between a pair of frosted microscope slides in 10mL of staining media on a Petri dish. Cells were then transferred to a 15 ml tube, spun down, supernatant removed and the pellet lysed and washed, then spun down again and resuspended in staining medium and counted, as indicated above.
Peripheral blood was collected from the tail vein of mice in a 5ml tube, containing 500μl of5mM EDTA in PBS. In order to separate peripheral blood mononuclear cells, two ml of 2% dextran was next added and incubated at 37°C for 30–45 minutes. The cloudy upper phase was transferred to a 5 ml tube and centrifuged (1500rpm for 5 min at 4°C), the supernatant removed and the pellet was resuspended in 1 ml of RBC lysing buffer and processed as described above. For analysis and sorting of hematopoietic progenitors, cells were stained with a mix of antibodies in staining media and incubated on ice for 45 minutes, followed by washing in staining medium. As a second step, cells were stained with streptavidin coupled to the fluorochromes of interest on ice for 45 minutes. Finally, cells were resuspended in staining medium containing 1μg/ml of PI. For FACS analysis of cultured cells, media was removed and washed 2× with PBS and incubated for 15 minutes at 37°C in 0.05% Trypsin in 0.53mM EDTA (Invitrogen) to dissociate the cells from the culture plates. Cells were then transferred to a 1.5mL tube and centrifuge (1500rpm for 5 mins at 4°C) and stained with antibodies as described.
Single cell sorting
Single cell sorting of isolated myeloid progenitor populations was performed using an Automated Cell Deposition Unit (ACDU) installed in the BD-FACS Aria cell sorter. Cells were plated at one cell per well into 96 well tissue culture plates. Single cells were cultured in conditions allowing their differentiation to macrophages, dendritic cells and osteoclasts, as described below.
Osteoclast progenitor cultures and quantification
Sorted OCP cells were spun down and resuspended in MEMα (GIBCO BRL, Carlsbad, CA) with 10% fetal bovine serum (FBS), penicillin, streptomycin, recombinant mouse M-CSF (R&D Systems, Minneapolis, MN) and RANKL, both at 30ng/mL. The cells were plated in five replicates per mouse in 96 well plates and incubated at 37°C with 5% CO2. Fresh medium was changed every 48 hours and osteoclast formation was evaluated after 5 to 7 days in culture. For single cell cultures, individual cells were first expanded with M-CSF (30ng/ml) for 18 days then media was removed and changed to media containing M-CSF and RANKL (both at 30ng/ml). Osteoclast formation was evaluated after an additional 5 to 7 days in culture. To visualize and quantify osteoclastogenesis, cells were fixed with 2.5% glutaraldehyde in PBS for 30 minutes and stained for TRAcP using the Leukocyte Acid Phosphatase kit (Sigma. St. Louis, MO) according to the manufacturer’s instructions. Osteoclast-like cells (OCLs) were identified as TRAcP+ cells with more than 3 nuclei.
Bone resorption assay
Cells (bulk or single sorted) were deposited onto bovine cortical bone slices in 96 well plates containing MEMα (GIBCO BRL) with 10% fetal bovine serum (FBS), penicillin, streptomycin, recombinant mouse M-CSF (R&D systems) and RANKL (both at 30ng/mL) for 12 days. As mentioned above, for single sorted cells, individual cells were first expanded with M-CSF followed by M-CSF and RANKL (both at 30ng/mL) and osteoclast formation was evaluated after 5 to 7 days. The bone slices were fixed with 2.5% glutaraldehyde in PBS, stained for TRAcP (as previously described) and visualized by microscopy to assure that osteoclasts had formed on the bone slices. To visualize resorption pits, bones were sonicated in 0.25M NH4OH solution (2× ~30sec to 1 min) to remove osteoclasts and the resorption pits were then stained with a solution of 1% toluidine blue (Sigma) in 1% sodium borate (Sigma) for 0.5 to 1 minute and washed with dH2O. To evaluate the ability of osteoclasts to resorb bone, we measured the area of individual pits for each group using the Via-160 video image measurement system (Boeckeler Instruments, Tucson, AZ).
Phagocytosis assay
Single cells were cultured for 14 days with M-CSF (30 ng/ml) and tested for their ability to phagocytose pHrodo E. coli bacterial particles (Invitrogen. Carlsbad, CA). These particles contain rhodamine, which is colorless at neutral pH and activated at the low pH inside vesicles of phagocytic cells. Cells derived from bone marrow progenitors or control RAW 264.7 cells were pre-treated for 4 hr at 37°C with DMSO (10μM) or Cytochalasin D (10μM) then washed 2× with PBS and cultured with Rodo E. coli bacterial particles for 1 hr at 37°C in media containing MEMα 10%FBS and pen/strep. Wells containing cells were washed 2× with PBS and phagocytosed particles were visualized by fluorescent microscopy, using a Zeiss Imager Z1 microscope (Carl Zeiss, Thornwood, NY) and detected using a TRITC (red) filter (Chroma Technology, Bellows Falls, VT).
Antigen presentation assay
Single cells were cultured for 8 days with GM-CSF (R&D Systems, Minneapolis, MN) and then media changed to media containing GM-CSF and IL-4 (R&D Systems, Minneapolis, MN) for an additional 4 to 6 days. All cytokines were used at a concentration of 30 ng/ml. Cells were then tested for their ability to process and present antigen to T cells, leading to their activation. BM-PIV induced DCs (20,000 cells) or DC2.4 (100,000 control DC line) cells were co-cultured with B3Z T cells. This T cell line recognizes the ovalbumin peptide SIINFEKL in the context of H-2Kb and expresses β-galactosidase upon activation by cognate antigen presenting cells (14). Ovalbumin (OVA, 300μg/mL) or SIINFEKL peptide (SHL8, 300μg/mL) was added to the cultures and incubated for 24 hr. Cells were then incubated with CPRG for 12 hours and supernatants were tested for B-galactosidase activity, through a colorimetric assay.
Statistical analysis
All data was analyzed using GraphPad Prism 5 software using unpaired Student’s t test.
RESULTS
Identification of a clonal population of bone marrow osteoclast progenitors
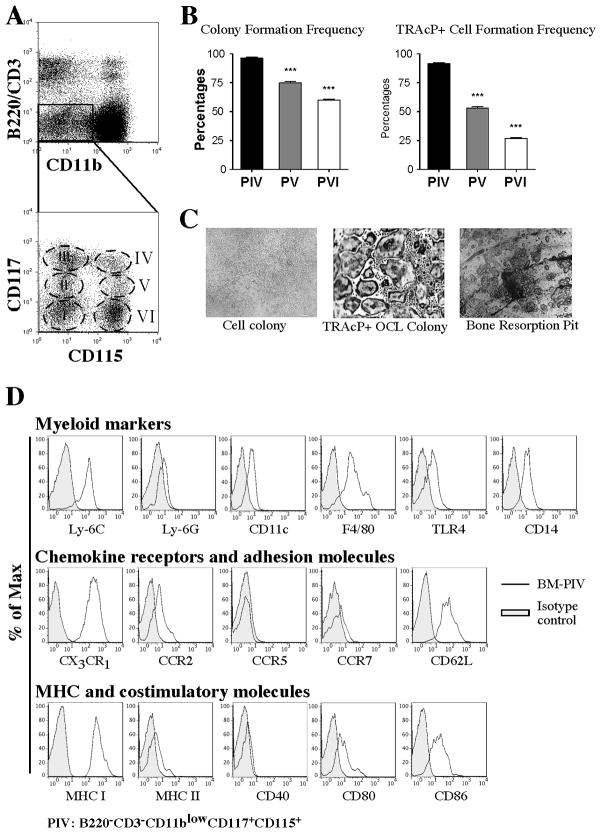
Our previous work found that osteoclast progenitor cell activity in the bone marrow was contained in three discrete populations (populations IV, V, and VI; Figures 1A) (12). Of these, population IV, bearing the phenotype B220− CD3− CD11b−/low CD115+ CD117+, contains the highest in vitro osteoclastogenic potential. As a progression from those studies, we evaluated the clonal ability of each of these populations to generate osteoclasts in vitro.
Figure 1. Single cell cloning and phenotypic analysis of bone marrow osteoclast progenitors.
A) Flow cytometry plots showing the dissection of bone marrow populations containing osteoclast progenitor activity. Population IV (B220−CD3−CD11b−/lowCD115+CD117+) contains the highest potential to generate osteoclasts in vitro. B) Evaluation of colony formation and osteoclast formation efficiency of populations IV, V and VI after single cell cloning by FACS. PIV contains the highest cloning efficiency (PIV=96.3 ± 0.9 versus PV=74.8 ± 1.3, p=0.0001, and PIV versus PVI=60±0.8, p=0.0001) and TRAcP+ cell formation efficiency (PIV=91.5 ± 0.7 versus PV=53.3 ± 1.2, p=0.0001, and PIV versus PVI=26.8 ± 1.0, p=0.0001) when compared to PV and PVI. Data represent the mean±SEM of 4 independent experiments. C) Pictures showing colonies of PIV from single cells after expansion with M-CSF for 14 days (left panel) and subsequently cultured with RANKL and M-CSF for 5 days to generate TRAcP+ osteoclasts (middle). Right panel shows bone resorption pits generated by osteoclasts generated from PIV after single cell cloning. D) Flow cytometric analysis of freshly isolated bone marrow PIV showing the expression of surface markers (histograms) common to monocyte populations.
We individually sorted each one of the three populations and subsequently plated them as a single cell per well in 96 well plates, using the single cell deposition unit available in our cell-sorting instrument. Our initial sorts reached over 95% of purity. This efficiency coupled with the subsequent cloning (Supplemental Figure 1A), which was equivalent to a second sort, ensured 100 % purity. After plating, single cells were expanded for 18 days in M-CSF (30 ng/ml) and the growth efficiency was evaluated by visual observation of expanded cells in each well. Subsequently, we replaced the media with one containing M-CSF and RANKL (both at 30 ng/ml) and cultured the expanded cells for an additional 5 to 7 days to stimulate the formation of multinucleated TRAcP+ osteoclasts.
To evaluate cloning efficiency, we quantified the percent of wells generating colonies (more than 5 cells per well) at the end of the experiment. This analysis rendered cloning efficiencies of 97%, 77% and 62% for populations IV, V, and VI respectively (Figure 1B).
More significantly, we found that the TRAcP+ cell formation efficiency of the three populations (defined as the number of colonies with TRAcP+ cells at the end of the experiment divided by the total number of plated wells) was equivalent to the cloning efficiency of the populations. For population IV, 92% of the clones formed TRAcP+ cells, while for population V and population VI these values were 55% and 28%, respectively (Figure 1B). Importantly, when single cells were sorted onto bovine cortical bone slices, and induced towards osteoclastogenesis as described, cells from population IV were able to form mature bone-resorbing osteoclasts as assessed by TRAcP staining and toluidine blue stained-pit formation (Figure 1C).
In summary, these data showed that population IV is a relatively homogeneous for osteoclast progenitors with the ability to generate bona fide osteoclasts at a clonal level. Interestingly, as depicted in Figure 1D, the general phenotypic characteristics of population IV match the phenotype of a clonogenic precursor population of macrophages and DCs that was described by Fogg et al. (15). Thus, using similar strategies, we investigated if population IV also has the ability to generate cells with characteristics of macrophages and DCs.
Differentiation of bone marrow osteoclast progenitors into functional macrophage- and dendritic- like cells in vitro
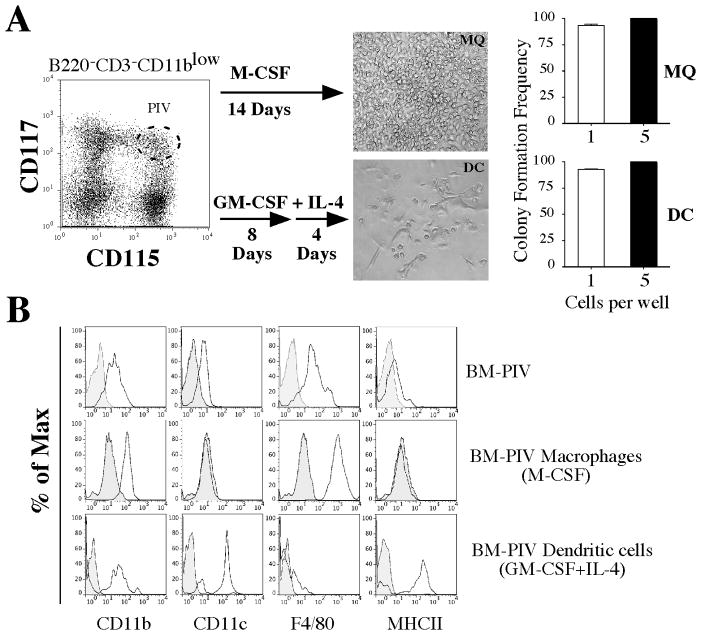
To test if population IV could correspond to a common progenitor for osteoclasts, macrophages and dendritic cells, we cultured single cells in 96 well plates in the presence of cytokines that favored the differentiation of monocyte progenitors into either macrophage- or DC-like cells. For macrophage cultures, single cells were cultured for 14 days with M-CSF (30 ng/ml). For dendritic cell cultures, single cells were initially expanded in media supplemented with GM-CSF (30 ng/ml) for 8 days, followed by media containing GM-CSF plus IL-4 (both at 30 ng/ml) for 4 additional days. For both conditions, we calculated the cloning efficiency, evaluated morphological characteristics and the defined cell surface phenotypes of the cells at the conclusion of the culture. Phagocytic properties were tested for cells that were cultured to become macrophages while cells that were cultured to become dendritic cells were tested for their ability to present antigen.
When cultures were initiated as single cells, the cloning efficiency of population IV to generate either macrophages or dendritic cells in vitro was 94% and 100% when cultures were plated at 5 cells/well. Characteristic morphological features observed for single colonies are shown in Figure 2A. Phenotypic analysis of the cultures at the conclusion of the experiment showed the upregulation of mature monocyte markers. In addition, each condition resulted in the differential expression of a number of lineage specific markers. For example, cultures progressing towards macrophages showed preferential expression of CD11b and F4–80, while cultures progressing under DC polarizing conditions showed preferential expression of CD11c and MHC class II (Figure 2B).
Figure 2. Evidence of a common bone marrow monocyte progenitor for osteoclast, macrophages and dendritic cells in vitro.
A) Single cell cloning of BM-PIV cultured with M-CSF for 14 days or with GM-CSF for 8 days and IL-4 for 4 additional days in 96 U-bottom well plates. Microphotographs represent cells derived from a single colony of macrophage-like cells (top) or dendritic-like cells (bottom). Bar graphs on right indicate colony formation efficiency of single cells (93.5 ± 1.92) cultured with M-CSF (top) and single cells (93.5 ± 1.92) cultured with GM-CSF and IL-4 (bottom). Plating 5 cells per well resulted in a 100% cloning efficiency and no statistically significant differences were found. Data represent the average ± SD of 4 independent experiments. B) Comparative phenotypic analysis of PIV with cells derived from cultures under macrophage inducing (M-CSF) and DC inducing (GM-CSF+IL-4) conditions. Macrophage-like cells expressed the phenotype CD11b+ CD11c− F4.80+ MHC II−, while DC-like cells expressed the phenotype CD11b+ CD11c+ F4.80− MHC II+.
We also isolated bone marrow population IV cells from C×3CR1 GFP transgenic mice, a reporter system in which GFP (green fluorescent protein) was inserted into the Cx3cr1 locus by homologous recombination. Results from these experiments showed that over 99.5% of these cells expresses C×3CR1 (data not shown), and the expression of C×3CR1 was maintained in cells after they differentiated into either macrophage-like or dendritic-like cells.
To test if the monocyte common progenitor generated functionally active macrophages and dendritic cells, we evaluated phagocytosis or antigen presentation, respectively (Figure 3). For phagocytosis assays, we used commercially available bacterial particles conjugated with rhodamine, a fluorochrome that fluoresces at low pH (i.e. inside macrophage phagocytic vesicles) but not at neutral pH. We incubated in vitro-differentiated macrophages, derived from single cell sorts from bone marrow population IV, with these particles and found that cells that were differentiated towards macrophages were able to ingest bacteria particles and activate the red fluorochrome at low pH inside vesicles. In contrast, cells treated with cytochalasin D, which interferes with microtubule polymerization and prevents phagocytosis, showed reduced phagocytic activity (Figure 3A). RAW 264.7 cells, used as positive control, produced equivalent results.
Figure 3. Bone marrow-derived monocyte progenitors generate functional macrophage-and dendritic-like cells in vitro.
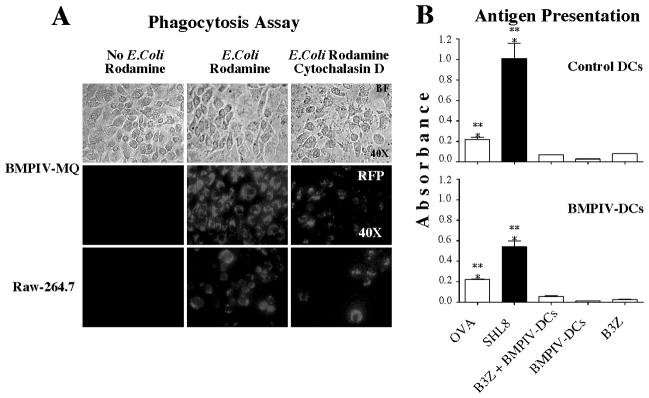
A) Phagocytosis assay. BM-PIV single cells were cultured with M-CSF for 14 days in 96 U-bottom well plates (top row) and were tested for their ability to phagocytose E. coli bacterial particles (pHrodo, Invitrogen). BM-PIV macrophages or RAW 264.7 cells (control) were pre-treated with Cytochalasin D (10μM for 4 hrs) and then cultured with E. coli particles (0.1μg/μL for 1hr) as indicated in upper panel. BM-PIV induced macrophages phagocytosed E. coli particles similarly to RAW 264.7 cells and Cytochalasin D prevented the phagocytosis of E. coli particles (middle and bottom row, respectively). Imaging was performed using a Zeiss Imager Z1 microscope (Carl Zeiss, Thornwood, NY) and detection of fluorescence using a TRITC (red) filter (Chroma Technology, Bellows Falls, VT). B) Antigen presentation assay. BM-PIV single cells were cultured with GM-CSF for 8 days and IL-4 for 4 additional days in 96 U-bottom well plates. BM-PIV induced DCs or DC2.4 control DC line, were co-cultured with B3Z T cells. Ovalbumin or SIINFEKL peptide was added to the cultures and incubated for 24 hr. Cells were then incubated with CPRG for 12 hours and supernatants were tested for B-galactosidase activity, measured by absorbance. Bar graphs represent one experiment of 2 independent experiments (top: control DC2.4 cells, bottom: BM-PIV DCs). Data represent the mean ± SEM of 1 out of 2 independent experiments. Statistical significant differences were found in OVA and SIINFEKL compared to controls, *** p≤0.005.
Dendritic cells are professional antigen-presenting cells with the ability to intake and process antigens and present antigen-derived peptides on their cell surface to T cells in the context of MHC class molecules. To test antigen presentation in DCs elicited from bone marrow population IV cloned single cell cultures, we incubated them with B3Z T cells, which are a modified T cell clone that recognizes OVA and has the ability to express β-galactosidase upon activation by cognate antigen (14). Experiments were performed in the presence of ovalbumin or the SIINFEKL peptide, which is processed from ovalbumin and specific for the T cell receptor on B3Z T cells. We tested the ability of in vitro-differentiated, single cell-derived DC cultures to activate T cells in the context of MHC class I molecules. The read-out for this experiment is a colorimetric assay that measures the activity of β-galactosidase. We found that with either ovalbumin or SIINFEKL peptide, B3Z T cells were activated to levels that were significantly above control values when they were incubated with cloned bone marrow population IV cells that were driven towards DCs (Figure 3B).
These results directly demonstrated a common developmental pathway for osteoclasts, macrophages and dendritic cells through a bone marrow common progenitor with the phenotype B220− CD3− CD11b−/low CD115+ CD117+ C×3CR1+. Significantly, this phenotype is similar to the common progenitor for macrophages and dendritic cells that was reported by Fogg et al (15).
Phenotypic characterization of peripheral osteoclast progenitors
Cells with osteoclastogenic potential exist in the periphery (13). However, the function and ultimate lineage fates of these putative peripheral osteoclast progenitors are not well established. It has also recently been appreciated that peripheral monocyte populations are widely heterogeneous and contain cells with multiple related phenotypes and functional abilities (1, 2). Therefore, we attempted to define where within this heterogeneity does the ability to generate osteoclasts reside. Using a strategy that was similar to the one we utilized to identify murine bone marrow osteoclast progenitor, we fractionated murine spleen and peripheral blood cells and tested their in vitro osteoclastogenic potential.
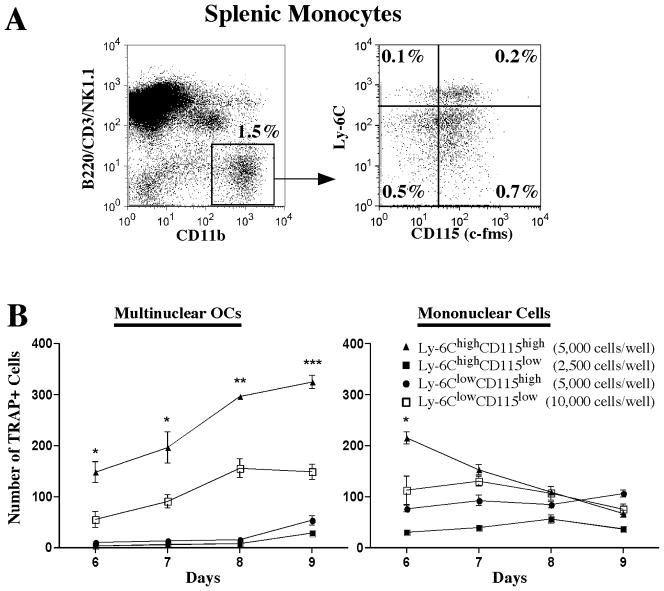
In spleen, we found that cells that were negative for lymphoid markers (LYM) identifying T cells, B cells and Natural Killer cells (CD3, B220/CD45R and NK1.1) and positive for CD11b generated a higher number of multinuclear TRAcP+ OCLs than did the CD11b negative fraction (113±6 vs. 9±7 p=0.0001, when 20,000 cells were plated under osteoclastogenic conditions). This population could be further dissected by its expression of Ly6C and CD115. We found that the population bearing the phenotype LYM− CD11b+ Ly6Chi CD115+ contained the highest potential to form OCLs in vitro (Figure 4). This phenotype is consistent with reported characteristics associated with peripheral circulating monocytes (16, 17). This population also was negative for the granulocyte marker Ly6G and expressed intermediate levels of CD117. Evaluation of cells from C×3CR1-GFP mice showed that this population was also positive for C×3CR1 (data not shown).
Figure 4. Identification and phenotypic characterization of spleen monocyte progenitors with osteoclastogenic activity.
A Phenotypic dissection of spleen monocytes by flow cytometry. Osteoclastogenic precursor candidates were identified based on the lack of expression of CD3, CD45R/B220, NK1.1 (LYM) and high expression of CD11b. Four populations contained among these cells were identified in the context of cell surface expression of Ly6C and CD115. B) In vitro osteoclastogenic assays. The four populations identified in A, were sorted and plated in 96-well plates with M-CSF (30ng/mL) and RANKL (30ng/mL) at the indicated density, cells were fed every 48 hrs. TRAcP assays were performed at different time points and positive cells were counted. Left graph shows multinuclear TRAcP+ osteoclasts (3 or more nuclei per cell) and right graph shows mononuclear TRAcP+ cells. Data represent the mean ± SEM of 1 out of 3 independent experiments. Statistical significant differences were found (LYM− CD11b+Ly-6ChighCD115high vs LYM−CD11b+Ly-6ClowCD115low at day 6, day 7, day 8 and day 9) * p≤0.05, ** p≤0.005, *** p≤0.0005.
When we tested the cloning and precursor frequency of this progenitor population by single cell plating, we found it to have a low colony formation efficiency compared with the progenitor populations from bone marrow, since 100% efficiency was reached only when 50 or more cells were plated per well (Supplemental Figure 2). This result indicates either that this population isheterogeneous or its progression in culture is dependent on density requirements.
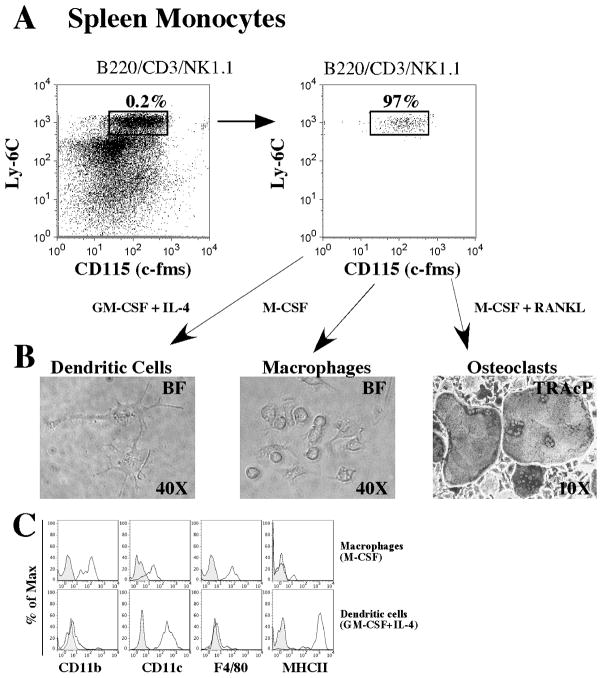
We next evaluated these splenic osteoclastogenic progenitors for their ability to generate macrophages and dendritic cells in vitro. As shown in Figure 5, we sorted the above described splenic monocyte progenitors, incubate them with M-CSF or GM-CSF plus IL-4 and tested their expression of surface markers that characterize macrophages or DCs. Spleen monocyte progenitors cultured with M-CSF for 8 days generate a population expressing high levels of CD11b and F4/80, low levels of CD11c and low levels of MHC class II. In contrast, when cultured with GM-CSF plus IL-4, the developed population expressed high levels of CD11c, low levels of CD11b and F4/80 and high levels of MHC class II. As observed in bone marrow progenitors, both in vitro generated cell types still retained the expression of C×3CR1 (data not shown).
Figure 5. Evidence of a common splenic monocyte progenitor for osteoclast, macrophages and dendritic cells in vitro.
A) Cells with the phenotype LYM− CD11b+ Ly6C+ CD115+ were sorted to homogeneity and plated at a density of 5,000 cells in GM-CSF (30ng/mL) plus IL-4 (30ng/mL) for 8 days, M-CSF (30ng/mL) for 8 days or M-CSF (30ng/mL) plus RANKL (30ng/mL) for 8 days in 96 well plates. B) Images of cells developed at the end of the culture period. DC- and macrophage-like cells are visualized by phase contrast (right and middle) and osteoclasts are visualized by light microscopy after TRAcP staining (right). C) After culture, cells were detached from 96 well plates and analyzed for their cell surface expression of signature cell surface markers for DC and macrophages. Cells cultured with GM-CSF plus IL-4 (top row, histograms) show preferential expression of cell surface molecules related to DCs (CD11c+MHCII+F4/80−CD11blow), and cells cultured with M-CSF (bottom row) show preferential expression of cell surface markers related to macrophages (CD11b+F4/80+CD11clowMHCII−).
Interestingly, we identified an osteoclastogenic progenitor with similar phenotypic characteristics to the spleen progenitor in peripheral blood (Figure 6). This progenitor was also Ly6G− Ly6Chigh and expressed high levels of CD115. Also, expansion and differentiation potential in limiting numbers showed equivalent results to those observed for the progenitor populations isolated from spleen (Supplemental Figure 2).
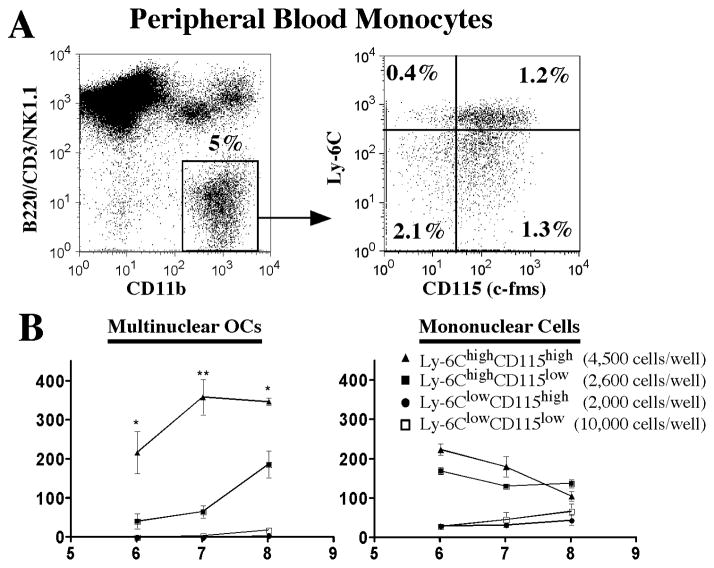
Figure 6. Identification and phenotypic characterization of peripheral blood monocyte progenitors common to osteoclasts, macrophages and dendritic cells.
A) Phenotypic dissection of peripheral monocytes by flow cytometry. As observed in spleen, osteoclastogenic precursor candidates were identified based on the lack of expression of CD3, CD45R/B220, NK1.1 (LYM) and high expression of CD11b. Four populations contained among these cells were segregated in the context of cell surface expression of Ly6C and CD115. B) In vitro osteoclastogenic assays. The four populations identified in A, were sorted and plated in 96-well plates with M-CSF and RANKL at the indicated density; cells were fed every 48 hrs. TRAcP assays were performed at different time points. Left graph shows multinuclear TRAcP+ cells and right graph shows mononuclear TRAcP+ cells. Data represents the mean±SEM of 1 out of 3 independent experiments. Statistical significant differences were found (LYM−CD11b+Ly-6ChighCD115high vs LYM−CD11b+Ly-6ChighCD115low at day 6, day 7 and day 8) * p≤0.05, ** p≤0.005.
In summary, these data indicate that there exists a phenotypically similar population of cells with high osteoclastogenic potential in spleen and peripheral blood. Their phenotype is indicative of a cell with characteristics of inflammatory monocytes (18) (Supplemental Table 1). These cells are homogeneous in their expression of multiple cell surface markers and, although not strictly proven at a clonal level, cells bearing this phenotype also had the ability to generate macrophage like and dendritic- like cells with high efficiency.
DISCUSSION
We have previously identified a bone marrow progenitor able to generate osteoclasts in vitro with high efficiency (12). This progenitor, characterized by the cell surface expression phenotype: B220− CD3−CD11b−/low CD115+ CD117+, shares characteristics with monocyte progenitors and for that reason we tested its ability to clonally differentiate into functional osteoclasts, dendritic cells and macrophages. Published studies of monocyte progenitors have focused mostly on their bipotential ability to generate macrophages and DCs, while analyses of osteoclastogenesis have not been considered (15, 19). We report here that single cells of this progenitor population can generate mature functional cells of these three types with high efficiency. Hence, this result demonstrates the existence of an isolatable common monocyte progenitor for osteoclasts, macrophages and DCs (OcMDC).
In addition to the studies identifying common progenitors for macrophages and DCs (2, 5), an independent study has reported a related population to the one we have identified, with the ability to serve as precursors of osteoclasts and DCs (20). We think that the population characterized in this report is not fundamentally different from the published ones, although detailed phenotypic analysis indicated some minor discrepancies. For example, Auffray et al. reported that the common macrophage/DC precursor falls within a population expressing CD135(19). Our analyses of CD135 expression showed that population IV can be dissected into two discrete populations. However, we found that both populations had similar abilities to generate osteoclasts (data not shown). We did not perform separate assays for macrophages and DCs from each isolated population, but considering the cloning efficiency observed for macrophage and DC formation, it is clear that, at least in vitro, the CD135 marker does not segregate lineage potential within our monocyte precursor population.
We have also identified and characterized monocyte progenitors in spleen and peripheral blood that can efficiently form osteoclasts in vitro. Like the bone marrow progenitors, these cells also express CD115 and C×3CR1, but in contrast, the peripheral osteoclastogenic activity was contained within the CD11b positive population. The overall phenotypic characteristics of these populations are consistent with cells belonging to a peripheral inflammatory type monocyte population. Comparing cloning frequency with osteoclast precursor frequency, we can conclude that the osteoclastogenic potential of clones expanded in culture is as high as the ones observed in bone marrow. These populations also have the capacity to generate macrophages and DCs in vitro. Compared with the bone marrow counterpart, these peripheral osteoclast precursor populations showed a lower frequency for colony formation, which implies that they are heterogeneous and some of their components cannot be expanded in vitro. Alternatively, these cells could be more dependent on density for their progression in culture.
Different groups have found that the peripheral monocyte populations are heterogeneous and that such heterogeneity could be explained by an inherent plasticity of these populations to generate different types of macrophages under the influence of differential microenvironmental cues (21–24).
The osteoclastogenic potential of peripheral monocyte progenitors is intriguing as their functional meaning is not evident. Especially, considering that the main inducing cytokines for osteoclastogenesis, M-CSF and RANKL, are expressed to various degrees in periphery without formation of osteoclasts at these sites. One possibility is that monocyte progenitors circulate to the periphery for additional maturation and then migrate back to bone where they are induced to terminally differentiate into osteoclasts because of a relatively unique osteoclastogenic microenvironment, which amplifies the response to M-CSF and RANKL. Specifically, recent work demonstrating that collagen proteins are ligands for OSCAR (20), a receptor expressed on osteoclasts that appears to amplify RANKL signaling (25, 26), may help explain the unique osteoclastogenic properties of the bone microenvironment. Alternatively, monocyte peripheral progenitors could maintain their osteoclastogenic potential, independent of the bone marrow counterparts, and after migrating to sites of bone remodeling, be induced to form osteoclasts. In a physiological or tissue repair scenario, inflammatory signals provided by micro-fractures, which need continuous remodeling for their resolution, could serve as the driving force to attract monocyte progenitors. Furthermore, these peripheral progenitors could be attracted during the generation of a fracture repair callus, serving as the main source of osteoclasts at this site (27, 28). Finally, in pathological inflammatory conditions, these progenitors may be important/crucial contributors to pathologic osteolytic processes that occur in rheumatoid arthritis or periodontal disease (29, 30).
Finally, we propose that circulating monocyte progenitors are derived from a common bone marrow osteoclasts/macrophage/dendritic cell progenitor (OcMDC), which we have now characterized at a clonal level. However, the lineage relationship between the bone marrow and peripheral monocyte progenitors is yet to be defined. Our phenotypic characterization indicates that the transition from bone marrow to peripheral progenitor is accompanied of phenotypic changes (acquisition of CD11b). As mentioned, the precursor frequency of peripheral populations is not clonal and the most plausible explanation is that these populations are still heterogeneous. We have revisited in peripheral progenitors the expression of multiple markers ascribed to bone marrow monocyte progenitors. In these, CD135, a marker proposed to be useful to define monocyte progenitors in the bone marrow, is not expressed. Interestingly, CD117 expressed at low levels in both spleen and peripheral blood monocyte progenitors, can dissect the latter in two populations, with one fraction accounting for 40% of the population expressing slightly higher levels of CD117. In the future, this could be used for further dissection in order to define better the clonality of this population in vitro.
Certainly, the definitive proof of relationship between progenitors should be established in in vivo studies. One requirement to study this aspect is the availability of bona fide visual reporters that allow the detection of terminally differentiated cells in vivo. Until recently, there existed less than ideal systems for doing this with osteoclasts. However, we have found that some forms of the collagen type I promoter can drive the expression of fluorescent reporter proteins in terminally differentiated osteoclasts in vitro and in vivo (31). More recently, the cathepsin K promoter has been proposed and validated for its expression in terminally differentiated osteoclasts(32, 33). Using these systems we are planning to study migration patterns of our various progenitors in vivo to more clearly define the exact phenotype of osteoclast progenitor cells that circulate, home to bone and differentiate into mature multinucleated resorbing cells in vivo during normal remodeling or in disease states.
Supplementary Material
Acknowledgments
Funding: This work was supported by funds from the grant AR048714 to JAL and HLA from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS).
We thank Mrs. Katie Lamothe and Mrs. Judith Kalinowski for technical support, and Mrs. Sierra Root for critical reading of the manuscript. This work was supported by grants from the National Institute of Health: R01 AR048714 to J.A.L and RC1HL100569-01 to H.L.A.
Footnotes
Supplemental Figures are included with the submission.
Disclosure
All authors state that they have no conflicts of interest.
Authorship contributions
C.E.J-G: performed most experimental aspects of the research, including data analysis and writing of the manuscript. S-K L.: performed experimental procedures and contributed to data analysis and discussion. J.A.L.: designed experimental strategies, performed data analysis and contributed with the writing of the manuscript. H.L.A.: designed and coordinated experimental strategies, performed data analysis, and contributed with the writing of the manuscript.
Contributor Information
Christian E. Jacome-Galarza, Email: jacome@student.uchc.edu.
Sun-Kyeong Lee, Email: slee@neuron.uchc.edu.
Joseph A. Lorenzo, Email: JLorenzo@nso2.uchc.edu.
Hector LeonardoAguila, Email: aguila@nso1.uchc.edu.
References
- 1.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–92. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 2.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins CS, Swirski FK. The multiple roles of monocyte subsets in steady state and inflammation. Cell Mol Life Sci. 2010;67(16):2685–93. doi: 10.1007/s00018-010-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan WY, Kohsaka S, Rezaie P. The origin and cell lineage of microglia: new concepts. Brain Res Rev. 2007;53(2):344–54. doi: 10.1016/j.brainresrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Geissmann F, Auffray C, Palframan R, Wirrig C, Ciocca A, Campisi L, et al. Blood monocytes: distinct subsets, how they relate to dendritic cells, and their possible roles in the regulation of T-cell responses. Immunol Cell Biol. 2008;86(5):398–408. doi: 10.1038/icb.2008.19. [DOI] [PubMed] [Google Scholar]
- 6.Hume DA. The mononuclear phagocyte system. Curr Opin Immunol. 2006;18(1):49–53. doi: 10.1016/j.coi.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11(11):788–98. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 8.Lorenzo J, Horowitz M, Choi Y. Osteoimmunology: interactions of the bone and immune system. Endocr Rev. 2008;29(4):403–40. doi: 10.1210/er.2007-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arai F, Miyamoto T, Ohneda O, Inada T, Sudo T, Brasel K, et al. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J Exp Med. 1999;190(12):1741–54. doi: 10.1084/jem.190.12.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20(3):345–57. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto T, Ohneda O, Arai F, Iwamoto K, Okada S, Takagi K, et al. Bifurcation of osteoclasts and dendritic cells from common progenitors. Blood. 2001;98(8):2544–54. doi: 10.1182/blood.v98.8.2544. [DOI] [PubMed] [Google Scholar]
- 12.Jacquin C, Gran DE, Lee SK, Lorenzo JA, Aguila HL. Identification of multiple osteoclast precursor populations in murine bone marrow. J Bone Miner Res. 2006;21(1):67–77. doi: 10.1359/JBMR.051007. [DOI] [PubMed] [Google Scholar]
- 13.Xing L, Schwarz EM, Boyce BF. Osteoclast precursors, RANKL/RANK, and immunology. Immunol Rev. 2005;208:19–29. doi: 10.1111/j.0105-2896.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 14.Shastri N, Gonzalez F. Endogenous generation and presentation of the ovalbumin peptide/Kb complex to T cells. J Immunol. 1993;150(7):2724–36. [PubMed] [Google Scholar]
- 15.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311(5757):83–7. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 16.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19(1):71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 17.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74–80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 18.Yona S, Jung S. Monocytes: subsets, origins, fates and functions. Curr Opin Hematol. 2010;17(1):53–9. doi: 10.1097/MOH.0b013e3283324f80. [DOI] [PubMed] [Google Scholar]
- 19.Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, et al. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009;206(3):595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrow AD, Raynal N, Andersen TL, Slatter DA, Bihan D, Pugh N, et al. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J Clin Invest. 2011;121(9):3505–16. doi: 10.1172/JCI45913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 22.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swirski FK. The spatial and developmental relationships in the macrophage family. Arterioscler Thromb Vasc Biol. 2011;31(7):1517–22. doi: 10.1161/ATVBAHA.110.221150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willenborg S, Lucas T, van Loo G, Knipper JA, Krieg T, Haase I, et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood. 2012 doi: 10.1182/blood-2012-01-403386. [DOI] [PubMed] [Google Scholar]
- 25.Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428(6984):758–63. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 26.Mocsai A, Humphrey MB, Van Ziffle JA, Hu Y, Burghardt A, Spusta SC, et al. The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRgamma) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci U S A. 2004;101(16):6158–63. doi: 10.1073/pnas.0401602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fazzalari NL. Bone fracture and bone fracture repair. Osteoporos Int. 2011;22(6):2003–6. doi: 10.1007/s00198-011-1611-4. [DOI] [PubMed] [Google Scholar]
- 28.Schindeler A, McDonald MM, Bokko P, Little DG. Bone remodeling during fracture repair: The cellular picture. Semin Cell Dev Biol. 2008;19(5):459–66. doi: 10.1016/j.semcdb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Graves DT. The potential role of chemokines and inflammatory cytokines in periodontal disease progression. Clin Infect Dis. 1999;28(3):482–90. doi: 10.1086/515178. [DOI] [PubMed] [Google Scholar]
- 30.Schett G, Teitelbaum SL. Osteoclasts and Arthritis. J Bone Miner Res. 2009;24(7):1142–6. doi: 10.1359/jbmr.090533. [DOI] [PubMed] [Google Scholar]
- 31.Boban I, Jacquin C, Prior K, Barisic-Dujmovic T, Maye P, Clark SH, et al. The 3.6 kb DNA fragment from the rat Col1a1 gene promoter drives the expression of genes in both osteoblast and osteoclast lineage cells. Bone. 2006;39(6):1302–12. doi: 10.1016/j.bone.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, et al. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 2007;130(5):811–23. doi: 10.1016/j.cell.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Fernandez MA, Sbacchi S, Correa-Tapia M, Naumann R, Klemm J, Chambon P, et al. Transgenic mice for a tamoxifen-induced, conditional expression of the cre recombinase in osteoclasts. PLoS One. 2012;7(5):e37592. doi: 10.1371/journal.pone.0037592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.