Abstract
Our innate immune system distinguishes microbes from self by detecting conserved pathogen-associated molecular patterns (PAMPs) 1. However, all microbes produce PAMPs, regardless of their pathogenic potential. To distinguish virulent microbes from ones with lower disease-causing potential the innate immune system detects conserved pathogen-induced processes 2, such as the presence of microbial products in the host cytosol, by mechanisms that are not fully resolved. Here we show that Nod1 senses cytosolic microbial products by monitoring the activation state of small Rho GTPases. Activation of Rac1 and Cdc42 by bacterial delivery or ectopic expression of a Salmonella virulence factor, SopE, triggered the Nod1 signaling pathway with consequent Rip2-mediated induction of NF-κB-dependent inflammatory responses. Similarly, activation of the Nod1 signaling pathway by peptidoglycan required Rac1 activity. Furthermore, constitutively active forms of Rac1, Cdc42 and RhoA activated the Nod1 signaling pathway. Our data identify activation of small Rho GTPases as a pathogen-induced process sensed through the Nod1 signaling pathway (Fig. S1).
One process that marks pathogens for recognition by the host is the delivery of microbial molecules into the host cell cytosol 2. For example, the enteric pathogen Salmonella enterica serotype Typhimurium (S. Typhimurium) utilizes a type III secretion system (T3SS-1) encoded by Salmonella pathogenicity island (SPI) 1 to deliver proteins, termed effectors, into epithelial cells 3,4. S. Typhimurium elicits pro-inflammatory responses by translocating four T3SS-1 effector proteins, termed SipA, SopE, SopB and SopE2 5–8. To investigate the mechanism of T3SS-1-dependent nuclear factor kappa B (NF-κB) activation reported previously 7–9, we used human cells transfected with a NF-κB luciferase reporter construct. A S. Typhimurium mutant lacking pro-inflammatory effector proteins (sipA sopE sopB sopE2 mutant) was deficient for NF-κB activation (P < 0.05) in epithelial HeLa cells (Fig. S2A). Inactivation of pro-inflammatory effector genes resulted in a partial inhibition of NF-κB activation in human embryonic kidney (HEK) 293 cells, presumably because endogenous TLR5 expression renders HEK293 cells responsive to stimulation with flagellin (Fig. S2B) 10–12. Compared to a mutant lacking pro-inflammatory effector proteins, NF-κB activation was significantly (P < 0.05) enhanced after infecting host cells with a mutant expressing SopE (sipA sopB sopE2 mutant) (Fig. S2A and S2B). A cytosolic localization of SopE in the absence of other bacterial molecules is sufficient for inducing NF-κB activation 5, as illustrated by ectopic expression of a green fluorescent protein (GFP)-SopE fusion protein in HEK293 cells (Fig. S2C).
In the host cell cytosol, SopE activates Rac1 and Cdc42 by serving as a nucleotide exchange factor that facilitates the transition from an inactive GDP-bound state to an active GTP-bound state of these small Rho GTPases 5. Transfection of HEK293 cells with plasmids encoding dominant negative forms of Cdc42 (Cdc42DN) or Rac1 (Rac1DN) 13 inhibited NF-κB activation elicited by ectopic expression of SopE (P < 0.05) (Fig. S2D and S2E), which was consistent with previous reports 5. In contrast, no significant inhibition was observed when HEK293 cells were transfected with a plasmid encoding a dominant negative form of the small Rho GTPase RhoA (RhoADN). A G168A amino acid substitution in SopE (SopEG168A) is known to abrogate its nucleotide exchange factor activity for Rac1 and Cdc42 14. Transfection of HEK293 cells with a plasmid encoding GFP-SopEG168A no longer resulted in NF-κB activation (Fig. 2F). Collectively, these observations raised the possibility that activation of Rac1 and Cdc42 might be a pathogen-induced process detected by host cells as a pattern of pathogenesis 15. However, the identity of the host’s pathogen recognition receptor (PRR) that activates NF-κB when it detects this pattern of pathogenesis remained elusive.
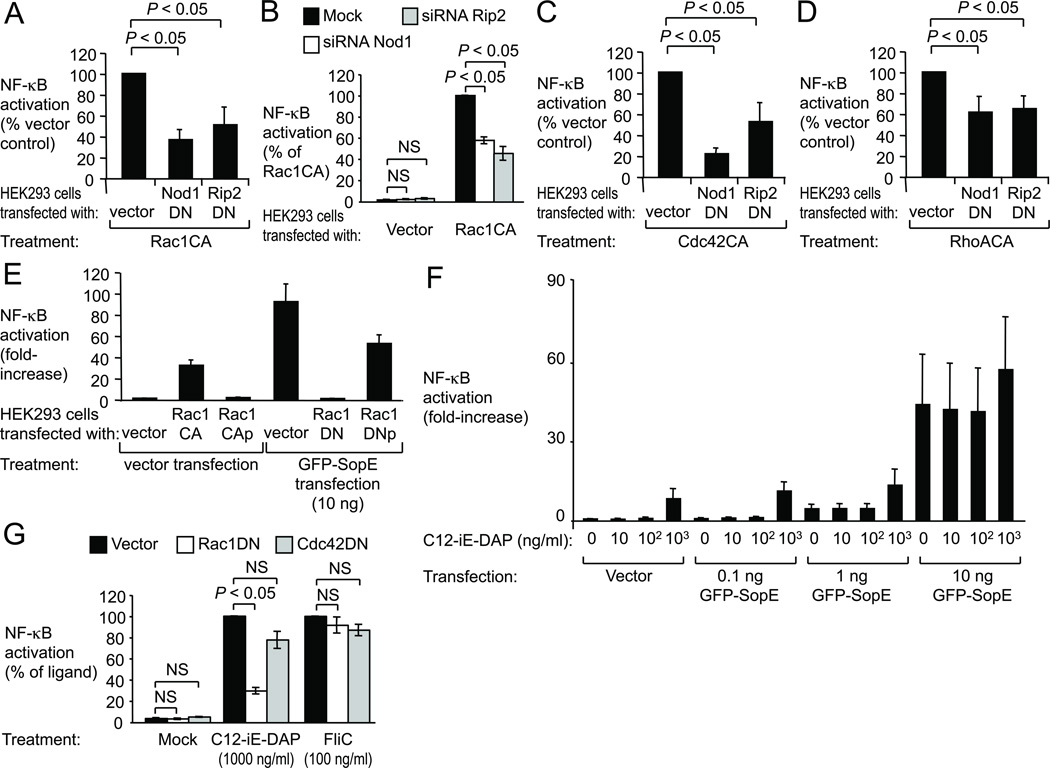
Figure 2. NOD1 senses the activation state of small Rho GTPases.
HEK293 cells were transfected as indicated, together with a NF-κB luciferase reporter. (A–E) Luciferase activity was measured 48 hours after transfection. (F–G) Cells were stimulated with C12-iE-DAP or FliC as indicated and luciferase activity was measured 5 hours thereafter. (A–E) Data are presented as mean ± s.e.m. from at least three independent experiments. Brackets indicate the significance of differences. NS, not significantly different.
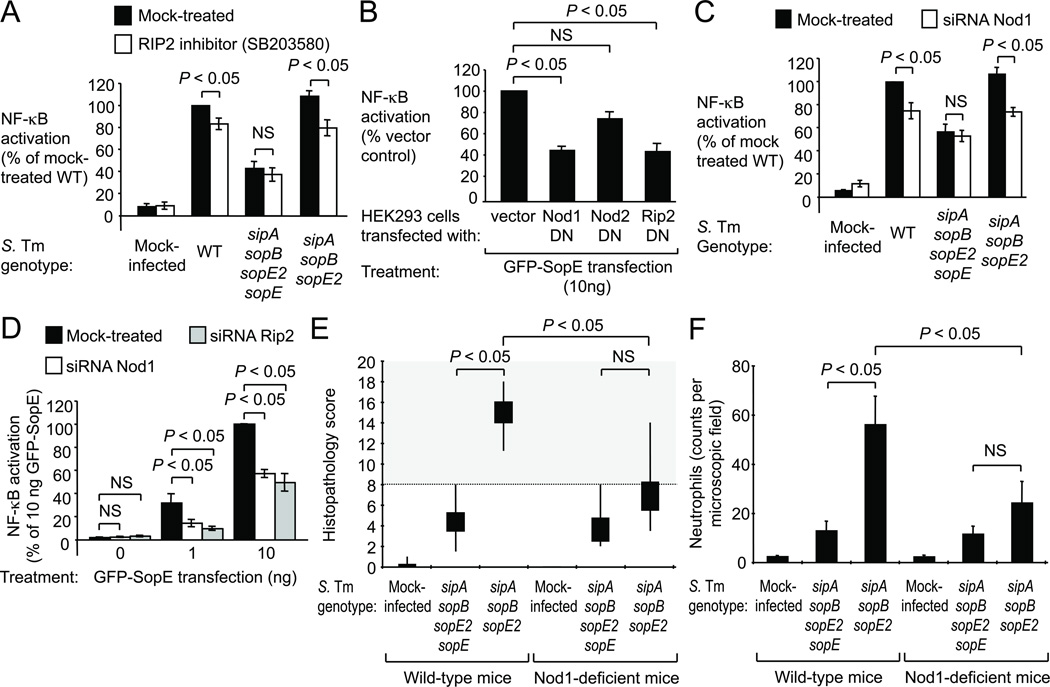
The induction of T3SS-1-dependent inflammatory responses during S. Typhimurium colitis is largely MyD88-independent 16. One MyD88-independent mechanism by which S. Typhimurium induces NF-κB activation is the induction of nucleotide-binding and oligomerization domain (Nod) 1 and Nod2, two cytosolic PRRs that sense cell wall fragments 12,17,18. Nod1 and Nod2 interact with the protein kinase receptor interacting protein-2 (Rip2) to mediate NF-κB activation 19,20. Remarkably, treatment of HEK293 cells with SB203580, a pyridinyl imidazole inhibitor of Rip2 activity (Fig. S2G)21, significantly (P < 0.05) reduced NF-κB activation elicited by bacterial delivery of SopE (i.e. infection with sipA sopB sopE2 mutant) (Fig. 1A). In this model, no SopE-independent NF-κB activation was observed when cells were infected with non-flagellated S. Typhimurium strains (Fig. S2H), suggesting that flagella were responsible for background levels of NF-κB activation elicited by a mutant lacking pro-inflammatory effector proteins (sipA sopE sopB sopE2 mutant) (Fig. 1A).
Figure 1. SopE-induced NF-κB activation requires Rip2 and Nod1.
(A–D) HEK293 cells transfected with a NF-κB-luciferase reporter were treated as indicated. (A and C) Cells were infected with S. Typhimurium (S. Tm) wild type (WT) or mutants. (A–D) Mean luciferase activity ± s.e.m., N = 3. (E–F) Streptomycin-pretreated Nod-1-deficient mice and wild-type littermates (for N see Figure S5B) were infected and the cecum collected after 24 hours. (E) Average histopathology score. Whisker plots represent the second and third quartiles (boxes) and the first and fourth quartiles (lines). (F) Average number of neutrophils per microscopic field. (A–F) NS, not significantly different.
We next investigated whether Nod1 and/or Nod2 were required for SopE-dependent NF-κB activation by transfecting HEK293 cells with plasmids encoding dominant negative forms of Nod1 (Nod1DN), Nod2 (Nod2DN) or Rip2 (Rip2DN) 12. In control experiments, expression of Nod1DN or Rip2DN inhibited NF-κB activation elicited by stimulation with the Nod1 ligand C12-iE-DAP (Fig. S3A), while expression of Nod2DN or Rip2DN inhibited NF-κB activation elicited by stimulation with the Nod2 ligand muramyl dipeptide (MDP) (Fig. S3B). Transfection of HEK293 cells with dominant negative forms of Nod1 (Nod1DN) or Rip2 (Rip2DN) inhibited NF-κB activation elicited by ectopic expression of GFP-SopE (P < 0.05) (Fig. 1B and S3C). In contrast, a dominant negative form of Nod2 (Nod2DN) did not reduce GFP-SopE-induced NF-κB activation.
To further investigate whether Nod1 is required for SopE-dependent NF-κB activation, we silenced Nod1 expression in HEK293 cells with small interfering RNA (siRNA). In a control experiment, Nod1-specific siRNA inhibited NF-κB activation elicited with C12-iE-DAP, but did not affect responses elicited by MDP or flagellin (FliC) (Fig. S3D). Importantly, Nod1-specific siRNA significantly (P < 0.05) reduced NF-κB activation elicited through bacterial delivery of SopE by flagellated (Fig. 1C) or non-flagellated S. Typhimurium strains (Fig. S3E). To further investigate whether Nod1 and Rip2 were required for SopE-dependent NF-κB activation, we silenced Nod1 and Rip2 expression in cells ectopically expressing SopE. In a control experiment, Rip2-specific siRNA inhibited NF-κB activation elicited by C12-iE-DAP and MDP, but not by flagellin (Fig. S3D). Remarkably, Nod1-specific siRNA and Rip2-specific siRNA inhibited NF-κB activation elicited by ectopic expression of GFP-SopE (P < 0.05)(Fig. 1D). Inhibition of NF-κB activation by Nod1DN, Rip2DN, Nod1-specific siRNA or Rip2-specific siRNA could be overcome when large quantities (100 ng) of the plasmid encoding GFP-SopE were used for transfection (Fig. S4A and S4B), which resulted in a loss of cells (Fig. S4C). These effects might explain why a contribution of Rip2 to SopE-induced host cell responses was missed in a previous study 7. Collectively, our data suggested that SopE-mediated NF-κB activation required both Nod1 and Rip2 activity.
To test the biological significance of our observations, we determined the role of Nod1 in SopE-mediated inflammation in vivo using the mouse colitis model of S. Typhimurium infection 22. To restrict our analysis to SopE-mediated mechanisms of inflammation, we compared a S. Typhimurium strain producing only SopE (sipA sopB sopE2 mutant) with an isogenic SopE-deficient mutant (sipA sopE sopB sopE2 mutant). Remarkably, the SopE-proficient S. Typhimurium strain triggered cecal inflammation in wild-type littermates, but not in Nod1-deficient mice (Fig. 1E, 1F and S5A). In contrast, a SopE-deficient mutant did not elicit marked cecal inflammation, although the strain was recovered from the cecum in numbers similar to those of the SopE-proficient strain (Fig. S5B). Thus, a SopE-proficient S. Typhimurium strain required Nod1 for eliciting intestinal inflammation in vivo.
Since SopE-mediated NF-κB activation was both Rac1-dependent (Fig. S2D) and Nod1-dependent (Fig. 1), we investigated whether Nod1 senses the activation state of Rac1 (Fig. 2). HEK293 cells were transfected with a plasmid encoding a Rac1-derivative carrying an amino acid substitution (Q61L) that abrogates its GTPase activity, resulting in a constitutively active, GTP-bound, form of Rac1 (Rac1CA) 13. Expression of Rac1CA induced NF-κB activation, which could be blunted by expressing Nod1DN or Rip2DN (Fig. 2A and Fig. S6A). Similarly, introducing siRNA specific for Nod1 or Rip2 blunted NF-κB activation induced by Rac1CA (Fig. 2B). Notably, constitutively active forms of Cdc24 (Cdc42CA) and RhoA (RhoACA) also induced NF-κB activation in a Nod1 and Rip2-dependent manner (Fig. 2C, 2D and S6A), suggesting that the Nod1 signaling pathway senses the activation of several small Rho GTPases. An intact membrane anchor of active Rac1 was important for NF-κB activation, because a GTP-bound form of Rac1 lacking its prenyl-group (Rac1CAp)23 did not activate NF-κB and a dominant negative form of Rac1 lacking its prenyl-group (Rac1DNp) was no longer capable of inhibiting SopE-induced NF-κB activation (Fig. 2E and S6B). Together, these data suggested that Nod1 detects bacterial-induced activation of small Rho GTPases in host cells.
Since Nod 1 is known to sense the presence of Gram-negative peptidoglycan in the cytosol 20, we wanted to investigate whether detecting this pattern of pathogenesis required small Rho GTPases. When we transfected cells with increasing amounts of plasmid encoding GPF-SopE and stimulated them with different concentrations of C12-iE-DAP, the effects on NF-κB activation were additive, not synergistic (Fig. 2F). Similarly, Rac1CA and C12-iE-DAP did not synergize in inducing NF-κB activation (Fig. S7). Remarkably, NF-κB activation elicited by treatment of cells with C12-iE-DAP was blunted when cells were transfected with Rac1DN, but not with Cdc42DN (Fig. 2G). In contrast, NF-κB activation elicited by treatment of cells with flagellin (FliC) was not affected by transfection with Rac1DN or Cdc42DN. In summary, our data suggested that Nod1 detects peptidoglycan as a pattern of pathogenesis by sensing the activation state of Rac1.
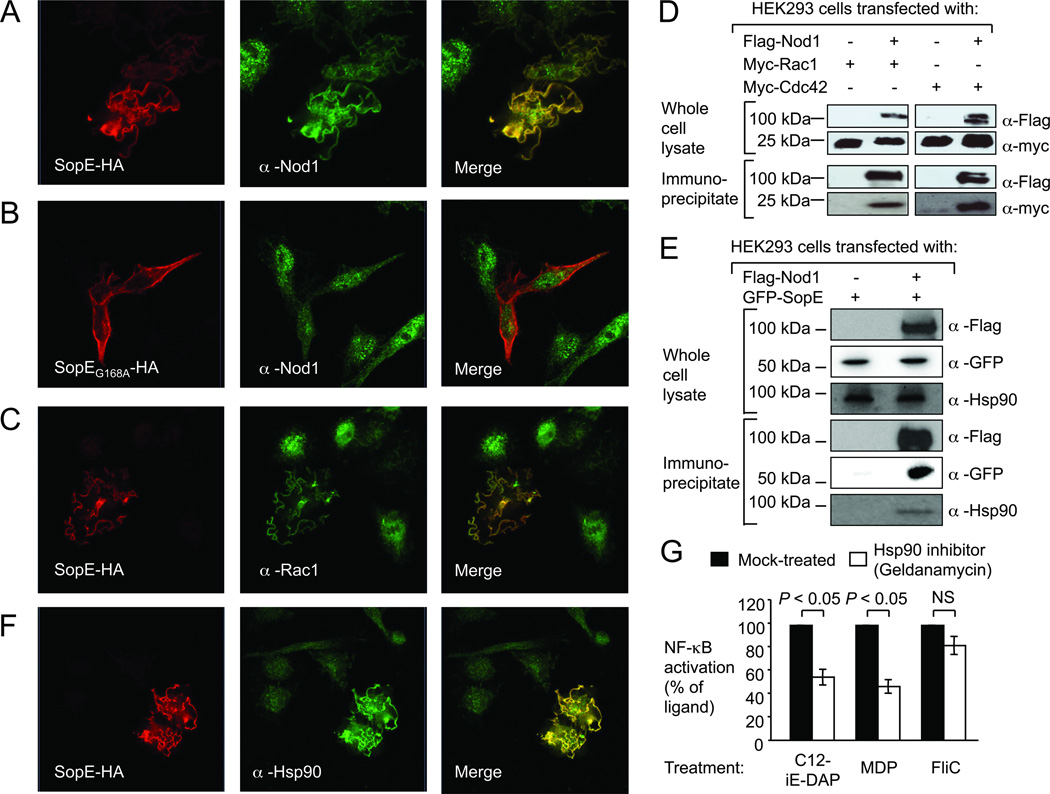
To further investigate a possible interaction of Nod1 with SopE and small Rho GTPases we performed confocal microscopy with HEK293 cells ectopically expressing HA-tagged SopE (SopE-HA) (Fig. 3). Expression of SopE-HA resulted in the formation of membrane ruffles and the recruitment of endogenous Nod1, which was detected with an anti-Nod1 antibody (α-Nod1) (Fig. 3A). In contrast, a SopE-derivative carrying an amino acid substitution that prevents activation of Rac1 and Cdc42 (SopEG168A-HA) did not induce the formation of membrane ruffles and no co-localization with endogenous Nod1 was observed (Fig. 3B). Endogenous Rac1, which was detected with an anti-Rac1 antibody (α-Nod1), also co-localized with SopE-HA in membrane ruffles (Fig. 3C). When HEK293 cells expressing Flag-tagged Nod1 (Flag-Nod1) were co-transfected with plasmids encoding either Myc-tagged Rac1 (Myc-Rac1) or Myc-Cdc42, a monoclonal anti-Flag antibody co-immuno-precipitated Myc-Rac1 and Myc-Cdc42 (Fig. 3D), suggesting that Nod1 is present in a protein complex containing these small Rho GTPases. Further analysis of immuno-precipitates by mass spectrometry identified heat shock protein 90 (Hsp90) as a possible component of Nod1-containing multi-protein complexes (Tab. S1). To confirm this interaction, cells expressing Flag-Nod1 and GFP-SopE were immuno-precipitated with a monoclonal anti-Flag antibody. GFP-SopE and endogenous cellular Hsp90 were detected in the immuno-precipitate (Fig. 3E). Furthermore, endogenous Hsp90 co-localized with SopE-HA in membrane ruffles (Fig. 3F). To investigate whether the presence of Hsp90 is required for signaling, we abolished Hsp90 activity in HEK293 cells by treatment with the specific Hsp90 inhibitor geldanamycin. Geldanamycin treatment inhibited NF-κB activation elicited by stimulation with C12-iE-DAP and MDP, but did not significantly reduce NF-κB activation induced by flagellin (Fig. 3G). Furthermore, treatment of HEK293 cells with geldanamycin significantly (P < 0.05) reduced NF-κB activation elicited by bacterial delivery of SopE (i.e. infection with sipA sopB sopE2 mutant) (Fig. S8).
Figure 3. SopE forms a multi-protein complex with Rac1, Cdc42, Nod1 and Hsp90.
(A–C and F) Ectopically expressed SopE-HA (A, C and F), SopEG168A-HA (B) and endogenous Nod1 (A–B), Rac1 (C), and Hsp90 (F) were detected by confocal microscopy. (D and E) Whole cell lysates and anti-Flag immunoprecipitates were separated by SDS-PAGE and subjected to immunoblotting. (G) HEK293 cells transfected with a NF-κB luciferase reporter were pretreated with the Hsp90 inhibitor geldanamycin (100 nm). Data are presented as mean ± s.e.m. of at least three independent experiments. Brackets indicate the significance of differences. NS, not significantly different.
We conclude that a multi-protein complex composed of small Rho GTPases, Hsp90 and Nod1 responds to pathogens gaining cytosolic access by activating NF-κB (Fig. S1). Escape of pathogens (e.g. Shigella flexneri) from the phagosome introduces peptidoglycan fragments into the cytosol 20 and our data suggest that a complex composed of active Rac1, Hsp90 and Nod1 detects this pattern of pathogenesis. In addition, pathogens commonly manipulate the actin cytoskeleton of host cells by injecting proteins into the cytosol that alter the activity of small Rho GTPases 24,15. Here we show that activation of small Rho GTPases by a bacterial virulence factor is a second pattern of pathogenesis detected through the Nod1 signaling pathway in mammalian cells. Alternatively, manipulation of small Rho GTPases by effector proteins could be viewed as a mechanism through which pathogens manipulate host responses to gain a growth advantage 25. Effector-triggered immune responses of invertebrates might represent an interesting parallel, because activation of the small Rho GTPase Rac2 in Drosophila by a bacterial toxin leads to activation of the innate immune adaptor IMD (immune deficiency), which shares homology to the mammalian Rip1 protein 26. Rip1 and Rip2 are important activators of NF-κB in response to cellular stress in mammals. Rip2 binds the caspase recruitment domains (CARDs) of Nod1 and Nod2 to initiate NF-κB activation 27. Our data suggest that Nod1 interacts with proteins in addition to Rip2, because this PRR was present in a multi-protein complex, which contained a bacterial effector (SopE), small Rho GTPases and HSP90. The term nodosome has been proposed for mammalian Nod1 and/or Nod2-containing protein complexes functioning in NF-κB activation 28. RhoA has previously been implicated in Nod1-dependent NF-κB activation induced by the S. flexneri effector protein OspB, however, this small Rho GTPase was proposed to act downstream of Nod1 29. The finding that Nod1 senses the activation state of small Rho GTPases acting upstream in the signaling cascade (Fig. S1) significantly changes our understanding of the mechanisms that trigger this signaling pathway.
METHODS
Bacterial strains, tissue culture cells, and culture conditions
The bacterial strains used in this study are listed in Table S2. E. coli and S. Typhimurium strains were routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB agar plates supplemented with antibiotics. HeLa57A and HEK293 cell lines have been described previously 12 and were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS) at 37°C in a 5% CO2 atmosphere.
Animal experiments
All mouse experiments were approved by the Institutional Animal Care and Use Committees at the University of California, Davis. Nod1+/−Nod2+/−, Nod1−/−Nod2+/− mice in a C57BL/6 background were bred and housed at the Center for Laboratory Animal Science at UC Davis. DNA was isolated from mouse-tails with the DNeasy Blood & Tissue kit (Qiagen) and used as a template for genotyping. Primers for genotyping are listed in Table S3. Streptomycin (20mg/mouse) (Sigma)-pretreated mice were orally inoculated with 0.1 ml of sterile LB broth or 1 × 109 CFU (in 0.1 ml of LB broth) of an S. Typhimurium sipAsopBE2E mutant or a sipAsopBE2 mutant. At 24 h after infection, mice were sacrificed and tissue samples were collected. Cecal contents were collected in PBS and serial dilutions were plated on LB agar containing the appropriate antibiotics to determine bacterial numbers. Formalin-fixed, hematoxylin and eosin (H&E)-stained cecal tissue sections were blinded for evaluation by a veterinary pathologist. The following pathological changes were scored: (i) neutrophil infiltration, (ii) infiltration by mononuclear cells, (iii) submucosal edema, (iv) epithelial damage, and (v) inflammatoroy exudate. The pathological changes were scored on a scale from 0 to 4 as follows: 0, no changes; 1, detectable; 2, mild; 3, moderate; 4, severe. Neutrophil counts were determined by high-magnification (×400) microscopy, and numbers were averaged from 10 microscopic fields for each animal. Images were taken using an Olympus BX41 microscope.
Construction of plasmids
Plasmids and primers used in this study are listed in Table S2 and S3. Standard cloning techniques were performed as described previously 30. A DNA fragment upstream of the sipA coding sequence was amplified from S. Typhimurium, cloned into pCR2.1 using the TOPO-TA cloning kit (Life Technologies) and subcloned into pRDH10 using EcoRI, yielding pSW244. To facilitate transduction of the unmarked ΔsipA mutation, pSW244 was introduced in this locus in the ZA10 chromosome by conjugation with S17-1 λpir as the donor strain, creating SW974. The sopE gene was PCR amplified from M30 and cloned into the mammalian expression vectors pEGFP-C1 and pCMV-HA (BD Biosciences Clontech). The small GTPase Rac1, Cdc42 and RhoA were PCR amplified from cDNA prepared from HeLa57A cells. SopEG168A was PCR amplified from pEGFP-SopE. The Dominant Negative (DN) forms of Rac1 (Rac1DN), Cdc42 (Cdc42DN) and RhoA (RhoADN), are mutated in the GTP binding site at positions 17, 17 and 19, respectively, from a threonine to an asparagine 13. The Constitutively Active (CA) forms of Rac1 (Rac1CA), Cdc42 (Cdc42CA) and RhoA (RhoACA), are mutated in the GTP hydrolysis site at positions 61, 61 and 63, respectively, from a glutamine to a leucine 13. The prenylation mutants of Rac1 are mutated at position 189, where a cysteine is replaced by a serine 23. SopEG168A and the DN and CA mutant forms of Rac1, Cdc42 and RhoA were engineered by overlap extension PCR. The two DNA fragments needed for the construction of these mutant forms were amplified by PCR, gel-purified, then fused in one PCR reaction before cloning into pEGFP-C1, pCMV-HA and pCMV-myc. All constructs were verified by DNA sequencing (SeqWright).
Transfections
Transfections were performed using FuGene HD (Roche) according to the manufacturer’s instructions. 48 hours post transfection, cells were lysed either without any treatment, or infected with the indicated bacterial strains or the NOD1, NOD2 and TLR5 ligands with or without pretreatment (30 min) of the cells with the inhibitors (SB203580 (10µM) and geldanamycin (100nM) (Invivogen)). The cells were infected with the indicated S. Typhimurium strains (106 CFU/ml) for 1 h, after which the cells were washed with DPBS, and incubated at 37°C for an additional 4 h in the presence of DMEM containing 10% FCS. After 5 hours of treatment the cells were lysed and analyzed for beta-galactosidase activity and luciferase activity 12.
Generalized phage transduction
Phage P22 HT int-105 was used for transductions 31. A lysate of ZA21 was used to separately transduce the sopB::MudJ and sopE2::pSB1039 mutations into SL1344, creating SW798 and SW800, respectively. The ΔsipA::pSW244 mutation from SW974 was separately introduced into SL1344 as well as the ΔsopE mutant SW976 by transduction and subsequent sucrose selection 32 was performed to create SW1007 and SW1009, respectively. To construct SW973 and SW1008, a P22 phage lysate of SW798 was used to transduce the sopB::MudJ mutation into SW1009 and SW1007, respectively. A P22 lysate of SW800 was employed to transduce the sopE2::pSB1039 mutation into SW973 and SW1008, generating SW868 and SW867, respectively.
To generate AMK457 and AMK456, the ΔfliC::pSPN29 mutation from SPN305 was separately introduced into SW1008 and SW973 by transduction and subsequent sucrose selection was performed to remove pSPN29. The fljB::MudCm mutation from SPN287 was then transduced into AMK457 and AMK456 to create AMK461 and AMK460, respectively. A P22 lysate of SW800 was used to introduce the sopE2::pSB1039 mutation into the AMK461 and AMK460 chromosome, thus creating AMK465 and AMK464, respectively.
Co-immunoprecipitation and Western Blotting
Transfected HEK293 cells were lysed and co-immunoprecipitation assays were performed with protein G beads (Invitrogen) coated with anti-Flag M2 antibody (Sigma) 12. Whole cell lysates and immunoprecipitates proteins were analyzed for protein expression by Western blot using antibodies raised against rabbit anti-GFP, mouse anti-Flag (Sigma), mouse anti-HA (Covance), rabbit anti-myc, rabbit anti-HSP90 or anti-tubulin (Cell Signaling Technology) 12.
Fluorescence microscopy
HeLa cells transiently transfected with expression plasmids were fixed with 3% paraformaldehyde as described previously 12. Cells were incubated with mouse anti-HA (Covance) and rabbit anti-Nod1, rabbit anti-HSP90 (Cell Signaling) or rabbit anti-Rac1 (Fisher). Cells were incubated with Alexa fluor 647 goat anti-mouse IgG and Alexa fluor 488 goat anti-rabbit IgG (Invitrogen). Samples were embedded in Fluorsave (Calbiochem) and analyzed with a LSM700 Confocal Microscope.
Mass spectometry of NOD1-Flag-interacting proteins
NOD1-Flag interacting proteins bound to Protein G beads were digested directly with trypsin (Promega, sequencing grade) overnight in 50 mM ammonium bicarbonate, pH 8. Extracted peptides were analyzed by LC-MS/MS on a Thermo-Finnigan LTQ with Michrom Paradigm LC and CTC Pal autosampler. MS and MS/MS spectra were acquired using a top 10 method with an MS survey scan obtained for the m/z range 375–1300. Tandem mass spectra were extracted with Xcalibur version 2.0.7. All MS/MS samples were analyzed using X!Tandem (The GPM, thegpm.org; version TORNADO (2010.01.01.4). X!Tandem was set up to search the uniprottaxon_20100804_YNtL3Y human database (44512 entries) appended to the cRAP database of common contaminants; samples were also searched with an identical but reversed database to calculate false discovery rates (FDR). Scaffold (version Scaffold_3_00_08, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm 33. Protein identifications were accepted if they could be established at greater than 95.0% probability. These parameters yielded a protein FDR of 0.6% and a peptide FDR or 0.1% for identification of interacting proteins.
Statistical analysis
Statistical differences were calculated using a paired Student’s t-test. To determine the statistically significant differences in the total histopathology scores, a Mann-Whitney U-test was used. A two-tailed p<0.05 was considered to be significant.
Supplementary Material
Acknowledgements
We would like to thank Sean Paul Nuccio for providing PCR primers for the construction of the bacterial strains. This work was supported by Public Health Service Grants AI044170 and AI076246. A.M.K. is supported by the American Heart Association Grant 12SDG12220022.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Contributions
A.M.K contributed to the experimental design, performed experiments and contributed to Figures 1B, 1C, 1D, 1E, 1F, 2A, 2B, 2C, 2D, 2E, 2G, 3A, 3B, 3C, 3F, 3G, S1, S2D, S3A, S3B, S3D, S4A, S4B, S4C, S7 and S8. M.G.W. performed experiments, constructed bacterial strains and contributed to Figures 1A, 1E, 1F, 3D, 3G, S2A, S2B, S2E, S2H, S2G, S3E, S4C, S5B and S8. J.J.A. constructed expression plasmids and contributed to Figures 2F, 3C and S7. S.P.F. constructed expression plasmids and contributed to Figures 2G, S2F, S3C, S6A and S6B. M.N.X contributed to Figures 1E, 1F and S5A. S.E.W. constructed bacterial strains, contributed to Figure S5B and critically read the manuscript. A.K. constructed expression plasmids and contributed to Figure S2C and S7. V.P. constructed bacterial strains. M.M.R. contributed to Figure S2D. J.W. constructed expression plasmids. R.A.E. performed mass spectrometry. A.M.K., A.J.B. and R.M.T. provided financial support for the study and contributed to the experimental design. A.M.K. and A.J.B. were responsible for the overall study design and for writing the manuscript.
Competing financial Interests
The authors declare no competing financial interests
Contributor Information
A. Marijke Keestra, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Maria G. Winter, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Josef J. Auburger, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Simon P. Fräßle, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Mariana N. Xavier, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Sebastian E. Winter, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Anita Kim, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Victor Poon, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Mariëtta M. Ravesloot, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Julian Waldenmaier, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Renée M. Tsolis, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Richard A. Eigenheer, Proteomics Core Facility, UC Davis Genome Center, University of California, Davis, CA, USA
Andreas J. Bäumler, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
REFERENCES
- 1.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y, Galan JE. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Molecular microbiology. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhou D, Mooseker MS, Galan JE. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc Natl Acad Sci U S A. 1999;96:10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 6.Hapfelmeier S, et al. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infection and immunity. 2004;72:795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruno VM, et al. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueiredo JF, et al. Salmonella enterica Typhimurium SipA induces CXC-chemokine expression through p38MAPK and JUN pathways. Microbes Infect. 2009;11:302–310. doi: 10.1016/j.micinf.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Hobbie S, Chen LM, Davis RJ, Galan JE. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 10.Smith MF, Jr, et al. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-kappa B activation and chemokine expression by epithelial cells. J Biol Chem. 2003;278:32552–32560. doi: 10.1074/jbc.M305536200. [DOI] [PubMed] [Google Scholar]
- 11.Lu W, et al. Cutting edge: enhanced pulmonary clearance of Pseudomonas aeruginosa by Muc1 knockout mice. J Immunol. 2006;176:3890–3894. doi: 10.4049/jimmunol.176.7.3890. [DOI] [PubMed] [Google Scholar]
- 12.Keestra AM, et al. A Salmonella Virulence Factor Activates the NOD1/NOD2 Signaling Pathway. mBio. 2011;2:e00266-00211. doi: 10.1128/mBio.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coso OA, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 14.Schlumberger MC, et al. Amino acids of the bacterial toxin SopE involved in G nucleotide exchange on Cdc42. J Biol Chem. 2003;278:27149–27159. doi: 10.1074/jbc.M302475200. [DOI] [PubMed] [Google Scholar]
- 15.Muller AJ, Hoffmann C, Hardt WD. Caspase-1 activation via Rho GTPases: a common theme in mucosal infections? PLoS Pathog. 2010;6:e1000795. doi: 10.1371/journal.ppat.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hapfelmeier S, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 17.Le Bourhis L, et al. Role of Nod1 in mucosal dendritic cells during Salmonella pathogenicity island 1-independent Salmonella enterica serovar Typhimurium infection. Infection and immunity. 2009;77:4480–4486. doi: 10.1128/IAI.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geddes K, et al. Nod1 and Nod2 regulation of inflammation in the Salmonella colitis model. Infection and immunity. 2010;78:5107–5115. doi: 10.1128/IAI.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inohara N, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 20.Girardin SE, et al. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Argast GM, Fausto N, Campbell JS. Inhibition of RIP2/RIck/CARDIAK activity by pyridinyl imidazole inhibitors of p38 MAPK. Mol Cell Biochem. 2005;268:129–140. doi: 10.1007/s11010-005-3701-0. [DOI] [PubMed] [Google Scholar]
- 22.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong KW, Mohammadi S, Isberg RR. The polybasic region of Rac1 modulates bacterial uptake independently of self-association and membrane targeting. J Biol Chem. 2008;283:35954–35965. doi: 10.1074/jbc.M804717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aktories K, Schmidt G, Just I. Rho GTPases as targets of bacterial protein toxins. Biol Chem. 2000;381:421–426. doi: 10.1515/BC.2000.054. [DOI] [PubMed] [Google Scholar]
- 25.Lopez CA, et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3 doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyer L, et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation. Semin Immunopathol. 2007;29:289–301. doi: 10.1007/s00281-007-0083-2. [DOI] [PubMed] [Google Scholar]
- 29.Fukazawa A, et al. GEF-H1 mediated control of NOD1 dependent NF-kappaB activation by Shigella effectors. PLoS Pathog. 2008;4:e1000228. doi: 10.1371/journal.ppat.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES FOR METHODS
- 30.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- 31.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 32.Lawes M, Maloy S. MudSacI, a transposon with strong selectable and counterselectable markers: use for rapid mapping of chromosomal mutations in Salmonella typhimurium. J Bacteriol. 1995;177:1383–1387. doi: 10.1128/jb.177.5.1383-1387.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.