Abstract
Hepatocyte nuclear factor (HNF)-4α is a key member of the transcription factor network regulating hepatocyte differentiation and function. Genetic and molecular evidence suggests that expression of HNF-4α is mainly regulated at the transcriptional level. Activation of HNF-4A gene involves the interaction of distinct sets of transcription factors and co-transcription factors within enhancer and promoter regions. Here we study the inhibitory effect of microRNAs (miRNA) on the 3′-untranslated region (3′-UTR) of HNF-4A mRNA. The potential recognition elements of a set of miRNAs were identified utilizing bioinformatics analysis. The family members of miR-34 and miR-449, including miR-34a, miR-34c-5p and miR-449a, share the same target elements located at two distinct locations within the 3′-UTR of HNF-4A. The over-expression of miR-34a, miR-34c-5p or miR-449a in HepG2 cells led to a significant decrease in the activity of luciferase reporter carrying 3′-UTR of HNF-4A. The repressive effect on reporter activity was partially or fully eliminated when one or two of the binding site(s) for miR-34a/miR-34c-5p/miR-449a were deleted within the 3′-UTR. The protein level of HNF-4α was dramatically reduced by over-expression of miR-34a, miR-34c-5p and miR-449a, which correlates with a decrease in the binding activity of HNF-4α and transactivation of HNF-4α target genes. These results suggest that the recognition sites of miR-34a, miR-34c-5p and miR-449a within 3′-UTR of HNF-4A are functional. The mechanism of down-regulation of the binding activity and transactivation of HNF-4α by the miRNAs involves the decrease in HNF-4α protein level via miRNAs selectively targeting HNF-4A 3’-UTR, leading to the translational repression of HNF-4α expression.
Keywords: HNF-4, microRNA, 3′-untranslated region, DNA binding activity, HepG2
1. Introduction
The liver-enriched transcription factors are part of a complex transcriptional network which is responsible for both determination and maintenance of the hepatic phenotype. This network is established by a number of auto-regulatory and cross-regulatory pathways securing balance and high-level expression of genes in the hepatocytes [1–4]. Hepatocyte nuclear factor-4α (HNF-4α) is considered to be a critical factor in the development and maintenance of the hepatic phenotype and to be at the top of the transcription factor cascade. The pivotal role of HNF-4α in the maintenance of the hepatic phenotype is highlighted by the severe metabolic defects in mice where HNF-4α was inactivated in the adult liver [5] and by the exceptionally high number of potential target genes revealed by genome-scale target search studies [6, 7]. In the adult human liver, HNF-4α was found to occupy approximately 12% of the genes represented in a 13K DNA microarray and approximately 42% of those bound by RNA polymerase II [7]. HNF-4α has been shown to contribute to human disease, such as, maturity onset diabetes of the young (MODY1), which results from haploinsufficiency of the HNF-4A gene [8, 9]. Previous work by our group has demonstrated that HNF-4α plays a role in the liver’s response to systemic injury and the development of the hepatic acute phase response phenotype [10, 11]. These studies have established that HNF-4α acts as a key regulator in liver function, which raises the importance of understanding how the activity and expression of HNF-4α are regulated.
The regulation of the HNF-4A gene has been well studied. The HNF-4A gene is regulated mainly at the level of transcription. Two main regulatory regions have been identified: the proximal promoter and a distant enhancer are located upstream of the transcription start site [12– 14]. It has been shown that activation of the HNF-4A gene requires the action of HNF-1α and HNF-6 on the proximal promoter, which communicates via a looping mechanism with a distant enhancer bound by HNF-1α, HNF-3β and C/EBPα [12, 13]. In addition, previous studies have shown that HNF-4α activity is also modified by post-transcriptional mechanisms, such as, phosphorylation [15–17], acetylation [18], and protein-protein interactions with other factors or co-regulators [19–21], suggesting that HNF-4α activity can be controlled by multiple pathways.
The functional elements within the 3’-untranslated region (3’-UTR) of various mRNAs have been shown to play an important role in translation and stability of these mRNAs [22] with significant impact on gene regulation and phenotype. However, the potential mechanisms of regulating HNF-4α expression and function by targeting the 3’-UTR of HNF-4A mRNA via microRNAs (miRNAs) remain to be further explored.
MiRNAs are endogenous non-coding RNA molecules that mainly mediate posttranscriptional regulation of gene expression by targeting sequence specific regions in the 3’- UTR of mRNA and repressing their translations into proteins [23, 24]. MiRNAs are the most abundant regulators of gene expression in the human genome, and have enormous regulatory potential. Hundreds of miRNAs have been identified in humans. MiRNAs are predicted to control approximately 30% of the genes within the human genome [25, 26], and participate in the regulation of various biological processes [27]. It has been recently reported that HNF-4α is a gene target of a number of miRNAs [28–31], which provides a potential regulatory mechanism by which HNF-4α function can be modulated by miRNAs. In the present study, we focus our work to validate and analyze the effect of several members of miRNA-34 and miRNA-449 on HNF-4α expression and the ability of HNF-4α to bind and transactivate its target genes. Our results indicate that miRNAs (miR-34a, miR-34c-5p and miR-449a) not only posttranscriptionally regulate the expression of HNF-4α, but also affect HNF-4α binding and transactivation of its target genes by a mechanism of selectively targeting of the HNF-4A 3’-UTR.
2. Materials and Methods
2.1. Cell culture
The human hepatoma cell line, HepG2, was obtained from ATCC (HB-8065), and maintained in Dulbecco’s modified Eagle’s medium, supplemented with 100 units/ml of penicillin, 100 µg/ml of streptomycin and 10% (v/v) heat-inactivated fetal bovine serum (Mediatech, Herndon, VA) at 37 °C in a humidified atmosphere with 5% CO2.
2.2. The miRNA target prediction
The miRNA target predictions were performed using TargetScan version 6.0 (http://www.targetscan.org), and miRanda (http://www.microrna.org) to identify potential miRNA targeting sequences in HNF-4A 3’-UTR (GenBank accession no. NM_000457.3). The parameters were set as default settings.
2.3. Luciferase reporter constructs and transfections
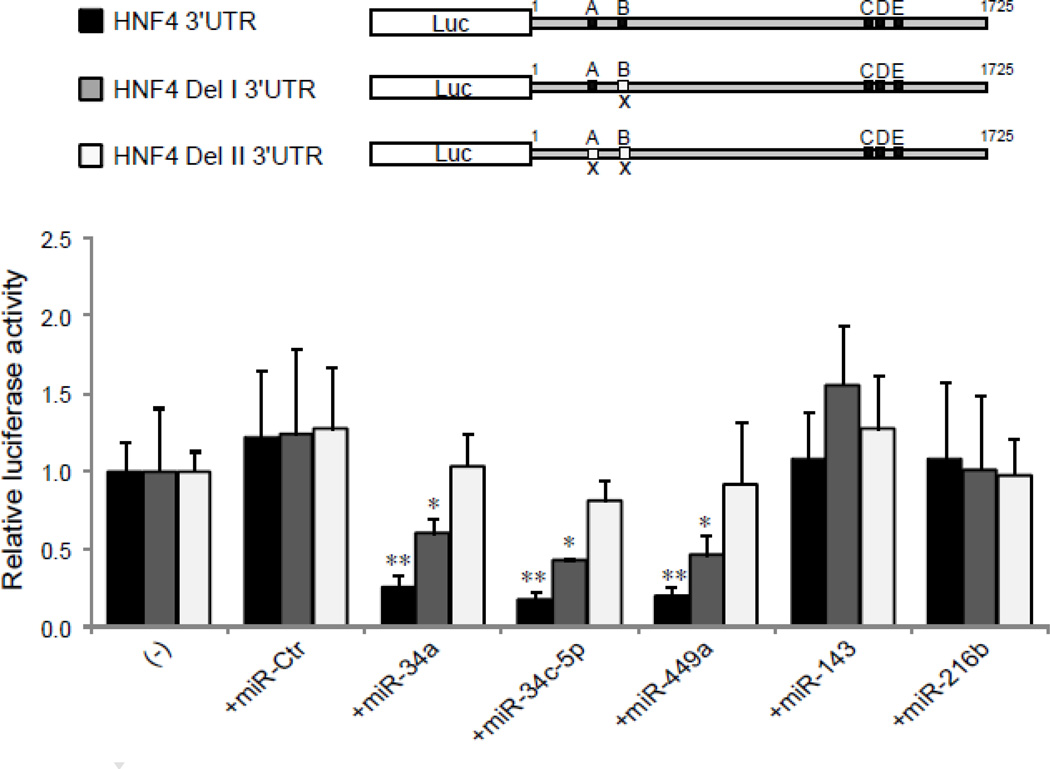
To construct a vector containing the HNF-4A 3’-UTR fused to the 3’ of a luciferase reporter, a dual luciferase pmirGLO vector (Promega, Madison, WI) was used. A 1.7-kb fragment of human HNF-4A 3’-UTR (NM_000457.3) was purchased from SwitchGear (Menlo Park, CA), and cloned into the NheI/XhoI sites of pmirGLO to generate pmirGLO-HNF4-3’UTR. In addition, two deletion variants were generated by PCR. Deletion I (deletion of the seed region for miR-34a/miR-34c-5p/miR-449a in Site B, Fig. 1 and Fig. 3) was created by using primers: 5’- GCTTAATTAAGTTTAAACAAGCCGCTGGGGCTTGGGGGCT-3’ and 5’- CTCGAGGCTAGCGCTTCAACATGAGAAAAGTT-3’. Deletion II (deletion of the seed regions for miR-34a/miR-34c-5p/miR-449a in both Site A and Site B, Fig. 1 and Fig. 3) was generated by using primers: 5’- GCTTAATTAAGTTTAAACAAGCCGCTGGGGCTTGGGGGCT-3’ and 5’- CTCGAGGCTAGCGCTTCAACATGAGAAAAGTTGTCCAAGGCAGTAGAGGTCTCCCC AAGTCAAAGTCTTGTTATCCAGAGCAGGGCGTCAAGGGTGGATGTGGCCCTTAGGC CAT-3’. The two PCR products of deletion I and II were cloned into the PmeI/NheI sites of pmirGLO to generate pmirGLO-HNF4 Del I 3’UTR and pmirGLO-HNF4 Del II 3’UTR, respectively. The basic vector pmirGLO served as an empty vector control. The sequences of all vectors were verified.
Fig. 1. The potential binding sites of miRNAs in the 3’-UTR of human HNF-4A mRNA.
Alignments and locations of the predicted target sequences of miRNAs (miR-34a, miR-34c-5p, miR-449a, miR-143 and miR-216b) within the 3’-UTR of HNF-4A mRNA are shown. The numbering refers to the 5′-end of the 3’-UTR as 1.
Fig. 3. Deletion of the target seed region(s) for miR-34a/miR-34c-5p/miR-449a in HNF-4A 3’-UTR eliminates the repressive effect induced by the corresponding miRNAs.
Three different luciferase constructs are shown: (1) HNF4 3’UTR (black color), a full length of HNF-4A 3’-UTR with the seed sequences of all miRNAs tested, Site A and B for miR-34a/miR-34c-5p/miR-449a, Site C for miR-143, Site D and E for miR-216b. (2) HNF4 Del I 3’UTR (grey color), HNF-4A 3’-UTR carrying only one binding site (Site A), for miR-34a/miR-34c-5p/miR-449a and Site C/D/E. (3) HNF4 Del II 3’UTR (white color), HNF-4A 3’-UTR with two binding sites (sites A and B) deleted for miR-34a/miR-34c-5p/miR-449a. The luciferase constructs with various miRNAs mimics as indicated were introduced into HepG2 cells. The relative luciferase activities were analyzed as described in Fig. 2 legend. *p<0.05 and **p<0.01 versus the cells transfected with luciferase reporter alone (−).
A HNF-4α responsive luciferase reporter construct was created utilizing the promoter region of mouse transthyretin (TTR) gene, a 196 bp DNA fragment, corresponding to −191 to +5 of the mouse TTR gene (nucleotide numbering relative to the transcriptional start site) (GenBank accession no M19524), was cloned into the pGL4.11 [luc2P] vector (Promega, Madison, WI, USA) at BglII/HindIII sites [32].
HepG2 cells, seeded into 48-well plates, were cotransfected with 100 ng of the reporter constructs and 25 pmol of miRIDIAN miRNA mimics or miRNA mimic negative control (Dharmacon, Chicago, IL), or 25 pmol of siDicer I (Applied Biosystems, Foster City, CA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The sequence of miRIDIAN miRNA mimic Negative Control #1 is UCACAACCUCCUAGAAAGAGUAGA. The cells were lysed at 48 h after transfection and analyzed by a dual reporter assay system, (Promega, Madison, WI). Each transfectant was assayed in triplicates. Activity of Firefly luciferase was normalized to Renilla luciferase.
2.4. Total RNA isolation and RT-qPCR analysis
At 48 h post-transfection of miRNA mimics, total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Quantitative RT-qPCR analysis was conducted on StepOnePlus™ Real-Time PCR System. Relative mRNA expression was quantified using the comparative Ct (ΔΔCt) method according to the ABI manual (Applied Biosystems, Foster City, CA). Amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used in each reaction as an internal reference gene. Assays were performed in triplicate. TaqMan probes were used for the human HNF-4A (Hs00230853_m1), transthyretin (TTR, Hs00174914_m1), α1-antitrypsin (α1-AT, Hs00165475_m1), apolipoprotein B (ApoB, Hs00181142_m1) and GAPDH (Hs99999905_m1) from the TaqMan®Gene Expression Assays (Applied Biosystems).
2.5. Isolation of nuclear extracts, Western blotting analysis, Electrophoretic mobility shift assay (EMSA) and Chromatin immunoprecipitation (ChIP) assay
The cells were harvested after 72 h transfection. The experimental procedures for nuclear extracts isolation, Western blotting analysis, EMSA and ChIP assays are given in Li, et al [10], and Wang, et al [11, 20]. The antibody directed against HNF-4α (sc-6556) or β-actin (A5541) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma (St. Louis, MO), respectively.
2.6. Statistical analysis
Data are expressed using the mean and the standard deviation of a minimum of three independent experiments. The statistical significance was evaluated with the Student t test. A p value of 0.05 or less was considered statistically significant.
3. Results
3.1. Bioinformatics analysis reveals HNF-4A as a potential miRNA target
To address whether cellular miRNAs are involved in the regulation of HNF-4α expression, we screened the 3’-UTR of HNF-4A mRNA. Several hundred possible miRNA target sites were identified by the TargetScan database. We focused our study on the miR-34 and 449 family members, because some members of the miR-34 and miR-449 families have shown by others to be able to regulate the expression of HNF-4α [29, 31]. The families of miR-34 and miR-449 are also structurally related and share the same seed sequences. In the HNF-4A 3’UTR, the target sequences (CACUGCC) for the members of miR-34/449 families (miR-34a, miR-34c-5p and miR-449a) are found in two separate locations at positions 164–171 (Site A) and 254–260 (Site B) of the 3’-UTR, respectively. In addition to miR-34/449 families, we also evaluated another two potential miRNAs, miR-143 and miR-216b, which are predicted by two algorithms (TargetScan and miRanda) to be able to interact with the HNF-4A 3’UTR (Fig. 1).
3.2. MiR-34a, miR-34c-5p and miR-449a target 3’-UTR of HNF-4A
To study the role of miRNAs in HepG2 cells, the expression levels of miRNAs were examined by RT-qPCR using specific TaqMan miRNA assays (Applied Biosystems). Both of miR-34a and miR-143 were modestly expressed, but others (miR-449a, miR-34c-5p and miR-216b) were expressed at low or very low levels. After the cells were over-expressed with 100 pmol of miRNA mimics for 48 h, the expression levels of the tested miRNAs all dramatically increased (Supplementary Fig. 1).
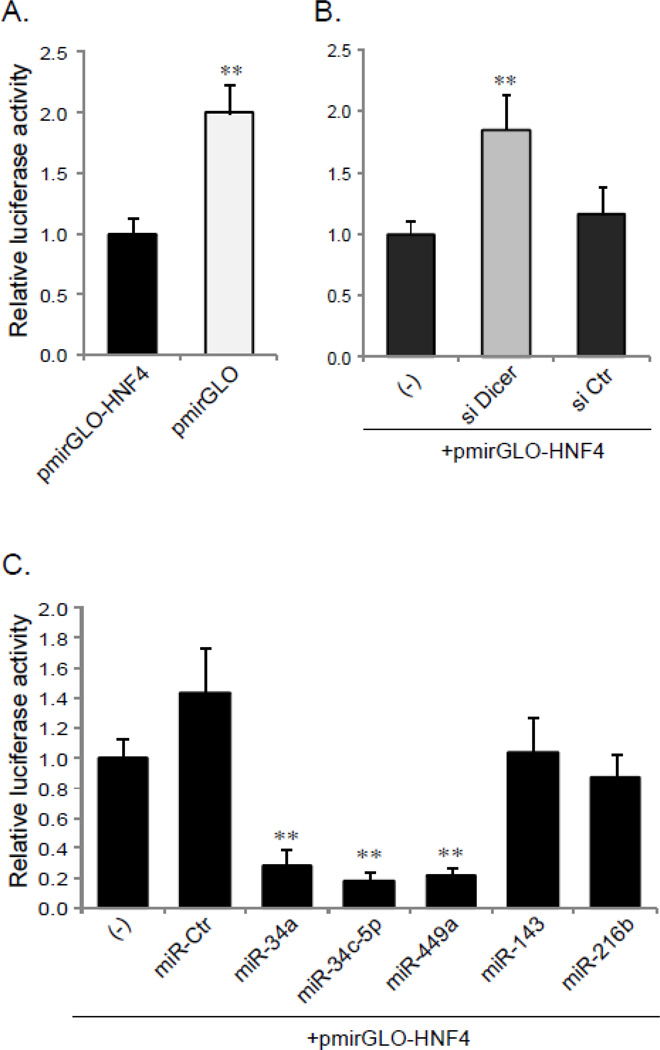
Next, we investigated whether these bioinformatics predicted miRNA targets in the 3´-UTR of HNF-4A affect HNF-4A transcriptional activity. HepG2 cells were transfected with the luciferase reporters containing either a full-length 3´-UTR of HNF-4A or an empty luciferase vector (pmirGLO). As shown in Fig. 2, the activity of the luciferase reporter was significantly higher in the cells transfected with the empty luciferase vector alone (pmirGLO) than with the reporter carrying the HNF-4A 3’-UTR (pmirGLO-HNF4) (p<0.01) (Fig. 2A). The similar result was observed when miRNA processing was impaired by the knock-down of Dicer I with siDicer I, the luciferase reporter activity greatly increased, comparing to the untreated control or siControl (p<0.01) (Fig. 2B). The efficient knock-down of Dicer I was verified by Western blot (Supplementary Fig. 2). Taken together, these data indicate that miRNAs may have a global repressive impact on the 3´-UTR of HNF-4A.
Fig. 2. Analyzing the role of miRNAs in the HNF-4A 3’-UTR with luciferase reporters.
A. HepG2 cells were transfected with pmirGLO-HNF-4A 3’UTR (pmirGLO-HNF-4) or luciferase reporter vector alone (pmirGLO). B. The reporter (pmirGLO-HNF4) was cotransfected into the cells with siDicer I, siControl (siCtr) or alone (−). C. Cotransfections were carried out with reporter (pmirGLO-HNF4) and various miRNAs mimics as indicated. At 48 h after transfection, the cells were lysed and analyzed for Firefly and Renilla luciferase activity using a dual reporter assay system. The data shown are the ratio of the Firefly luciferase activity to the Renilla luciferase activity, and represent the mean ± SD obtained from three individual wells. The experiments were performed in triplicate. **p<0.01 versus pmirGLO-HNF4 (A), and versus the cells transfected with pmirGLO-HNF4 alone (−) (B and C).
To evaluate the direct interaction between specific miRNAs and the predicted binding sites within the 3´-UTR, the pmirGLO-HNF4-3’UTR reporter was cotransfected with the various synthetic miRNA mimics into HepG2 cells. The synthetic miRNA mimics (miR-34a, miR-34c-5p and miR-449a) led to a dramatic reduction in luciferase reporter activity compared to controls transfected only with pmirGLO-HNF4-3’UTR reporter (p<0.01, about 20% of controls). A slight increase in the reporter activity was observed when the negative control miRNA mimic (miRCtr) was introduced, but no significant difference was found between the control (without miRNA mimic treatment) and miR-Ctr (p>0.05). The over-expression of miR-143 and miR-216b did not show any significant influence either positively or negatively on the activity of the HNF4-3’UTR luciferase reporter (p>0.05) (Fig. 2C).
To further validate the role of the miR-34a, miR-34c-5p and miR-449a binding sites in regulating HNF-4α expression, we generated two variants of the pmirGLO-HNF4-3’UTR constructs by deleting either one or both of the binding sites for miR-34a/miR-34c-5p/miR-449a. Deleting one of the seed sequences of miR-34a/miR-34c-5p/miR-449a (position of 254–260 in the 3’-UTR, Site B) partially diminished the repressive effect induced by over-expressed the corresponding miRNAs, but did not abrogate the effect, implying that another site may be also functional. Indeed, when both sites (site A and B, positions of 164–171 and 254–260 within the 3’-UTR) were deleted, the reporter activities were no longer affected by miR-34a, miR-34c-5p or miR-449a. The deletion of either or both of the sites had no effect on the reporter activity by over-expression of miR-143 and miR-216b (Fig. 3). These data suggest that specific members of miR-34 and miR-449 family modulate HNF-4α expression by binding to the specific sites in the 3’-UTR of its mRNA, and that multiple but the specific miRNA target sites in the 3´-UTR are involved in the regulation of HNF-4α expression.
3.3. HNF-4α expression is negatively regulated by miR-34a, miR-34c-5p and miR-449a
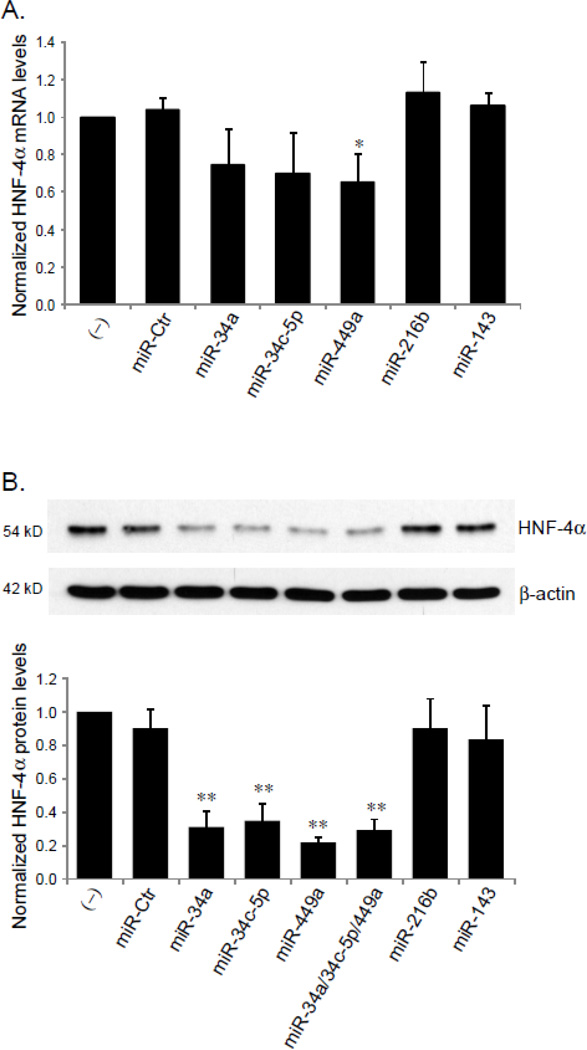
To determine whether specific miRNAs could silence HNF-4α expression, HepG2 cells were transfected with miRNA mimics, total RNA and nuclear protein were isolated at 48 h and 72 h after transfections. The mRNA and protein levels of HNF-4α were quantified by RT-qPCR and Western blot analyses, respectively. The results showed that in the cells transfected with the miR-34a, miR-34c-5p or miR-449a mimic, HNF-4A mRNA level slightly decreased compared to the controls (untransfected or transfected with the control miRNA mimic) (Fig. 4A). In contrast, HNF-4α protein levels were dramatically reduced by the exogenous expression of miR-34a, miR-34c-5p or miR-449a (p<0.01), but not by miR-216b and miR-143 (p>0.05) (Fig. 4B). The protein levels of β-actin remained unchanged in all miRNAs tested (Fig. 4B). Since multiple miRNAs can simultaneously interact with a target mRNA [33], a synergistic effect of miR-34a, miR-34c-5p and miR-449a on HNF-4α protein level was investigated by cotransfection of these 3 miRNAs into HepG2 cells. However, no increase in repressive impact on HNF-4α protein level was detected in combination of the 3 miRNA mimics than seen for the individual miRNA mimic transfections (Fig. 4B). The lack of additional or synergistic effect may be due to competition among miRNAs, as they are predicted to share a common target site.
Fig. 4. The effect of miRNAs on HNF-4α expression.
HepG2 cells, seeded into 6-well plates, were transfected with 100 pmol of miRIDIAN miRNA mimics, negative control miRNA mimic (miR-Ctr) or were untreated (−). At 48 h or 72 h after transfection, mRNA and nuclear extracts were prepared, the expression levels of HNF-4α mRNA (A) and protein (B) were determined by RT-qPCR and Western blot, respectively. The bar graph in panel B summarizes the densitometric analyses of the effects of miRNAs on HNF-4α protein expression normalized by β-actin levels. The experiments were performed in triplicate, and presented as mean ± SD. *p<0.05 and **p<0.01 versus untreated control (−) or miR-Ctr.
3.4. MiR-34a, miR-34c-5p and miR-449a affect HNF-4α binding ability to its target genes
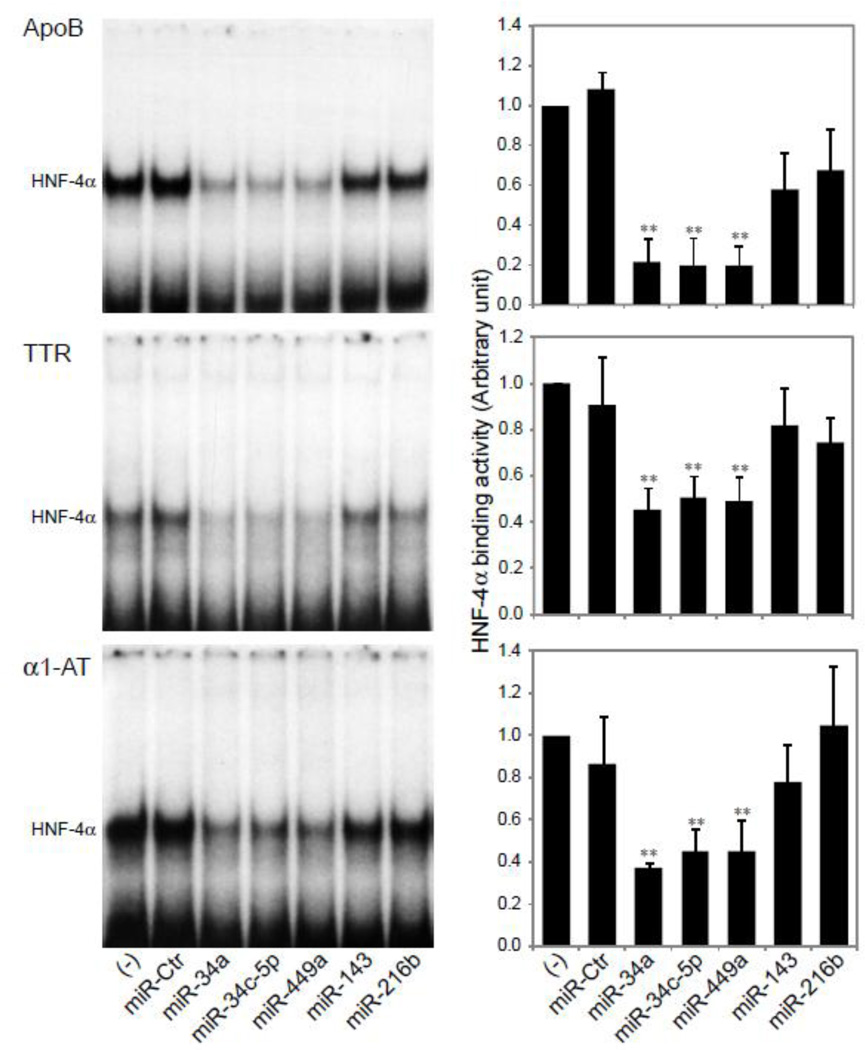
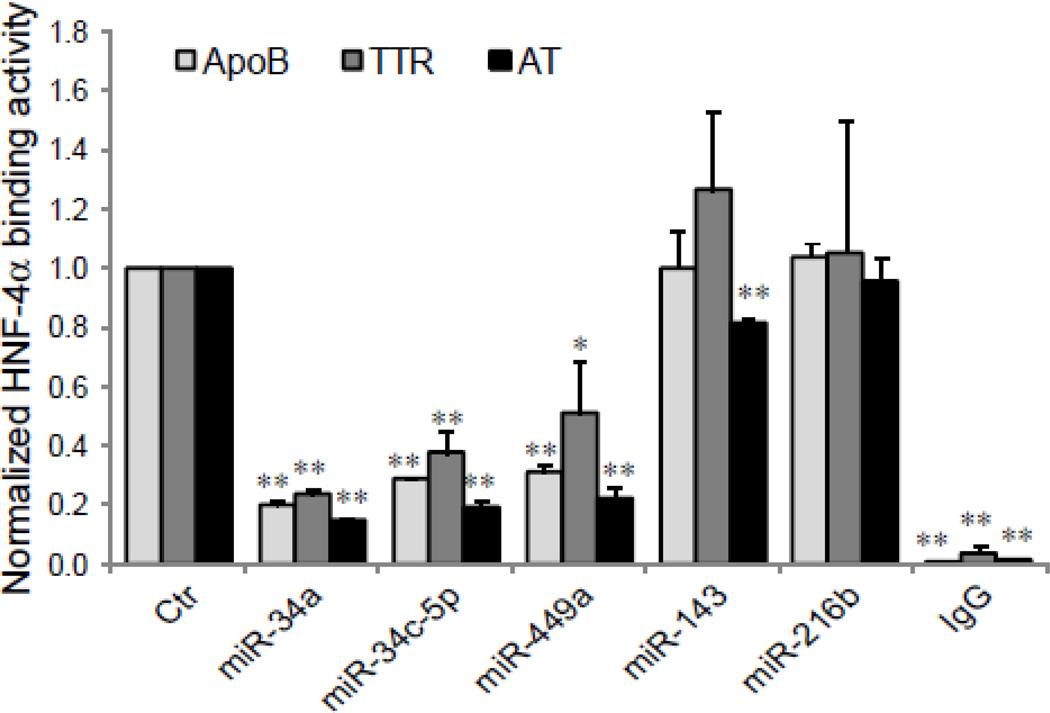
To assess functional consequences of the miRNAs-driven down-regulation of HNF-4α expression, we evaluated HNF-4α-DNA binding activity by EMSA and ChIP assays following the transfection of HepG2 cells with miRNA mimics. Our previous studies [11] [20] have demonstrated that HNF-4α, extracted from nuclear proteins in HepG2 cells, can specifically bind to a oligonucleotide sequence corresponding to the HNF-4α binding site in the promoter region of HNF-4α target genes, transthyretin (TTR), apolipoprotein B (ApoB), or α1-antitrypsin (α1-AT). The results of EMSA (Fig. 5) show that while a strong binding was seen in control cells untransfected or those transfected with a negative control miRNA mimic (miR-Ctr), or with miR-143 or miR-216b, the over-expression of miR-34a, miR-34c-5p or miR-449a resulted in a significant reduction in DNA binding, declining approximately by 20–50% of control levels, which is consistent with the reduction seen in the protein levels of HNF-4α in HepG2 cells treated with these miRNA mimics (Fig. 4B). Utilizing ChIP assays, changes in HNF-4α binding activity affected by over-expressed miRNAs in intact cells were also determined. Similar findings as those seen with EMSA were observed (Fig. 6). Our data from Fig. 5 and 6 suggest that miR-34a, miR-34c-5p and miR-449a specifically suppress HNF-4α binding activity.
Fig. 5. Over-expression of miR-34a, miR-34c-5p and miR-449a reduces HNF-4α binding to its target genes, ApoB, TTR, and α1-AT.
EMSAs were performed to detect the effect of miRNAs on HNF-4α DNA binding activity. HepG2 cells were transfected with miRNA mimics as indicated. Nuclear extracts were prepared. The 32P-labelled oligonucleotide probes based on the HNF-4α-specific binding site in the promoter of ApoB, TTR or α1-AT genes were used [11]. Representative EMSAs of HNF-4α binding are shown. Histograms (right-hand panels) show densitometric analyses of HNF-4α-DNA complexes. Values represent mean ± SD of three separate experiments. ** p<0.01 versus untreated control (−).
Fig. 6. The inhibitory effect of miR-34a, miR-34c-5p and miR-449a on HNF-4α binding activity in intact cells.
HepG2 cells were transfected with miRNA mimics as indicated. The abilities of HNF-4α to bind to the HNF-4α-specific binding site in the promoter of ApoB, TTR or α1-AT gene were determined by ChIP assay with either antibody against HNF-4α or mouse IgG (IgG, negative control). Chromatin-immunoprecipitated DNA was analyzed by qPCR with primers and probes specific for the HNF-4α-binding sites in ApoB, TTR or α1-AT gene promoter [20]. The control samples were set at 1. The results are mean ± SD (n =3) assayed in triplicate by qPCR. * p<0.05 and ** p<0.01 indicate that the value is significantly different from control.
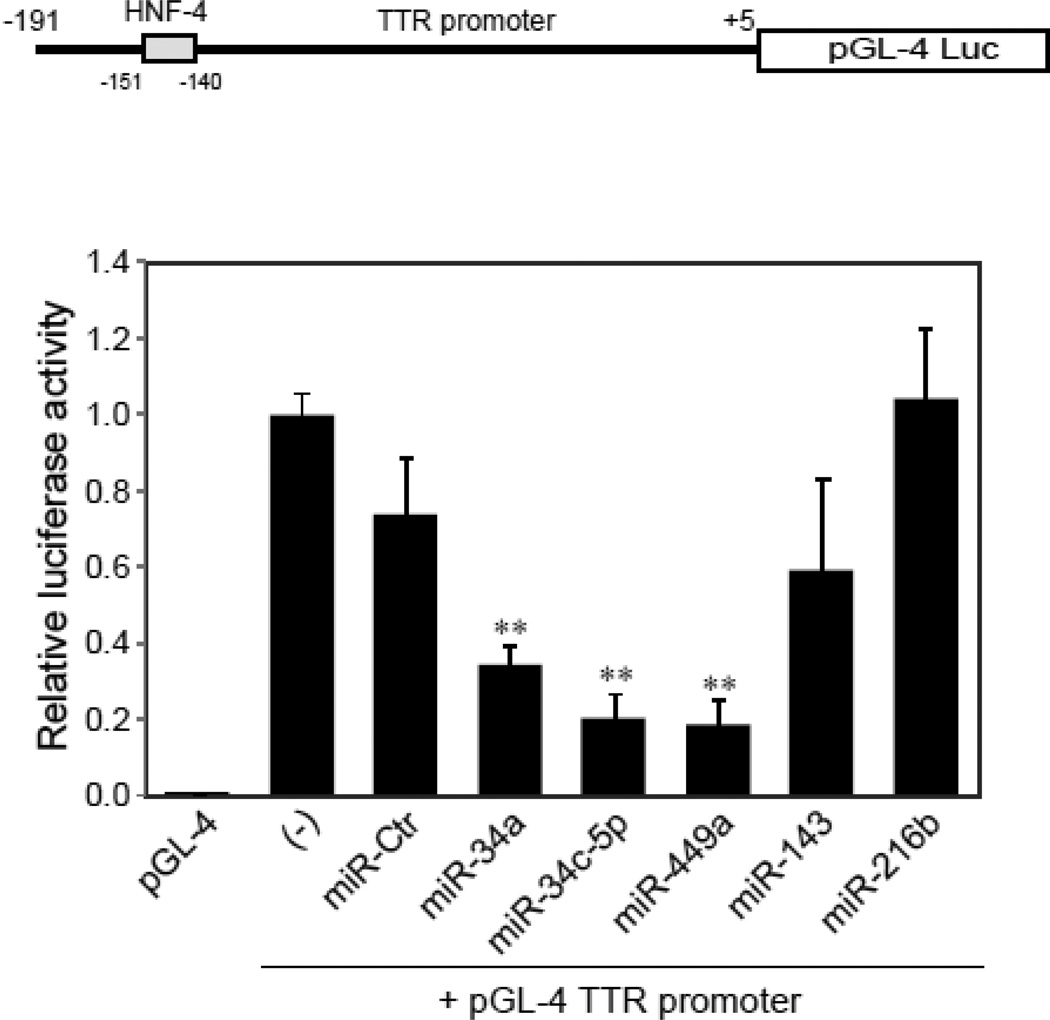
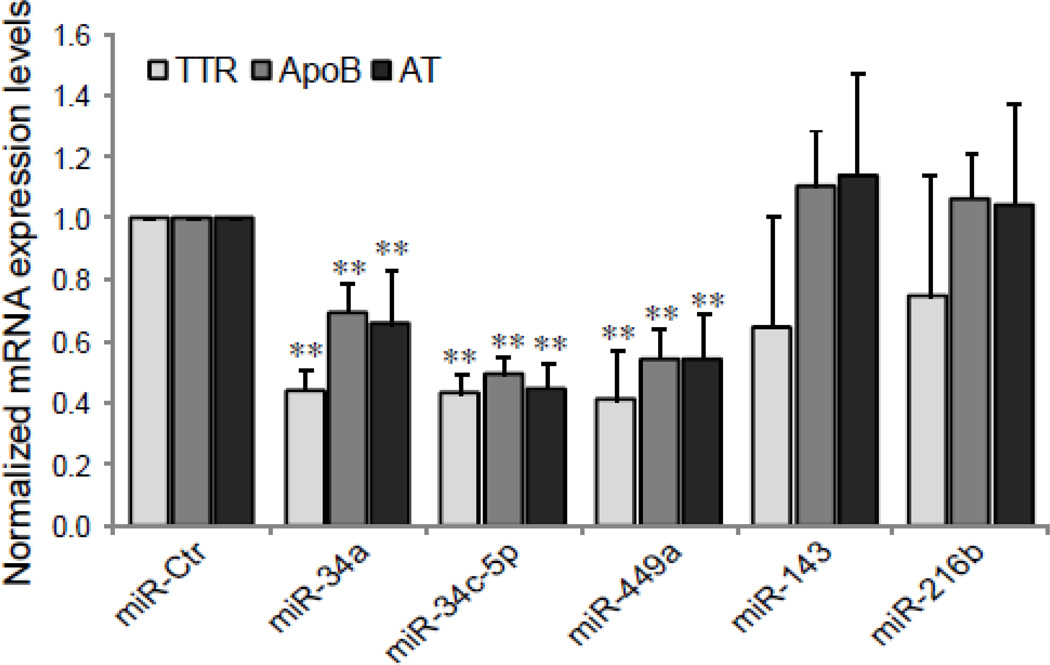
To explore the effect of miRNAs on HNF-4α transactivation of its target genes, we tested whether the over-expression of miRNAs influences the activity of a luciferase reporter driven by the TTR promoter, carrying a HNF-4α binding site. A dramatic decrease in the luciferase activity was found in the cells treated with miR-34a, miR-34c-5p and miR-449a mimics but not with miR-Ctr, miR-143 or miR-216b (Fig. 7). Next, we further examine the impact of miRNAs on the expression of HNF-4α target genes. As shown in Fig. 8, the over-expression of miRNAs (miR-34a, miR-34c-5p and miR-449a) significantly decreases the mRNA expression levels of TTR, ApoB and α1-AT genes compared to controls (p<0.01). Because these HNF-4α target genes have been known [34] to play an important role in the acute phase response, miR-34a, miR-34c-5p and miR-449a may be involved in hepatic acute phase response via regulation of HNF-4α expression.
Fig. 7. The transactivation potential of a HNF-4α target gene, TTR, is inhibited by miR-34a, miR-34c-5p and miR-449a.
The miRNA mimics were cotransfected into HepG2 cells with a luciferase construct containing the promoter region of TTR spanning nucleotides (191 to +5) or empty pGL4.11[luc2P] vector (pGL4). The data shown are the relative luciferase activity, the ratio of the Firefly luciferase activity to Renilla luciferase activity, and represent the mean ± SD of three independent experiments. **p<0.01 indicates a significant difference compared to controls without mimic treatment (−).
Fig. 8. Down-regulation of HNF-4α target genes by miR-34a, miR-34c-5p and miR-449a.
The mRNA expression levels of HNF-4α target genes, TTR, ApoB and α1-AT in HepG2 cells, transfected with indicated miRNA mimics as described in Fig. 4 legend, were examined by RTqPCR. Each column represents the mean ± SD of three independent experiments. The data are relative to that transfected with siControl (miR-Ctr), ** p<0.01.
4. Discussion
HNF-4α is a key member of a complex regulatory network that defines and maintains the hepatocyte phenotype. The central role of HNF-4α in liver function is highlighted by its regulation of a large number of liver specific genes [6, 7]. HNF-4α is also known to be directly and indirectly related with a number of human diseases [9], and plays a role in the hepatic response to injury [10]. The expression and activity of HNF-4α can be regulated at multiple levels. The mechanisms controlling the transcriptional regulation of HNF-4α have been well known, but less has been known about the post-transcriptional control of HNF-4α, especially about a role for miRNA targeting the HNF-4A 3’-UTR in regulating the level and function of HNF-4α. Recently, miRNA has emerged as a regulator of many of the diverse functions of HNF-4α, including modulating the expression of metabolic enzymes and the cell cycle [29], hepatocellular oncogenesis [30], and drug metabolism [31]. Additionally, it has also been reported that the tumor suppressor p53 can repress HNF-4α mediated transactivation [35], and that p53 directly targets members of miR-34 family (miR-34a, miR-34b, and miR-34c)[36, 37], which illustrate another potential and important role for HNF-4α regulation via p53 and miRNA network.
To get more insight into the significance of miRNAs in the regulation of HNF-4α function, especially the effect of miRNAs on the binding activity and transactivation of HNF-4α, we have cloned the human HNF-4A 3’-UTR regulatory region into a luciferase reporter, and analyzed the general and specific impacts of a number of miRNAs on the activity of HNF-4A 3’-UTR. To look for a general role of miRNAs in the 3’-UTR, the elimination of the target region of miRNAs in the 3’-UTR or the reduction of endogenous expression levels of miRNAs was carried out by utilizing the empty reporter (pmirGLO) or selectively knocking-down of Dicer I, a key enzyme that regulates miRNA biogenesis [23], respectively. As shown in Fig. 2, a significant increase in reporter activity was observed from either the absence of the miRNA target region (Fig. 2A) or the depletion of miRNA levels (Fig. 2B), suggesting that miRNAs target the 3’-UTR region of HNF-4A and down-regulate HNF-4α expression.
Several lines of evidence in our study support the hypothesis that specific miRNAs (miR-34a, miR-34c-5p and miR-449a) are able to modulate HNF-4α expression and function. First, the luciferase activity of the reporter carrying a full length of the 3’-UTR of HNF-4A (pmirGLOHNF4-3’UTR) was significantly reduced by over-expression of miR-34a, miR-34c-5p and miR-449a (Fig. 2C). After a single binding site (position of 254–260 in HNF-4A 3’-UTR, Site B) for the miRNAs (miR-34a/miR-34c-5p/miR-449a) were deleted within the 3’-UTR region, the repressive effect induced by the over-expression of these miRNAs was partially reversed. When both binding sites (position of 164–171 and 254–260, Site A and B) were eliminated, the effect was completely lost (Fig. 3). Secondly, the presence of the miRNAs (miR-34a, miR-34c-5p and miR-449a) was able to greatly repress the level of HNF-4α protein with only a small decrease in the level of its mRNA (Fig. 4). These findings indicate that these specific miRNAs (miR-34a, miR-34c-5p and miR-449a) negatively regulate the expression of HNF-4α, and predominantly function at a translational level. Finally and most importantly, we found that the repressive effect of miR-34a, miR-34c-5p and miR-449a on the HNF-4α protein expression led to a significant alteration in the transcriptional control of the HNF-4A target genes, TTR, ApoB and α1-AT (Figs. 5–8). The transcription of these HNF-4A sensitive genes have also been shown to be altered by the injury response [10, 11, 34]. Our previous work [10, 11, 17] has demonstrated that injury results in a significant change in binding activity of HNF-4α without significant alteration in the concentration of HNF-4α protein, and the mechanism for this change involves posttranscriptional modifications, such as phosphorylation, rather than a decrease in the level of HNF-4α protein.
Our experiments have successfully validated the computational predictions of miR-34a, miR-34c-5p and miR-449a targeting of HNF-4A 3’-UTR. However, other predicted interactions for the miRNAs (miR-143 and miR-216b) did not show a significant inhibitory effect on HNF-4α expression in HepG2 cells. These findings indicate that the prediction algorithms need to be used with caution and the predicted miRNA interactions need to be confirmed experimentally.
In summary, the specific miRNAs (miR-34a, miR-34c-5p and miR-449a) negatively regulate HNF-4α expression by directly targeting the 3’-UTR of HNF-4A mRNA. This miRNA-dependent regulation of HNF-4α expression affects transactivation and the activity of downstream HNF-4α regulated genes, which may have important impacts on liver phenotype.
Supplementary Material
Acknowledgements
This work was supported by NIH grant (R01DK064945).
Abbreviations
- HNF-4
hepatocyte nuclear factor (HNF)-4
- miRNA
microRNA
- 3′-UTR
3′-untranslated region
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- EMSA
electrophoretic mobility shift assays
- TTR
transthyretin
- ApoB
apolipoprotein B
- α1-AT
α1-antitrypsin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- 2.Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Jr, Crabtree GR. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 3.Watt AJ, Garrison WD, Duncan SA. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology. 2003;37:1249–1253. doi: 10.1053/jhep.2003.50273. [DOI] [PubMed] [Google Scholar]
- 4.Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Bishop EP, Burke PA. Expression profile analysis of the inflammatory response regulated by hepatocyte nuclear factor 4alpha. BMC Genomics. 2011;12:128. doi: 10.1186/1471-2164-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryffel GU. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 9.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Salisbury-Rowswell J, Murdock AD, Forse RA, Burke PA. Hepatocyte nuclear factor 4 response to injury involves a rapid decrease in DNA binding and transactivation via a JAK2 signal transduction pathway. Biochem J. 2002;368:203–211. doi: 10.1042/BJ20020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Burke PA. Effects of hepatocyte nuclear factor-4alpha on the regulation of the hepatic acute phase response. J Mol Biol. 2007;371:323–335. doi: 10.1016/j.jmb.2007.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 13.Hatzis P, Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol Cell Biol. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailly A, Torres-Padilla ME, Tinel AP, Weiss MC. An enhancer element 6 kb upstream of the mouse HNF4alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors. Nucleic Acids Res. 2001;29:3495–3505. doi: 10.1093/nar/29.17.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang G, Nepomuceno L, Yang Q, Sladek FM. Serine/threonine phosphorylation of orphan receptor hepatocyte nuclear factor 4. Arch Biochem Biophys. 1997;340:1–9. doi: 10.1006/abbi.1997.9914. [DOI] [PubMed] [Google Scholar]
- 16.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Salih E, Burke PA. Quantitative analysis of cytokine-induced hepatocyte nuclear factor-4alpha phosphorylation by mass spectrometry. Biochemistry. 2011;50:5292–5300. doi: 10.1021/bi200540w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 19.Dell H, Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Burke PA. Modulation of hepatocyte nuclear factor-4alpha function by the peroxisome-proliferator-activated receptor-gamma co-activator-1alpha in the acute-phase response. Biochem J. 2008;415:289–296. doi: 10.1042/BJ20080355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JC, Stafford JM, Granner DK. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzybowska EA, Wilczynska A, Siedlecki JA. Regulatory functions of 3'UTRs. Biochem Biophys Res Commun. 2001;288:291–295. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 28.Wirsing A, Senkel S, Klein-Hitpass L, Ryffel GU. A Systematic Analysis of the 3'UTR of HNF4A mRNA Reveals an Interplay of Regulatory Elements Including miRNA Target Sites. PLoS One. 2011;6:e27438. doi: 10.1371/journal.pone.0027438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285:4415–4422. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramamoorthy A, Li L, Gaedigk A, Bradford LD, Benson EA, Flockhart DA, Skaar TC. In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4alpha expression. Drug Metab Dispos. 2012;40:726–733. doi: 10.1124/dmd.111.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Burke PA. Hepatocyte nuclear factor-4alpha interacts with other hepatocyte nuclear factors in regulating transthyretin gene expression. Febs J. 2010;277:4066–4075. doi: 10.1111/j.1742-4658.2010.07802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Baumann H, Gauldie J. The acute phase response. Immunol Today. 1994;15:74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 35.Maeda Y, Seidel SD, Wei G, Liu X, Sladek FM. Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol. 2002;16:402–410. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]
- 36.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.