Abstract
The study aim was to describe the emergency of carbapenem resistance and clonal complexes (CC), defined by multilocus sequence typing (MLST), in Acinetobacter baumannii in a surveillance system for meningitis. Starting in 1996 at an urban setting of Brazil, surveillance detected meningitis by Acinetobacter sp for the first time in 2002. Until 2008, 35 isolates were saved. Carbapenem resistance emerged in 2006, reaching 70% of A. baumannii isolates in 2008, including one colistin-resistant. A. baumannii belonged to CC113/79 (University of Oxford/ Institute Pasteur schemes), CC235/162 and CC103/15. Dissemination of infections resistant to all antimicrobial agents may occur in the future.
Keywords: Acinetobacter baumannii, bacterial meningitis, carbapenem-resistance, multilocus sequence typing, clonal complexes
Acinetobacter baumannii has become increasingly recognized as a cause of multidrug resistant central nervous system infections [1]. A. baumannii clones are classified by multilocus sequence typing (MLST) by protocols hosted at Institut Pasteur (IP, www.pasteur.fr) and the University of Oxford (UO, www.pubmlst.org), and grouped in clonal complexes (CC). Typing an isolate by both schemes is useful as there is no link between IP and UO databases. To date, little is known about the population structure of A. baumannii from cases of meningitis worldwide [2]. In 1996, a hospital-based active-surveillance for bacterial meningitis was established at Hospital Couto Maia, a state infectious disease reference hospital in Salvador, Brazil [3]. The main purpose of this system was to investigate classical pathogens Haemophilus influenzae, Neisseria meningitidis and Streptococcus pneumoniae. Non-classical agents were also sought since all CSF specimens from public hospitals in the city are processed at this hospital. The aim of the present study was to describe the emergency of carbapenem resistance in Acinetobacter spp and the distribution of A. baumannii CCs in isolates recovered from meningitis in this system. A case of culture-proven bacterial meningitis was a patient with typical symptoms and Acinetobacter sp isolated from CSF. From 2002 to 2008, 57 cases of hospital-acquired Acinetobacter sp meningitis were detected; 35 isolates (one/per patient) were saved. Species were identified by sequence analysis of 350-bp rpoB gene fragments [4] and defined by at least 97% similarity with one in a set of reference strains and by BLAST [5].
Antimicrobial susceptibility was determined by disk diffusion [6] for: amikacin, gentamicin, tobramycin, ampicillin-sulbactam, cefepime, ceftazidime, ciprofloxacin, imipenem, meropenem, minocycline, tetracycline, piperacillin-tazobactam, trimethoprim-sulfamethoxazole. Minimum inhibitory concentrations (MICs) of cefepime, imipenem, meropenem and tigecycline were defined by Etest following the manufacturer’s instructions (bioMérieux, Solna, Sweden). Colistin MICs were determined by broth microdilution [7]. Susceptibility to all agents was interpreted as recommended by CLSI [8] except for tigecycline, interpreted as proposed by the US Food and Drug Administration (FDA) for Enterobacteriaceae. Isolates were classified as multidrug-resistant (MDR) or extensively drug-resistant (XDR) [9]. Metallo-β-lactamase production was screened by a double-disk test [10]. The following carbapenemase encoding genes was investigated by PCR: blaOXA-23-like,blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaOXA-143, blaKPC, blaNDM, blaGIM-1, blaIMP-type, blaSIM-1, blaSPM-1, blaVIM-type, blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9 and blaCTX-M-25 [11–15]. Isolates were typed by pulsed-field gel electrophoresis (PFGE) [16] and included within a pulsotype if band profiles had ≤ five differences. UO and IP MLST schemes were performed [17,18]. CCs were formed by STs with five or more identical alleles by goeBURST (goeburst.phyloviz.net). STs and CCs are here referred by UO/IP scheme.
From 2001 to 2008, 1,398 meningitis cases were detected among ~ 3,000 patients, and 931 (67%) were caused by classical agents. Acinetobacter sp, identified for the first time in 2002, increased significantly (R2= 0.94) from 0.9% in 2001–2002 to 4.3% in 2007–2008. Median age of patients was 25 ± 21.3 (range 3–82) years and 71.4% were men. From 57 stored Acinetobacter spp isolates (one per patient), 35 (61%) were available for further characterization. Most (31) were A. baumannii, two Acinetobacter nosocomialis, and one each Acinetobacter ursingii and Acinetobacter genomic species 15TU. Non-A. baumannii isolates were susceptible to all drugs or resistant only to sulfamethoxazole-trimethoprim. All A. baumannii isolates were susceptible to minocycline and tigecycline. One isolate from 2008 was colistin resistant (MIC = 64 mg/L), and susceptible only to minocycline, tetracycline, tigecycline and tobramycin. MICs50/ MICs90 were 32/ >256 mg/L for cefepime, 1/ >32 mg/L for imipenem, 4/ >32 mg/L for meropenem, 0.5/ 1 mg/L for colistin, and 0.38/ 1 mg/L for tigecycline. Thirteen A. baumannii isolates were MDR and fourteen XDR. Carbapenem resistance emerged in May 2006 and became endemic (Figure 1). All carbapenem-resistant isolates carried the blaOXA-23-like gene, and the natural blaOXA-51-like gene, detected in all A. baumannii isolates. blaCTX-M-2 was detected in one MDR A. baumannii from 2004. No other carbapenemase encoding-genes or metallo-β-lactamase production was observed.
Figure 1.
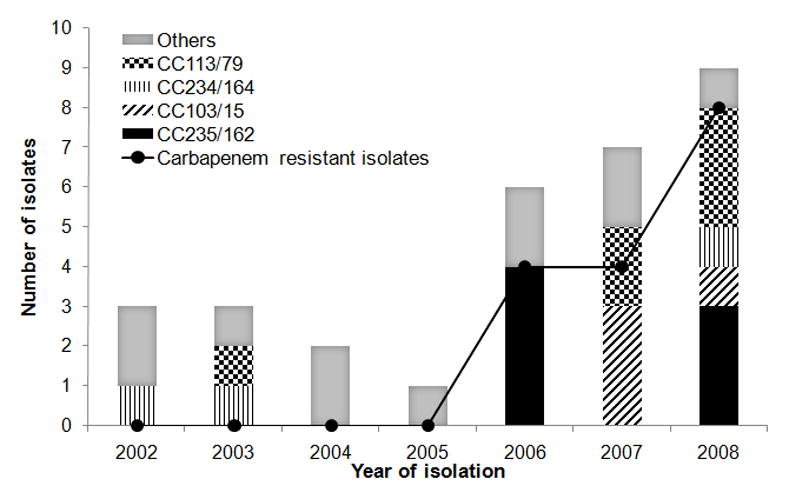
Temporal distribution of Acinetobacter baumannii clonal complexes (CCs) and carbapenem-resistant A. baumannii isolates over seven years of study. “Others” include single pulsotypes and one not typeable isolate not selected for MLST analysis. CCs are described according to University of Oxford / Institute Pasteur schemes.
A. baumannii formed fifteen pulsotypes, and one isolate was not typeable. Nineteen of 30 typeable A. baumannii isolates were included in four pulsotypes (A–D). Fourteen isolates of main pulsotypes were selected for MLST. Ten STs (all new) were identified by UO scheme and five (three new) by IP scheme (Table 1). STs formed four CCs by UO, and three by IP scheme, unrelated to international clones I, II and III. ST164 by IP scheme was not assigned to a CC because this is a double locus variant (DLV) of just one other ST. Each of the seven CCs was isolated over more than twenty months during the six years surveillance (Figure 1). CC113/79 included the colistin resistant isolate (ST237/79). Although 22 of the 57 cases detected in the surveillance system could not have the species determined, we believe all these cases were indeed caused by the genus Acinetobacter since such identification is simple. No further data about Acinetobacter infections in the study hospital are available, however, these infections are likely part of hospital dissemination of this pathogen, previously noted in Brazil [19].
Table 1.
Characteristics of Acinetobacter baumannii isolates from 31 patients with meningitis
| MLST-UO | MLST-IP | Characteristic (number of isolates) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| CC | ST (isolates sequenced) | CC | ST (isolates sequenced) | Pulsotype | blaOXA-23 | Susceptibility phenotype |
| 113 | 237a (2) | 79 | 79 (5) | A (6) | + (3) | COL MIN TET TGC TOB (1) COL MIN TGC TOB (1) MIN TET TGC TOB (1) |
| 258a (1) | + (1) | COL MIN TET TGC TOB (1) | ||||
| 259a (1) | − (1) | AMS COL GEN IPM MEM MIN TET TGC TOB (1) | ||||
| 233a (1) | − (1) | AMS COL GEN IPM MEM MIN TET TGC TOB (1) | ||||
| 235 | 235a (3) | 162 | 162a (4) | B (6) | + (3) | COL MIN TET TGC (1) COL MIN TGC (2) |
| − (1) | AMI AMS COL GEN IPM MEM MIN SXT TGC TOB (1) | |||||
| 415a (1) | + (2) | COL MIN TET TGC (1) COL MIN TGC (1) |
||||
| 416a (1) | 163a (1) | L (1) | + (1) | AMS COL GEN MIN TET TGC TOB (1) | ||
| 103 | 236a (1) | 15 | 15 (1) | C (4) | + (4) | COL GEN MIN TET TGC TOB (1) COL MIN TET TGC (1) COL MIN TGC (2) |
| 234 | 234a (2) | NA | 164a,b (3) | D (3) | − (3) | AMS CIP COL FEP IPM MEM MIN PTZ TET TGC (1) AMS COL IPM MEM MIN TET TGC (1) |
| 232a (1) | AMS CIP COL FEP IPM MEM MIN PTZ TET TGC (1) | |||||
| ND | ND | ND | ND | E – K, M - N (9) | − (9) | Variousc |
| O (1) | + (1) | AMS COL MIN TGC TOB (1) | ||||
| Not typeable (1) | + (1) | AMI AMS CAZ COL GEN MIN SXT TGC TOB (1) | ||||
CC: clonal complex, ST: sequence type, NA: not assigned, ND: not determined, AMI: amikacin, AMS: ampicillin-sulbactam, CAZ: ceftazidime, CIP: ciprofloxacin, COL: colistin, FEP: cefepime, GEN: gentamicin, IPM: imipenem, MEM: meropenem, MIN: minocycline, PTZ: piperacillin-tazobactam, SXT: trimethoprim-sulfamethoxazole, TET: tetracycline, TGC: tigecycline, TOB: tobramycin.
ST described in the present study.
ST164 was not assigned to neither clonal complex because this one is DLV of other published ST;
Includes susceptibility to all drugs (3 isolates); 1 isolate each of AMI AMS CAZ CIP COL FEP GEN IPM MEM MIN PTZ TET TGC TOB, AMS CIP COL FEP IPM MEM MIN TET TGC, COL GEN IPM MEM MIN TET TGC TOB, COL CIP IPM MEM MIN TET TGC TOB, AMS COL IPM MEM MIN TGC and COL IPM MEM MIN TET TGC.
Carbapenem resistance was first detected by the system in 2006 and increased over time to affect 16 of the 31 study A. baumannii isolates, associated with the presence of the blaOXA-23 gene. Alarmingly, 14 of A. baumannii isolates were XDR. High susceptibility to colistin, minocycline and tigecycline was observed. Colistin has been recommended for meningitis by carbapenem resistant A. baumannii [20], but resistance should become increasingly frequent. Use of tigecycline has been described as effective in a few case reports [20], however, the pharmacodynamic profile of this drug does not seem adequate for this purpose [20].
CC113/79, CC235/162 and CC103/15 were important causes of meningitis in the present study and prone to develop resistance to multiple agents. Except for ATCC 17978 strain, no other isolate from patients with meningitis could be related to the CCs of the present study. This finding suggests meningitis is not caused preferentially by isolates with a specific tropism for the central nervous system, but by clones circulating in hospitals.
Acknowledgments
Financial support: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)/ Comissão Fullbright-Brasil, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) of Brazil and Fogarty International Program in Global Infectious Diseases (TW006563) of the National Institute of Health. This publication made use of the Acinetobacter baumannii MLST website (http://pubmlst.org/abaumannii/) developed by Keith Jolley and sited at the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust. We thank also platform Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for coding MLST alleles and profiles and making them available at www.pasteur.fr/mlst.
Footnotes
Transparency declarations
No conflicts of interest to declare.
References
- 1.Kim BN, Peleg AY, Lodise TP, et al. Management of meningitis due to antibiotic-resistant Acinetobacter species. Lancet Infect Dis. 2009;9:245–255. doi: 10.1016/S1473-3099(09)70055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayramoglu G, Kaya S, Besli Y, et al. Molecular epidemiology and the clinical significance of Acinetobacter baumannii complex isolated from cerebrospinal fluid in neurosurgical intensive care unit patients. Infection. 2012;40:163–172. doi: 10.1007/s15010-011-0215-4. [DOI] [PubMed] [Google Scholar]
- 3.Lima JB, Ribeiro GS, Cordeiro SM, et al. Poor clinical outcome for meningitis caused by Haemophilus influenzae serotype A strains containing the IS1016-bexA deletion. J Infect Dis. 2010;202:1577–1584. doi: 10.1086/656778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemec A, Krizova L, Maixnerova M, et al. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU) Res Microbiol. 2011;162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Nemec A, Musílek M, Maixneróva M, et al. Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov. haemolytic organisms isolated from humans. Int J Syst Evol Microbiol. 2009;59:118–124. doi: 10.1099/ijs.0.001230-0. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. Performance standards for antimicrobial disk susceptibility tests; approved standard–eleventh edition, M02-A11. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 7.CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard–ninth edition, M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 8.CLSI. Performance standards for antimicrobial susceptibility testing; twenty-second information supplement, M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 9.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 10.Picão RC, Andrade SS, Nicoletti AG, et al. Metallo-β-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol. 2008;46:2028–2037. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain A, Ferraro MJ, Pino RM, et al. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob Agents Chemother. 2004;48:4438–4440. doi: 10.1128/AAC.48.11.4438-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Mendes RE, Kiyota KA, Monteiro J, et al. Rapid detection and identification of metallo-beta-lactamase-enconding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007;45:544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Revathi G, Bernabeu S, et al. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother. 2011;55:934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XR, Chen JC, Kang Y, et al. Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J. 2012;125:894–900. [PubMed] [Google Scholar]
- 16.Seifert H, Dolzani L, Bressan R, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartual SG, Seifert H, Hippler C, et al. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemec A, Krízová L, Maixnerová M, et al. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J Antimicrob Chemother. 2008;62:484–489. doi: 10.1093/jac/dkn205. [DOI] [PubMed] [Google Scholar]
- 19.Sader HS, Gales AC, Pfaller MA, et al. Pathogen frequency and resistance patterns in Brazilian hospitals: summary of results from three years of the SENTRY antimicrobial surveillance program. Braz J Infect Dis. 2001;5:200–214. doi: 10.1590/s1413-86702001000400006. [DOI] [PubMed] [Google Scholar]
- 20.van de Beek D, Brouwer MC, Thwaites GE, et al. Advances in treatment of bacterial meningitis. Lancet. 2012;380:1693–1702. doi: 10.1016/S0140-6736(12)61186-6. [DOI] [PubMed] [Google Scholar]


