Abstract
Metabolic homeostasis and interventions that influence nutrient uptake are well-established means to influence lifespan even in higher eukaryotes. Until recently, the molecular mechanisms explaining such an effect remained scantily understood. Sirtuins are a group of protein deacetylases that depend on the metabolic intermediate NAD+ as a cofactor for their function. For this reason they sense metabolic stress and in turn function at multiple levels to exert proper metabolic adaptation. Among other things, sirtuins can perform as histone deacetylases inducing epigenetic changes to modulate transcription and DNA repair. Recent studies have indicated that beyond sirtuins, the activity of other chromatin modifiers, such as histone acetyl transferases, might also be tightly linked to the availability of their intermediate metabolite acetyl-CoA. We summarize current knowledge of the emerging concepts indicating close crosstalk between the epigenetic machineries able to sense metabolic stress, their adaptive metabolic responses and their potential role in longevity.
Keywords: Acetylation, HDAC, Sirtuin, Aging, Metabolism, Senescence
Epigenetics, senescence and lifespan
The term epigenetics derives from the Greek epi- (επί- over, above, outer) and genetikos (γενετικός, from γένεσις genesis, origin), and indicates changes in gene expression due to chromatin and histone modifications rather than changes in the DNA sequence. Even though a proper definition is yet to be agreed upon, it is safe to consider “epigenetic” those changes, both heritable (genomic imprinting) and acquired, that can be propagated through meiosis and mitosis [1]. These changes include DNA methylation as well as histone modifications, such as acetylation and methylation.
Modifications of DNA and histone tails are specifically recognized by chromatin-remodeling complexes, such as members of the SWI/SNF family or the Polycomb group (PcG) proteins, and eventually transcription factors that would influence activation or silencing of chromatin regions.
Chromatin modifiers
Chromatin has been canonically divided into euchromatin and heterochromatin, which contain respectively expressed “open” and silenced “closed” regions. More recently it has become clearer that chromatin actually exists in three states: active, poised and inactive [2]. Each state is determined by specific histone modifications, combined in a particular manner, usually referred to as the histone code. Typically, trimethylation of lysine 9 on histone H3 (H3K9me3) acts as a repressive mark and can be found on both regulatory elements and at gene bodies. Also H3K9 dimethylation (H3K9me2) can be present at gene bodies and enhancers, while promoters exhibit trimethylation of H3K27 (H3K27me3) to maintain a repressed status. Together with these histone methylation marks, DNA methylation is found at regulatory elements as well. The active state of chromatin is instead characterized by DNA hypomethylation, di- and trimethylation of H3K4 at the promoters, H3K36 trimethylation (H3K36me3) and H3K79 dimethylation (H3K79me2) at the gene bodies, and H3K4 mono- and dimethylation (H3K4me1 and me2) at the enhancers together with H3K27 acetylation (H3K27ac) [2]. In between these two states, there are regions in the chromatin that are “poised” to be activated, and this is of particular importance during development and tissue differentiation. Poised promoters present both activating and repressing marks, such as H3K4me3, H3K4me2 and H3K27me3 [3]. More recently, poised enhancers have also been identified. These enhancers, particularly important in early developmental stages, are characterized by the presence of H3K27me3 and H3K4me1 [4]. Recent work by Zhao et al. [5] has shown that H3K5 acetylation is another important histone mark for regulating gene expression, marking those genes whose transcription is repressed just before mitosis, but that are needed to be immediately re-expressed once mitosis is complete.
A general concept appears to be emerging, in which it is becoming clear that acetylation and methylation compete for the same residue on histones. Therefore, gene expression of a particular cell at a particular moment results from the concerted action of multiple enzymes, including histone deacetylases (HDACs), acetyltransferases, demethylases and methyltransferases.
Histone deacetylases
Histone deacetylases (HDACs) include 18 family members divided into 4 classes (class I, IIA/IIB, III and IV). Classes I, II and IV comprise the 11 canonical HDACs (HDAC 1–11) which are sensitive to the inhibitor trichostatin A (TSA), while class III includes sirtuins, NAD+-dependent deacetylases that are not inhibited by TSA [6, 7].
HDACs
Canonical HDACs were first identified as histone deacetylases. Their activity is mostly associated with condensed and therefore inactive chromatin. Nonetheless, in recent years many nonhistone targets have been identified. In this context, not all the HDACs are exclusively nuclear. For instance, HDAC6 (class IIB) is normally cytoplasmic, while several other HDACs are both nuclear and cytoplasmic [6]. So far HDAC1, HDAC2 and HDAC3 are the only ones found exclusively in the nucleus. Among the nonhistone targets of HDACs are several important transcription factors, such as p53, Stat3, Gata1 and Hif-1α [6]. It is not surprising that many tumors show an aberrant expression of these enzymes, and to date several HDAC inhibitors (HDACi) are being tested in clinical trials for cancer therapy ([8], see also the section Cancer).
HDACs in metabolism
With regard to metabolism, multiple studies have linked HDACs to glucose metabolism. HDAC1/4/5 inhibit expression of the GLUT4 glucose transporter [9, 10], thereby lowering insulin-induced glucose uptake. HDAC4 and HDAC5 are also involved in glucagon response via recruitment of HDAC3 and concomitant FOXO deacetylation at promoters of gluconeogenic genes [11]. Importantly, inhibition of class II HDACs in mouse models of type II diabetes have been shown to reduce glycemia [11]. Recent studies have also shown that HDAC1 promotes HNF4 expression and FOXO1 activity in the HepG2 liver cell line, in turn increasing expression of gluconeogenic genes [12]. In this context, it remains unclear how these usually “repressing” enzymes cause activation of these genes. In addition, recent studies in cancer cells have shown that HDAC4 deacetylates Hif-1α, increasing its stability and therefore the expression of a subset of Hif-1α target genes [13]. Considering that many glycolytic genes are among Hif-1α targets, it may be possible that HDACs participate in the metabolic changes observed in cancer cells, as described in detail below.
HDACs, senescence and aging
Recent studies have shown that reduced expression of HDAC1 and HDAC2 is associated with senescence of human multipotent stem cells (MSCs) [14], whereas HDAC1, HDAC5 and HDAC6 are downregulated upon aging in hematopoietic stem cells (HSC) [15]. Cellular senescence has been postulated as a potential mechanism underlying organismal aging [16]. However, the role of HDAC1 in senescent differentiated cells seems quite opposite to what has been reported for MSCs and HSCs. In fact, in both human melanocytes and fibroblasts, HDAC1 activity increases upon senescence [17, 18]. Further, overexpression of HDAC1 in human cervical adenocarcinoma cells (HeLa) induces senescence by deacetylating the transcription factor Sp1, which in turn interacts with p300. The p300/Sp1 complex is recruited to genes such as p16INK4, leading to activation of the pRb tumor suppressor gene, in this way triggering senescence [19]. A recent study by Miller et al. [20] showed that in human primary fibroblasts, senescence is also accompanied by an increase in HDAC1 and HDAC2 activity, with a concomitant decrease in H3K56 acetylation. This histone mark appears to play a role not only in replicative senescence, but also in oncogene-induced senescence (OIS), since hypoacetylation of H3K56 is observed upon overexpression of oncogenic Ras in primary fibroblasts. In contrast, H4K16ac is increased during OIS, but is decreased upon replicative senescence. This difference might be explained by the observation that H4K16ac is rapidly lost at DNA damage sites but then it increases during DNA repair, leading the authors to propose that during OIS, cells try actively to repair their DNA, while spontaneously senescing cells fail to do so [20]. This would be in agreement with one of the current theories of aging, which proposes that during aging there is a decline in the ability of these senescing cells to repair DNA damage [21].
Other possible roles have been described for HDACs in aging and senescence, as shown by studies on the regenerative potential of the liver in young and old mice. The liver is a very peculiar organ, with the ability to regenerate itself upon partial hepatectomy [18]. The regenerative potential of the liver decreases during aging, a decline that has been linked to increased levels of the c/EBPα/Brm complex in liver from old mice [18]. This complex includes HDAC1, which causes H3K9 deacetylation on E2F-target gene promoters, silencing these genes and inhibiting proliferation in liver from old mice [18]. These findings could also explain previous observations by Wang et al., who showed that liver-specific HDAC1 overexpression leads to hepatic steatosis, a hallmark of aging in the liver, by modulating expression of lipid metabolic genes [18]. Despite this clear link between HDAC1 and senescence, surprisingly HDAC1 is actually overexpressed in human cancer [22, 23]. One possibility could be that, in the context of tumors, upregulation of HDAC1 leads to the repression of tumor suppressor genes, such as cEBP/α, in turn increasing cell proliferation [18]. Overall, the precise role for HDACs in senescence and cell proliferation appears to be dependent on cell type and tissue context, an important point when considering HDAC modulators as therapeutic agents, as discussed further below.
Sirtuins
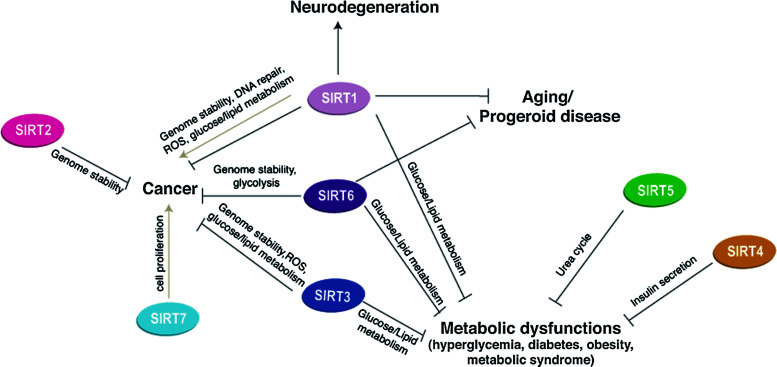
Sirtuins are class III HDACs. There are seven mammalian sirtuins (SIRT1–7), all homologs of the yeast protein Silent Information Regulatory 2 (Sir2) [7]. Sir2 is a NAD+-dependent deacetylase with a crucial role in regulation of both yeast metabolism and lifespan. It was actually first identified in a screening for silencing factors [24], and later shown to be required for extension of replicative lifespan in yeast upon calorie restriction (CR) [25]. Sir2 in Saccharomyces cerevisiae forms an active complex with Sir4 and binds acetylated H4K16 [26, 27]. Following Sir2-mediated deacetylation of H4K16, Sir3 is recruited to this residue, causing compaction and silencing of the region [28]. The seven mammalian sirtuins share partial homology at the catalytic domain. Based on the phylogenetic classification of sirtuins described originally by Frye [29], all the Sir2 homologs fall into four classes (I–IV), with mammalian sirtuins divided as follows: SIRT1, SIRT2 and SIRT3 (class I); SIRT4 (class II); SIRT5 (class III); and SIRT6 and SIRT7 (class IV). The deacetylation reaction catalyzed by sirtuins is NAD+-dependent, and leads the formation of O-acetyl-ADP ribose (AADPR), which can be used as a donor group in ADP-ribosylation reactions [30]. For this reason some of the sirtuins could also exhibit ribosyl transferase activity. Overall these proteins differ among themselves for localization and activity. SIRT1, SIRT2, SIRT3 and SIRT7 in vivo have only deacetylase activity, SIRT4 is mostly an ADP-ribosyl transferase, while SIRT6 exhibits both activities [7]. Strikingly, a recent study has demonstrated that SIRT5 appears to function as a desuccinylase and demalonylase in vitro and probably in vivo [31, 32]. Such activities have never been described before in mammals, and therefore future studies addressing their physiological role will likely draw much attention. In terms of localization, SIRT1, SIRT6 and SIRT7 are mostly nuclear, SIRT2 is cytoplasmic and SIRT3, SIRT4 and SIRT5 are mainly mitochondrial [7]. The role of mammalian sirtuins has been mostly elucidated through generation of knockout mice [33]. Loss of SIRT1 leads in the majority of cases to perinatal lethality with associated retinal, bone and cardiac defects [33]. SIRT2 knockout mice have been shown to develop tumors in several tissues upon aging, in a phenotype linked to genome instability associated with increased mitotic defects [34]. SIRT3 germline knockout mice show metabolic defects in several tissues (e.g. liver and muscle) associated with mitochondrial protein hyperacetylation [33]. Interestingly, conditional knockout mice lacking SIRT3 in either muscle or liver show the molecular but not the metabolic defects observed in the germline knockout mice [35]. Also mice deficient in SIRT4 and SIRT5 are born normally but develop metabolic defects [33]. SIRT6 knockout mice die within 4 weeks of birth due to severe hypoglycemia [36]. SIRT7-deficient mice exhibit cardiac defects and reduced lifespan in a strain-specific manner [33], and recent studies have indicated that SIRT7 deacetylates H3K18, enhancing proliferation in the context of tumor cells [37]. Overall, these observations strongly support a major role for sirtuins in modulating metabolism and potentially lifespan in mammalian organisms (Fig. 1). Given the focus of this review on epigenetics, we discuss below in detail some of the major functions of the two main sirtuins in the nucleus, SIRT1 and SIRT6.
Fig. 1.
Summary of the inhibitory and activating roles of the different mammalian sirtuins in metabolism, cancer, neurodegeneration and aging
Sirtuins in metabolism
SIRT1 is the closest mammalian homolog of ySir2. PGC1α and FOXOs proteins have been extensively characterized as SIRT1 targets. Their deacetylation leads to increased mitochondrial respiration and lipid oxidation through regulation of genes such as ERRα (mitochondria regulatory gene), IDH3α (TCA cycle), Cyt-c and COXVα (respiratory chain genes), MCAD, CPT-1β and pyruvate dehydrogenase kinase 4 (PDK4; fatty acid and glucose utilization) and PCG1α itself [38]. This activity of SIRT1 is particularly relevant when higher energy levels are needed (i.e. exercise) or during CR. It has been reported that SIRT1 protein levels increase upon CR in several tissues [39, 40]. However, in other tissues, such as skeletal muscle, SIRT1 activity increases without noticeable changes in protein content [41–43]. Cantó et al. [41] suggested that hyperactivation of SIRT1 in skeletal muscle in low nutrient/exercise conditions depends on the crosstalk with another key metabolic enzyme, AMPK. Both CR and exercise cause higher consumption of ATP in cells, which determines an increase in the AMP/ATP ratio. AMPK is a holoenzyme that can bind both ATP and AMP, the latter increases its catalytic activity. Therefore, during CR or upon exercise, the higher AMP/ATP ratio activates AMPK, pushing the use of lipids as an energy source in order to satisfy the cellular energy demands. In addition, activation of AMPK causes increased expression of nicotinamide phosphoribosyltransferase (Nampt). Nampt is the rate-limiting enzyme in NAD biosynthesis and its increase leads to higher NAD+ production, which in turn activates SIRT1 (discussed by Posavec et al. in this same review series). In addition, SIRT1 deacetylation and activation of PGC1α increases the biogenesis of mitochondria in muscle [43]. Overall, under conditions of nutrient scarcity or higher energy demands, AMPK and SIRT1 start a positive loop aiming to sustain metabolic adaptations required to survive under these conditions. Another important protein in the response to metabolic stresses is mTOR, a master regulator of cell growth and metabolism. It is well established that AMPK activation leads to mTOR downregulation, in turn shutting down energy-consuming processes, such as protein and glycogen synthesis, both stimulated by mTOR. Recent work suggests that SIRT1 could regulate mTOR as well, inhibiting its activity in a TSC2-dependent manner [44]. Interestingly, recent studies have indicated that SIRT1 stimulates fatty acid oxidation in muscle and adipose tissue in response to activation of the cAMP-PKA pathway, rather than to changes in NAD+ levels [45]. cAMP-PKA activates SIRT1 through phosphorylation of a highly conserved residue—serine 434—and this response is also part of a mechanism underlying temperature control in mice. Indeed, cAMP-PKA-dependent phosphorylation of SIRT1, and therefore its activity, is increased upon cold challenge, and SIRT1 transgenic mice have a better adaptation to cold [45].
SIRT1 exerts profound effects in liver metabolism as well. Under conditions of nutrient stress, SIRT1 deacetylation of PGC1α and FOXO1 causes increased mitochondrial biogenesis and gluconeogenesis, in part through upregulation of the gluconeogenic genes phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) [46]. In addition, SIRT1 increases fatty acid oxidation and inhibits lipogenesis and glycolysis in the liver by modulating expression of another sirtuin, SIRT6 [46]. SIRT1 also deacetylates and inhibits the transcription factor HIF-1α, thereby downregulating the expression of glycolytic genes [46]. Notably, the role of SIRT1 in gluconeogenesis has recently been challenged, with a study supporting negative regulation of gluconeogenesis by SIRT1, secondary to upregulation of Rictor, a component of the mTORC2 complex [47]. Further confirmation of a role of SIRT1 in regulating lipid metabolism comes from the analysis of liver-specific SIRT1 knockout mice. These animals develop increased insulin resistance, decreased fatty acid oxidation and hepatic steatosis, secondary to PGC1α inhibition [46], suggesting that SIRT1 is a potential target against obesity and obesity-associated diseases. In support of this theory, treatment of mice with resveratrol (RSV), a SIRT1 activator, improved their mitochondrial function, reducing the occurrence of diet-induced obesity and its associated insulin resistance [38]. More recently, a clinical study in 11 obese men with no other medical condition showed that 30 days of RSV treatment was sufficient to improve glucose homeostasis, mitochondrial function and systolic pressure [48]. These results support previous studies showing that SIRT1 overexpression prevents diabetes in mice [46]. Even though the clinical study with RSV was performed in a small cohort of human patients, it suggests that sirtuin activators could provide potential therapeutic benefit in the context of diabetes and obesity.
Another nuclear sirtuin with a very strong effect on metabolism is SIRT6. SIRT6-null mice die within 28 days of birth because of severe hypoglycemia [36]. SIRT6 deficiency leads to a dramatic increase in glucose uptake in culture cells as well as in brown adipose tissue and skeletal muscle in mice, explaining the hypoglycemic phenotype in these animals [49]. At the molecular level, SIRT6 functions as a histone H3K9 deacetylase to repress expression of multiple glycolytic genes, acting as a corepressor of the transcription factor Hif1α [49]. Among the genes affected, SIRT6 deficiency causes upregulation of the glucose transporter Glut-1 (responsible for the increase in glucose uptake), phosphofructokinase 1, lactate dehydrogenase A and B (LDHA and B) and PDK1 and PDK4. From a biological standpoint, SIRT6 deficiency triggers a shift towards aerobic glycolysis, with increased production of lactate, and reduced mitochondrial respiration. The metabolic changes observed in SIRT6-deficient cells are reminiscent of the metabolic shift observed in cancer cells, which in fact rely on lactate glycolysis rather than mitochondrial respiration for their ATP production even in the presence of oxygen, the so-called Warburg effect. In agreement with these considerations, recent studies have demonstrated that SIRT6 is indeed a powerful tumor suppressor, acting as a corepressor of both Hif1α and Myc to modulate glycolysis and protein synthesis [50]. SIRT6 has also been specifically deleted in the liver. These animals developed fatty liver with increased triglyceride synthesis, secondary to higher expression of genes involved in lipid metabolism, including fatty acid translocase. As shown previously for glycolytic genes, SIRT6 appears to control H3K9 acetylation at the promoters of these genes. These results are in line with the observation that SIRT6 transgenic mice exhibit lower levels of cholesterol and triglycerides, compared to control animals [51]. Overall, the above results indicate that SIRT6 plays a major role in controlling cellular metabolism, adapting glucose and lipid metabolism to promote survival under conditions of nutrient stress and counteracting the metabolic switch observed in cancer cells. Altogether, these observations support a critical role for SIRT6 in the context of normal as well as transformed cells. Future studies should provide further mechanistic details on its function, in order to translate this information into therapeutic and/or diagnostic tools, as further discussed below.
Sirtuins in aging
The role of the yeast Sir2 protein in lifespan may depend on its ability to compact chromatin, a function of particular importance at subtelomeric regions. Work by Dang et al. [52] showed that H4K16 acetylation plays a key role in regulating lifespan of yeast cells. They reported an age-associated increase in H4K16 acetylation, in particular at subtelomeric regions, which is accompanied by reduction of Sir2 protein levels. More importantly, Sir2 deletion in yeast and H4K16 mutation are epistatic in reducing cellular lifespan. It is important to consider that telomere dysfunction is associated with aging and genome instability both in yeast and higher eukaryotes. Telomeres are repetitive sequences at the end of chromosomes that are actively organized into structures that avoid recognition of such ends by DNA damage signaling pathways. Failure to maintain such structures leads to activation of DNA repair pathways and consequently p53-mediated senescence [53]. Together with changes in H4K16 acetylation, Dang et al. observed an age-dependent reduction of H3K56 acetylation at the same subtelomeric regions. Loss of this histone mark has been previously associated with genome instability [54], supporting the hypothesis that changes in histone acetylation at subtelomeric region might contribute to telomere-mediated aging.
The role that Sir2 homologs play in lifespan modulation in other species is as yet controversial. In Caenorhabditis elegans, overexpression of Sir2.1 has been reported to induce a 15–50 % increase in lifespan [55]. However, recent studies have challenged this notion, indicating that most of the effect was linked to a mutation in another locus. Indeed, when such a mutation was eliminated, the effect of Sir2.1 was either completely lost or minimal [56, 57]. Other studies in C. elegans have shown that CR extends lifespan in worms; this effect depends on Sir2.1 activity since Sir2.1 mutations abolished the CR-mediated lifespan extension [56, 57]. Overall, future studies will be important in deciphering the precise role of sirtuins in modulating lifespan in worms, and whether such discrepancies were due to differences in the assays or the strains utilized.
A similar controversy arises when we consider the effect of dSir2 on lifespan in Drosophila melanogaster. Rosenberg and Parkhurst showed that dSir2 is a HDAC playing an important role in heterochromatic silencing, interacting with Deadpan (Dpn), a member of the family of bHLH euchromatic repressors that control sex determination and segmentation in Drosophila. For these reasons, loss of dSir2 led to sex specificity lethality, and the heterozygous flies showed both segmentation and sex ratio defects [58]. Further, dSir2 also participates in maintenance of heterochromatin silencing through its interaction with the PcG-complex histone methyltransferase E(Z) [7, 59]. However, other groups were unable to demonstrate these effects [60]. It also remains controversial whether dSir2 influences lifespan in flies. One study has shown that overexpression of dSir2 in several fly strains extends lifespan [61], but other groups failed to observe such a phenotype [57, 62].
In mammals, the precise role of sirtuins in lifespan regulation is yet to be fully defined. Although a direct effect for SIRT1 in lifespan extension is yet to be described, SIRT1 regulates multiple age-related pathways that suggests a beneficial healthspan effect for this sirtuin. For instance, SIRT1 deficiency in the liver has been associated with an increased production of reactive oxygen species (ROS) [47]. ROS accumulation leads to oxidation of macromolecules in cells, including DNA, structural protein and lipids. Notably, oxidative damage and impaired redox control are among the factors that has been linked to aging in several organisms, suggesting that SIRT1 might play a role in modulating lifespan through regulation of ROS. In support of this hypothesis, SIRT1 deficiency is associated with increased replicative senescence in human primary fibroblasts [63], and SIRT1 protein levels decrease in cultured cells upon senescence, at least in part due to a negative feedback loop between PPARγ and SIRT1 [64]. A separate study evaluated how SIRT1 positioning on chromatin changes during aging. Oberdoerffer et al. observed that during aging, SIRT1 redistributes on chromatin, moving from its gene targets towards sites of DNA breaks, in this way affecting not only gene expression but also genome stability. Interestingly, the changes SIRT1 undergoes during aging overlap with those occurring during DNA damage, in particular those observed upon oxidative damage [62]. The role of SIRT1 in DNA damage is also supported by other studies showing that SIRT1 is involved in the repair of double strand breaks (DSBs) through deacetylation of NBS1, in nucleotide excision repair through deacetylation of XPA and XPC and in telomere maintenance by modulation of histone modifications [65]. In vivo findings have also demonstrated that SIRT1 overexpression in the context of p53 deficiency in mice improves survival after irradiation and protects against tumorigenesis, supporting a role of SIRT1 in maintenance of genome stability upon oxidative damage [62]. These results suggest a role for SIRT1 as a tumor suppressor acting on multiple levels, although such a role has recently been disputed, as discussed further below.
Another important protein that plays critical roles in oxidative stress responses and aging is the mitochondrial protein p66Shc. Inactivation of p66Shc is associated with increased resistance to oxidative stress, protection from aging-associated vascular disease, and increased lifespan [66]. Recent work has demonstrated that SIRT1-overexpressing transgenic animals are protected from age-related endothelial dysfunctions [66]. Moreover, when diabetes was induced in both control and transgenic mice by administration of streptozotocin, the transgenic mice exhibited less oxidative stress. Both these effects depend on SIRT1-mediated regulation of p66Shc. SIRT1 binds to the promoter of the p66Shc gene, deacetylating histone H3 and downregulating expression of this gene [66]. In the context of vascular diseases, it has also been shown that SIRT1 appears to stimulate angiogenesis by deacetylating Foxo1 and Notch1 [67]. In contrast to these results, SIRT1 overexpression was shown in another study to increase triglyceride and cholesterol synthesis in the liver, sustaining proatherogenic changes in lipid metabolism [68]. Overall, SIRT1 might influence angiogenesis and atherosclerosis acting on numerous substrates, with the precise outcome likely depending on which of the phenotypes dominates over the others.
However, it is important to consider that SIRT1 could also affect age-related phenotypes through modulation of other transcription factors. For instance, SIRT1 deacetylates and inhibits p53—the first target described for this sirtuin—reducing genotoxic stress and DNA damage-induced apoptosis [69]. SIRT1 also binds, deacetylates and inhibits p65/RelA, a component of the NF-κB complex [69], in this way protecting against NF-kB-induced apoptosis and senescence. Considering that the same stimuli that activate SIRT1 (genotoxic and oxidative stress) activate NF-κB as well, it is possible that under stress conditions SIRT1 tries to counteract NF-κB action. Consistent with the proposed stimulatory effect of CR on SIRT1 and its putative beneficial consequences on lifespan [25, 40, 70], CR has also been shown to inhibit NF-κB [71]. However, it remains to be determined whether such an effect on NF-κB is SIRT1-dependent. Recent work from the de Cabo’s laboratory has shown that in Rhesus monkeys CR does not increase lifespan, although it delays the onset of several age-related diseases, suggesting the possibility that CR may extend healthier aging without affect lifespan per se [72]. Whether SIRT1 plays a role in these phenotypes remains to be determined.
Remarkably, the first mammalian sirtuin to be linked to lifespan was SIRT6. In a recent study, Kanfi et al. demonstrated that male transgenic mice overexpressing SIRT6 live longer, but the mechanisms behind these observations remain unclear [73]. Original data suggested a role for SIRT6 in base excision repair, yet how SIRT6 modulates base excision repair at the molecular level remains unclear [36]. More recently, SIRT6 has also been shown to be involved in nonhomologous end joining (NHEJ) and homologous recombination (HR) pathways [70, 74]. These two pathways play critical roles in repairing DSBs [75]. Kaidi et al. demonstrated that SIRT6 is recruited to sites of damage to deacetylate the end-processing factor CtIP, an event necessary for recruitment of downstream effectors. Consistent with this observation, HR and resistance to DSB-generating agents are severely impaired in cells lacking SIRT6 [70]. More recently, it has been shown that SIRT6 overexpression improves not only HR but also NHEJ efficiency, through a mechanism that involves SIRT6-specific mono-ADP ribosylation of PARP1 [74]. Finally, SIRT6 has been implicated in the maintenance of telomere structures in human cells, by keeping H3K9 in a deacetylated state in telomere regions. Cells lacking SIRT6 show abnormal telomere structures, resembling the phenotypes observed in other progeroid diseases, such as Werner syndrome [76].
Even though the precise function of SIRT6 in DNA repair remains to be fully established, it is clear that this chromatin factor plays an important role in multiple DNA repair pathways. Whether the progeroid phenotypes in SIRT6-deficient mice and the extension of lifespan in transgenic SIRT6 animals are due to a protective role of SIRT6 against genomic instability, depend on its function in glucose metabolism, or a combination of both, remains to be established as well. Interestingly, SIRT6 can also mediate H3K9 deacetylation at NF-κB target gene promoters, repressing transcription of genes that have been previously linked to aging [69]. Future work is required to decipher how SIRT6 coordinates in a concerted fashion the metabolic adaptations and DNA repair activities required for cells to survive in the face of nutrient and oxidative stress.
Acetyltransferases
Cellular deacetylases are counteracted by the activity of acetyltransferases (HATs). In mammalian cells, several HATs have been identified, including CBP/p300, GCN5/pCAF and the MYST acetyltransferases such as TIP60. Deletion of GCN5, p300 and CBP in mice is embryonic lethal, indicating a critical role for these enzymes during development. A recent study has shown that deletion of GCN5/pCAF in mammalian cells dramatically reduces H3K9ac, while CBP/p300 deletion affects mainly H3K18 and H3K27 acetylation [77]. On the other hand, in vitro studies have suggested H3K9, H3K27 and H3K36 as targets for p300 [78]; acetylation at these residues induces opening of chromatin, and as such these modifications have been associated with increased accessibility of RNA polymerase and concomitant increased gene expression.
Acetyltransferases in metabolism
Recent studies have demonstrated a critical role for GCN5 in metabolic homeostasis. GCN5 interacts with and acetylates PGC1α [79], counteracting SIRT1 in modulating glucose homeostasis. GCN5 overexpression in hepatocytes reduces the expression of the gluconeogenic genes PEPCK and G6Pase, triggering the same response as observed in SIRT1 knockout hepatocytes [80]. These results suggest that GCN5 can induce metabolic changes depending on nutrient availability. This idea is further supported by recent studies in yeast. When exposed to low-glucose conditions, budding yeast undergo a cycle of growth/stand-by that can be divided into three phases: oxidative (OX), reductive building (RB) and reductive charging (RC) [81]. During the OX phase, cells are actively synthesizing proteins involved in growth—for instance ribosomal genes—and they exhibit high levels of acetyl-CoA and enhanced mitochondrial respiration. During the RB phase, mitochondrial respiration in yeast cells slows down, DNA replicates and the cells divide. Lastly, during the RC phase, mostly survival and stress genes are expressed. In other terms, these cells undergo phases of growth (OX), cell division (RB) and quiescence (RC). Each of these phases is characterized by a specific gene expression profile. Interestingly, the GCN5/SAGA complex is essential to trigger the expression of the growth-related genes in the OX phase [81]. Analysis of histone acetylation throughout the cell cycle has revealed a peak of histone acetylation during the OX phase, which persists into the RB phase. Acetylation of these histone residues occurs in growth-related genes, which were the ones for which the GCN5/SAGA complex peaked [81]. These observations demonstrate a strong link between mitochondrial metabolism and cell growth mediated by epigenetic mechanisms.
Acetyltransferases in aging
As has been shown for HDACs, the expression of HATs also changes upon senescence. In fact, levels of p300 decrease in human melanocytes as the number of cell passages increases [82]. In this context, downregulation of p300 leads to stable repression of the cyclin E promoter, with consequent induction of senescence. Interestingly, it has also been shown that p33ING levels are increased in high passage cells, leading to the binding of p300 to p53 [83]. p300 acetylates and activates p53, thereby inducing replicative senescence. Similarly, CBP also activates p53 by acetylation, a step required for OIS [84]. Taken together, these observations reinforce the concept that a fine balance between these chromatin enzymes is essential for controlling cellular homeostasis and growth.
Histone acetyltransferases are also involved in DNA repair and genome stability maintenance. Das et al. observed increased CBP/p300-dependent H3K56 acetylation at site of DNA breaks [85], although Miller et al. reported an opposite observation [20]. The dynamics of H3K56 acetylation upon damage remains still to be fully elucidated. An intriguing hypothesis is that changes in levels and distribution of H3K56ac are linked to repositioning of nucleosomes upon damage in order to prevent further breakage and to allow repair. For example, GCN5-mediated H3K56ac has been reported to be an essential step in nucleosomes assembly during DNA replication, and efficient nucleosome reassembly is required for maintaining genome stability [86]. GCN5 maintains genome stability also by stabilizing TRF1, a component of the shelterin complex, thereby contributing to telomere protection [87]. As we discuss further below, maintenance of telomere structure is a crucial feature for preventing genome instability and senescence. Work on yeast supports the idea that GCN5 may link metabolism, DNA repair and lifespan through modulation of the retrograde response [88]. Whether similar mechanisms take place in higher eukaryotes remains to be determined.
Although a direct role for the MYST family of acetyltransferases in aging remains to be established, these enzymes have also been involved in control of DNA repair and replication processes, and defects in these enzymes have been associated with cancer [89]. For instance, Tip60 controls the expression of several oncogenes and tumor suppressors and is a crucial player in the repair of DSBs. Tip60 in fact acetylates and activates both p53 and ATM, enhancing the DNA damage signaling cascade [90]. Recent studies indicate that Tip60 also mediates the recruitment of ribonucleotide reductase to sites of damage, inducing a local increase in dNTPs pool, required for efficient repair [91].
Methyltransferases
Another important epigenetic modification, which can occur both on DNA and proteins, is methylation. As acetylation, DNA and histone methylation play critical roles in multiple biological processes, including DNA replication, DNA repair, gene transcription and chromatin accessibility. This modification is regulated by a fine balance between the activities of methyl transferases and histone demethylases (and the newly emerging group of hydroxylases/carboxylases). Emerging evidence supports a role for these enzymes in metabolism, senescence and aging, as described elsewhere in this review series (see Chiacchiera et al.).
Epigenetic cross-talk and regulation of senescence
As discussed above, gene expression or silencing is influenced by multiple epigenetic modifications, including histone marks and DNA methylation. Although we treat them separately in the previous section, it is clear that chromatin-remodeling enzymes act in a concerted manner to activate or repress gene transcription. Indeed, these enzymes form part of large multiprotein complexes, such as NuRD and NoRC [92, 93]. These complexes are usually recruited to chromatin through recognition of particular histone and DNA methyl marks already in place. Once bound, the different complexes might either maintain the status quo or determine further changes in those epigenetic marks, thereby activating or repressing gene transcription. In order to better understand the dynamics and the interactions between these different modifications, we consider a model gene locus that plays a key role in cellular senescence: the INK4-ARF locus (Fig. 2).
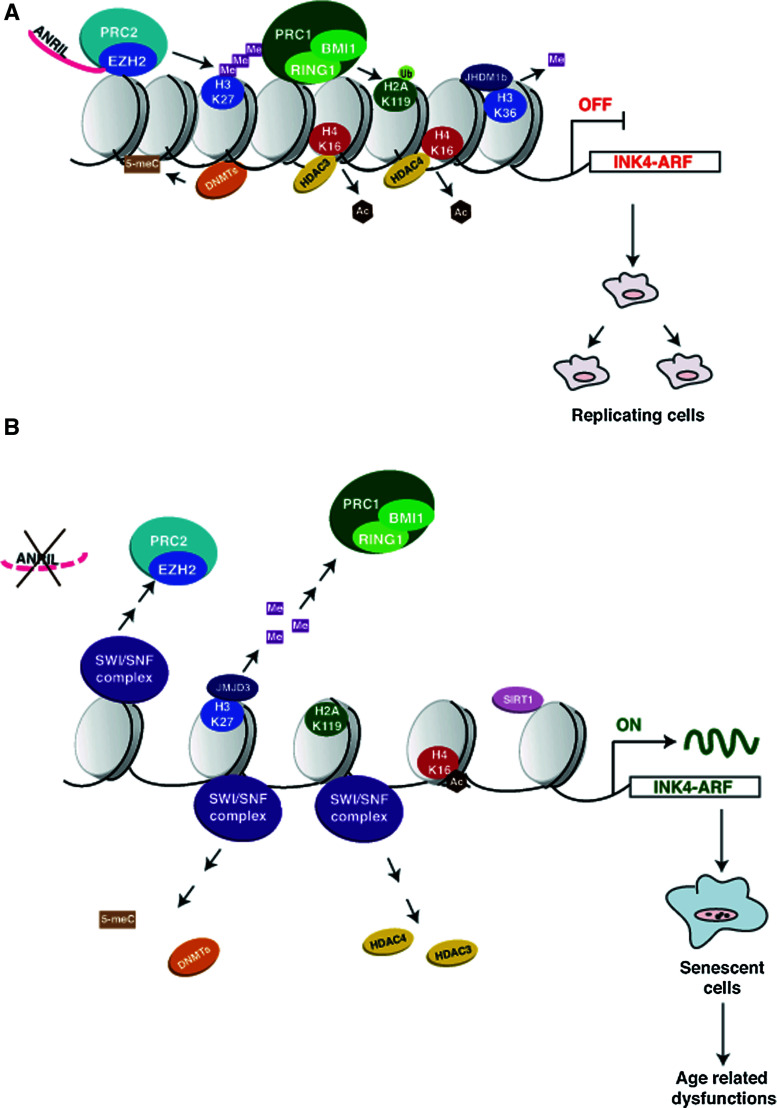
Fig. 2.
Epigenetic modifications at the INK4-ARF locus. In replicating cells (a) ANRIL noncoding RNA mediates PRC2 recruitment to the gene locus. The PRC2 subunit EZH2 trimethylates H3K27. H3K27me3 recruits PRC1, which in turn ubiquitinates H2AK119. Other factors recruited to the INK4-ARF locus are DNMTs (which methylate the DNA), HDAC3, HDAC4 (deacetylate histones) and JHDM1b (demethylates H3K4). These histones and DNA marks are necessary to maintain the locus in a repressed state. In senescent cells (b) ANRIL degradation and the recruitment of the SWI/SNF complex displaces the DNMTs, the HDACs and the PRC1 complex. JMJD3 is recruited at the site and demethylates H3K27, removing the signal for PRC1 recruitment and therefore H2AK119ub is lost. SIRT1 is recruited, but its action is not fully defined. The net result is the activation of p16INK4a, p15INK4b and p19ARF expression and induction of senescence, which at the organismal level will lead to age-related dysfunction
The INK4-ARF locus encodes three proteins: p16INK4a, p15INK4b and p19ARF. p16INK4a and p15INK4b are cyclin-dependent kinase inhibitors of CDK6, essentially blocking cell cycle progression by preventing phosphorylation of pRB; p19ARF sustains cell cycle arrest and apoptosis mainly by stabilizing p53 [94]. The expression of these three proteins increases during senescence—both physiological and OIS—triggering cell cycle arrest as a protective mechanism against stress [95]. Considering the importance of this locus, it is not surprising that it is tightly regulated during cellular lifespan, being one of the most common silenced loci in cancers [96].
In normally dividing cells, the INK4-ARF locus is usually repressed due to binding of the Polycomb repressive complex 2 (PRC2) at the locus. PRC2 contains the lysine methyltransferase EZH2, which maintains high levels of H3K27me3. H3K27me3 in turn is recognized by the Polycomb repressive complex 1 (PRC1), which binds to this modified residue with high specificity. PRC1 contains the RING protein RING1b and BMI1, both ubiquitin ligases that monoubiquitinate H2A on lysine 119. Together with PRC1 and PRC2, DNMTs are also recruited to the locus to methylate DNA. Overall, H3K27me3, H2AUb and DNA methylation provide a stable, heritable lock, maintaining the INK4-ARF locus in a repressed state for as many generations as necessary. Further studies have shown that HDAC3 and HDAC4 and the histone demethylase JHDM1b also participate in silencing this locus by guaranteeing, respectively, deacetylation of H3 and H4 and demethylation of H3K36 [97, 98]. While repression of this locus is faithfully maintained through many cellular divisions, multiple signals (oxidative stress, oncogenes, replicative stress) can cause de-repression of the INK4-ARF locus, with concomitant cellular senescence. One of the first steps in this process is displacement of the PRCs, a process mediated in part by downregulation of ANRIL, a noncoding RNA important for the recruitment of the PRC components to the INK4-ARF locus. At the same time, the SWI/SNF chromatin remodeling complex is recruited, which helps evict the PRC from the locus [94]. Removal of the PRC complex in turn induces CpG demethylation and acetylation of H4K16, an active chromatin mark. In addition, the H3K27me3 demethylase JMJD3 is activated, removing this key PRC recognition mark, further enhancing activation of the locus (Fig. 2) [94].
Notably, the SIRT1 deacetylase also binds to this locus, but it appears to play a dual role: it represses p16INK4a expression while activating p19ARF [99, 100]. How SIRT1 causes such phenotypes remains unclear. Original data on SIRT1 deletion show that knockout mouse embryonic fibroblasts compared are more resistant than wildtype cells to replicative senescence but not OIS [100]. This is supported by the observation that telomerase immortalized human fibroblasts and mouse HSCs also exhibit increased growth capacity upon SIRT1 silencing, through a mechanism involving the nutrient stress kinase AMPK [101]. Even though the above results would suggest that reduced levels of SIRT1 would be beneficial to avoid cellular senescence, other results are inconsistent with such an effect. In fact SIRT1 transgenic mice, compared to controls, exhibit a lower degree of age-associated phenotypes, such as osteoporosis, impaired glucose tolerance and accumulation of DNA damage. These phenotypes are at least in part ascribable to lower levels of p16INK4a [102], suggesting that SIRT1 repression of p16INK4a might dominate over the other functions. Consistent with these results, overexpression of SIRT1 has been shown to rescue PML-mediated premature senescence in mouse cells, through deacetylation and inactivation of p53 [103]. In addition, SIRT1 may also inhibit replicative senescence by deacetylating H1, reducing the formation of heterochromatin [104] and through activation of the Erk-S6K1 signaling pathway [99]. Such discrepancies might reflect different experimental conditions; however, they raise a note of caution with regard to modeling beneficial effects of putative sirtuin activators.
The accumulation of senescent cells in tissues and organs is now considered an integral part in the mechanisms contributing to organismal aging, as we discuss above. Most importantly, it appears to also play a role in several degenerative diseases associated with aging [105]. Strikingly, a recent study has shown a marked delay in the onset of age-related phenotypes, such as lordokyphosis and cataracts, in a progeroid murine model in which p16INK4a-expressing cells were selectively killed [106]. Further, clearance of senescent cells caused a beneficial effect even in mice that already showed age-related dysfunction. This study strongly supports the causal link between senescence and age-related diseases, and further suggests that modulation of p16INK4a could be sufficient to reverse some of these degenerative phenotypes.
Chromatin topology and aging
Beyond epigenetic modifications, chromatin could also be classified as euchromatin and heterochromatin, historically considered as the open/active and the close/repressed configurations, respectively. From a structural point of view, “euchromatin” is formed from nucleosomes assembled in the familiar “beads-on-a-string” configuration. Each nucleosome encompasses 147 bp of DNA wrapped around an octamer of histones, in turn formed by two H3/H4 dimers and two H2A/H2B dimers. In between nucleosomes, linker DNA adds up to 80 bp. The addition of other proteins, including the linker histone H1, enables the primary conformation to coil into 30-nm fibers or a higher-order, compacted conformation, as seen in heterochromatin [107]. In order for cells to either duplicate or transcribe their DNA, histones need to be evicted from the DNA to allow the passage of the replication and transcription machineries and then promptly reassemble on DNA. This requires a very fast and effective “histone exchange” process. Even though the precise mechanisms governing this process remain to be elucidated, several lines of evidence suggest that such histone dynamics may play a role during aging as well, as discussed below.
Histone expression and aging
Recent studies have shown that during aging, as well as during chronic exposure to damaging agents, histone levels in yeast are reduced [108–110]. In addition, deletion of ASF1, a histone chaperone involved in histone exchange, reduces replicative lifespan in yeast cells. A similar effect has been observed in cells lacking H3K56 acetylation, a chromatin mark in newly synthesized histones. This mark is recognized by ASF1, allowing this chaperone to transfer H3-H4 dimers to the CAF1 chaperone, which in turn incorporates them into nucleosomes [108]. Cells lacking ASF1 or with impaired acetylation of H3K56, exhibited lower levels of histones than control cells, suggesting that deficiency in total histone levels is sufficient to drive aging. Consistent with this idea, overexpression of histones cells was sufficient to dramatically extend lifespan in yeast [108], clearly indicating that at least in this organism, histone levels play a crucial role during aging.
Similar to what happens in yeast, human primary fibroblasts exhibit reduced histone levels following replicative senescence [109]. This process is also accompanied by significant changes in histone modifications. Indeed, H3K56ac, H3K9me2, H3K9me3 and H4K20me3 levels decrease, whereas H3K9me1 and H4K20me2 levels increase in cells undergoing senescence [109]. Due to these changes, overall nucleosome number decreases [111], causing major gene expression changes and increased susceptibility to DNA damage, explaining at least in part why senescent cells exhibit higher levels of pH2AX and increased genomic instability. Moreover, DNA damage itself can also affect histone expression, generating a vicious cycle with cellular senescence as the endpoint, as we discuss below [109]. Recent studies have shown that during aging, learning decline is associated with a specific decrease in H4K12 acetylation. Remarkably, the phenotype is rescued upon intrahippocampal injection of the SAGA acetyltransferase [112]. These compelling results suggest the tantalizing possibility that correcting epigenetic changes may suffice to restore function in the aging brain. Overall, these studies demonstrate that maintenance of histone content in a cell is an actively regulated process that plays a critical role in modulating proper gene expression and maintaining genome integrity, a process that seems to go awry with age.
Senescence associated heterochromatic foci
In 2003, Narita et al. observed that human senescent fibroblasts accumulate distinct heterochromatic structures, termed senescence-associated heterochromatic foci (SAHF). SAHF exhibit highly methylated DNA, binding of the heterochromatin protein HP1, hypoacetylated histones and high levels of H3K9me3 and H4K20me3 [94, 113]. Most importantly, these foci present a very peculiar histone variant: histone macroH2A. Deletion of macro H2A in senescent cells induces disappearance of SAHF. Work by Zhang et al. has shown that the histone chaperone ASF1 and HIRA are required to form SAHF and that these structures are not simply a consequence of senescence. Notably, cells expressing mutant forms of HIRA and/or ASF1 still undergo senescence, despite the fact that SAHF are not formed [113]. It was thought that SAHF formation contributes to inducing and/or maintaining senescence, activating pRB-mediated silencing of E2F genes [113, 114]. However, recent work indicates that SAHF are not observed in all cell types upon senescence. Further, their appearance depends on the type of insult triggering senescence [115]. For example, MRC5 fibroblasts develop SAHF both in OIS and DNA damage-induced senescence, while BJ fibroblasts exhibit SAHF only following oncogene insult. Taken together, these studies indicate that SAHF is an integral characteristic of at least some forms of senescence, but their precise role in this process, and whether they play a role during normal aging remain to be determined.
Role of telomeres structure in senescence
Telomeres are segments of repetitive DNA at the extremities of each chromosome. During S phase, the DNA polymerase machinery can fully replicate the leading DNA strand, while the lagging strand poses a serious challenge. In order to prevent, or at least slow down, telomere shortening, cells take advantage of telomerase, an enzyme capable of adding a repetitive sequence at the end of the chromosome using RNA as a template [116]. In 2003, Masutomi et al. reported that growing and presenescent fibroblasts exhibit telomerase activity, while senescent cells show almost undetectable activity [117]. These results prompted these authors to propose that incomplete replication at the end of the lagging strand, in association with a very low telomerase activity, could cause telomeres shortening in human cells, activating a checkpoint that causes replicative senescence [118]. These studies have now been extended to clearly demonstrate a critical role for telomere structure in preventing senescence. In this context, telomeres are fully covered by the shelterin complex [53], which mainly avoid telomeres being recognized as DNA breaks, preventing in this way the activation of the ATM signaling cascade. Indeed, mice with any of the shelterin complex proteins deleted exhibit signs of tissue degeneration and neoplastic transformation, even in the presence of telomeres of normal length, suggesting that persistent activation of DNA damage signaling pathways, rather than telomere length per se, is crucial in promoting senescence and transformation [119].
From an epigenetic point of view, telomeres are characterized by heterochromatic markers: CpG methylation, H3K9me3 and H4K20me3. Recent studies indicate that during aging, DNA hypomethylation and changes in levels of deacetylases and methyltransferases can affect these marks, therefore leading to telomere dysfunction [94, 120]. Furthermore, telomere shortening has also been associated with downregulation of H3 and H4 histone levels and destabilization of the histone chaperone ASF1 [109]. Reduction in ASF1 levels causes chromatin assembly defects, which in turn will further reduce H3 and H4 levels, starting a vicious cycle that could affect telomere structure [109]. When telomere shortening passes a certain threshold, it activates ATM signaling, leading to cell cycle arrest and cellular senescence. Interestingly, recent observations indicate that telomere dysfunction also influences the metabolic and mitochondrial changes that usually accompany the organismal decline observed with aging. Indeed, ATM-mediated p53 activation in telomerase-deficient mice leads to repression of PGC1α, with a concomitant decrease in mitochondrial biogenesis and function, decreased gluconeogenesis, cardiomyopathy, and increased ROS [121]. This contradicts the findings of previous studies showing a positive action of p53 on mitochondrial respiration [122]. It is possible that the effects of p53 on mitochondrial respiration depend on the specific context of the cell and interaction of the different signaling pathways.
Overall, it is clear that telomere biology plays a key role in cellular senescence and as such it represents an essential component of human aging. Remarkably, the progeroid phenotype observed in mTERT-deficient animals is basically reversed by inducing the re-expression of mTERT in vivo [123]. This study provides evidence that aging may actually be a reversible process and could represent a milestone in the field of regenerative medicine.
Mitochondrial metabolism affects histone acetylation
Acetylation of histones relies on the availability of acetyl-CoA as metabolite donor, as well as on the presence of NAD+ as cofactor for deacetylases. Given that these factors play critical roles in cellular metabolism and their availability is subject to nutrient availability, food intake and cellular energy status can directly impinge on epigenetic processes, as we discuss in this section.
Acetate freely diffuses through cell membranes. Therefore in order to maintain a discrete acetate pool in different cellular compartments, it is necessary to conjugate acetate to coenzyme-A to form acetyl-CoA. The conversion of acetate to acetyl-CoA is catalyzed by acetyl-CoA synthetase (AceCS in mammalian cells, ACS in bacteria), while the opposite reaction requires acetyl-CoA hydrolase. AceCS is present in two isoforms in mammalian cells: cytosolic AceCS1 and mitochondrial AceCS2. Both enzymes are regulated by acetylation and are targets of sirtuins [124, 125]. Acetyl-CoA in the mitochondria is mainly generated from pyruvate as part of glucose metabolism, but under fasting or hypoxic conditions citrate is generated from lipids or glutamine rather than glucose [126, 127]. Once acetyl-CoA is formed, it will enter the TCA cycle to be converted into citrate. Citrate will provide either the first step in the TCA cycle to form ATP, or else it will be shuttled outside the mitochondria, where ATP-citrate lyase (ACL) will convert it back to acetyl-CoA for lipid synthesis in the cytoplasm [128].
The acetyl-CoA pool is necessary not only to sustain many metabolic pathways in the cells, but also as acetate donor in the acetylation of proteins. Therefore mutations in pathways producing acetyl-CoA may affect protein acetylation. In this context, Wellen et al. showed that silencing of ACL causes a dramatic reduction on H2B, H3 and H4 acetylation, which can be rescued by adding acetate to the culture medium. Most importantly, depletion of ACL and the resulting hypoacetylation of histones impair both cell growth and differentiation [129], indicating that this crosstalk between mitochondria and chromatin plays an important homeostatic role. Cells lacking ACL also exhibit impaired lipogenesis and glucose-dependent growth, a phenotype that confers on these cells resistance to PI3K-induced tumorigenesis [128]. In support of these findings, chemical inhibition of ACL can suppress in vitro and in vivo tumor growth [130]. ACL activity might itself be affected by the availability of citrate, therefore changes in the metabolism of mitochondria can lead to changes in cytosolic acetyl-CoA, in turn influencing histone acetylation.
These observations support the idea that cell metabolism modulates the epigenetic states in cells [131]. Moreover, these changes may influence levels of metabolic genes, in turn affecting metabolism in another example of a feedback cycle. This represents a very important loop that may be relevant, for example, in cancer and metabolic syndrome. Undoubtedly, a better understanding of the crosstalk between these mechanisms could open new therapeutic and diagnostic horizons, and its potential we are only starting to grasp.
Impact of epigenetics in age-related diseases
Aging is a complex process associated with a progressive decline in the organism and its ability to react to external stresses. Many factors contribute to aging and among these increased oxidative stress, accumulation of DNA damage and changes in gene expression appear to have a particular impact [105]; as we discuss above, chromatin dynamics seems to play key roles in modulating these factors.
Not only might DNA and histone modifications participate in physiological aging, but they also might also contribute to the occurrence of age-related diseases, such as diabetes, neurodegeneration, cancer and progeria-like syndromes. Indeed, many trials investigating the potential of drugs regulating HDACs, sirtuins and DNMTs in the treatment of these diseases are ongoing. In the case of cancer, HDACi have already been approved for particular cases. In this section, we discuss in detail the link between epigenetic changes and age-related degenerative diseases. Considering that diabetes and progeroid diseases have been extensively discussed above, we focus on neurodegeneration and cancer.
Neurodegeneration
As mentioned above, changes in H4K12 acetylation may play an important role in learning decline during aging [112]. Recent studies have also shown that HDAC1 protects neurons in a mouse model of neurodegeneration [132]. Neurotoxic stimuli, as in Alzheimer’s disease (AD) and ischemia/stroke, induces calpain-mediated cleavage of p35, in turn forming the active form p25. p25 forms a complex with CDK5 and among other things, it inhibits HDAC1. HDAC1 inhibition leads to accumulation of DNA DSBs and expression of cell cycle genes [132], leading these postmitotic neurons to undergo apoptosis [133]. These observations partially contradict the findings of previous studies in which CBP was shown to protect neurons from apoptosis in neurodegeneration mouse models [134]. In addition, SIRT1 activation can also protect neurons from apoptosis in several models of neurodegeneration, including AD, amyotrophic lateral sclerosis, and primary neurons exposed to neurotoxic insults [135]. A different study has suggested instead that SIRT1 inhibition in mice restores cognition in AD by stabilizing tau protein [136]. According to Liu et al. such discrepancy might be linked to cellular NAD+ status [137]. They proposed that during cerebral hypoxia/ischemia, levels of ATP and oxygen in neurons are reduced, leading to a higher release of glutamate. Glutamate receptor overstimulation determines both bioenergetic and oxidative stress, leading to DNA breaks and SIRT1 activation. In this context, SIRT1 activation, instead of protecting cells, further reduces the NAD+ pool, worsening the bioenergetic defects and triggering apoptotic programs. With such a background, SIRT1 inhibition might actually protect cells from apoptosis. In other words, it appears that SIRT1 could protect neurons from neurotoxic insults when cells are not presensitized by severe bioenergetic impairments. For all these reasons, even if sirtuin and HDAC modulators represent putative therapeutic tools for neurodegenerative diseases, their use might need to be adapted to specific cases and bioenergetic contexts. Of note, deacetylase inhibitors affect not only neurons but also astrocytes and glia cells. For example, it has been shown that both HDAC3 and SIRT1 inhibit NF-κB transcription with opposing outcomes: enhancement of the inflammatory response in response to HDAC3 and diminished release of inflammatory molecules—such as iNOS—in response to SIRT1 [138]. Again, such results raise a note of caution, indicating that the net effect of these modulators might be context- and cell-dependent.
It is important to note that DNA damage accumulation is a common feature in the pathogenesis of many neurodegenerative disorders [75]. Given that chromatin also affects the susceptibility of cells to DNA damage, it is likely that the epigenetic changes that occur with age, as described above, may contribute to the development of neurodegenerative diseases through their effects on genome stability.
Cancer
Human tumors rely on the acquisition of multiple “hallmarks”: sustained proliferative signals, the ability to escape growth suppression, apoptosis and immune response; activation of angiogenesis and invasion/metastasis; immortalization and a metabolic shift towards glycolysis and lactate production, a phenomenon known as the “Warburg effect” [139]. Epigenetic changes may be associated with many, if not all, of these characteristics, as we have discussed.
SIRT1, 2, 3, 6 and 7 have all been shown to modulate tumorigenesis in mouse models. Haploinsufficiency of SIRT1 leads to genome instability and when associated with haploinsufficiency of p53 leads to a higher rate of spontaneous tumors [140]. Wang et al. also showed that SIRT1 is downregulated in human tumor samples. In contrast, other studies have shown that knockdown of SIRT1 in pancreatic tumor cells induces senescence and apoptosis, reducing invasiveness while enhancing chemosensitivity [141]. It is important to note that the latter was an in vitro study, and in this system, persistent p53 activation (secondary to SIRT1 knockdown) might have a dominant effect. Considering that the in vivo evidence collected so far supports mostly an antitumor role for SIRT1 [142], it is not surprising that several SIRT1 activators are in clinical trials for cancer therapy [143, 144]. As discussed above, other sirtuins, mainly SIRT6, also play a role in preventing tumorigenicity. In this context, overexpression of SIRT6—at least in vitro—induces apoptosis selectively in cancer cells [145]. Moreover, as indicated before, in recent studies SIRT6 was shown to work as a tumor suppressor, inhibiting the Warburg effect in a mouse model of colon cancer, and higher levels of SIRT6 strongly correlate with a better prognosis in human colorectal cancer patients [50], suggesting that SIRT6 could represent not only a therapeutic target, but also a prognostic tool.
Several cancers express high levels of HDACs. Indeed, in vitro experiments have shown that inhibition or silencing of HDACs induces cell cycle arrest and apoptosis in cancer cells [6]. In vivo, HDACi exhibit selective cytotoxicity towards tumor cells. This effect might be explained by the different cell cycle dynamics and checkpoints between normal and tumor cells, but also by the recent finding that HDACi seem to increase the expression of antigens recognized by cells of the immune system [146, 147]. However, if administered systemically, HDACi instead reduce the inflammatory response and the innate immunity against the tumor, through mechanisms that remain unclear [146]. Further studies are required to optimize the use of HDACi. Yet, they are emerging as a valid therapeutic option against this disease, and for this reason there are many ongoing trials with HDACi in cancer therapy, both alone and in combination with other drugs [6, 143]. In this context, the first such a drug, the HDACi Vorinostat (SAHA), has already been approved for treatment of T cell lymphoma [148], and it is conceivable that it will be the first of many to come.
Concluding remarks
Chromatin dynamics appear to play key roles in virtually every cellular process. In this context, the activity of histone and DNA modifiers seems closely intertwined with cellular metabolism, and this is evident at least at three levels: (1) epigenetic modifications affect expression of metabolic genes; (2) multiple histone modifiers have, among their nonhistone targets, metabolic proteins; (3) metabolic intermediates and cofactors influence the activity of histone and DNA modifiers. Changes in epigenetic marks can be dictated by changing metabolic conditions (i.e. nutrient availability and oxidative stress). In turn, defects in chromatin dynamics could impinge on cellular metabolic pathways, influencing processes beyond their obvious effects in the nucleus. Misregulation of histone and DNA modifications appear to play causal roles in aging and age-related diseases such as diabetes mellitus, neurodegeneration and cancer. As such, drugs that modulate chromatin enzymes provide promising therapeutic alternatives in such diseases. Considering that many of these enzymes act in a highly concerted manner, a better understanding of the precise mechanisms governing epigenetic dynamics may help in the design of better therapeutic strategies against these devastating diseases.
References
- 1.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2010;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 3.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13(11):1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Toiber D, Sebastian C, Mostoslavsky R (2011) Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. In: Yao T-P, Seto E (eds) Histone deacetylases: the biology and clinical information. Handbook of experimental pharmacology, vol 206. Springer Berlin, pp 189–224 [DOI] [PubMed]
- 8.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri N, Raychaudhuri S, Thamotharan M, Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283(20):13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen D, Dahllöf M, Lundh M, Rasmussen DN, Nielsen MD, Billestrup N, Grunnet LG, Mandrup-Poulsen T. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Mol Med. 2011;17(5-6):378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud P-D, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145(4):607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oiso H, Furukawa N, Suefuji M, Shimoda S, Ito A, Furumai R, Nakagawa J, Yoshida M, Nishino N, Araki E. The role of class I histone deacetylase (HDAC) on gluconeogenesis in liver. Biochem Biophys Res Commun. 2011;404(1):166–172. doi: 10.1016/j.bbrc.2010.11.086. [DOI] [PubMed] [Google Scholar]
- 13.Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, Qian DZ. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286(44):38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung J-W, Lee S, Seo M-S, Park S-B, Kurtz A, Kang S-K, Kang K-S. Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell Mol Life Sci. 2010;67(7):1165–1176. doi: 10.1007/s00018-009-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5(8):e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollina EA, Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30(28):3105–3126. doi: 10.1038/onc.2011.45. [DOI] [PubMed] [Google Scholar]
- 17.Soliman MA, Berardi P, Pastyryeva S, Bonnefin P, Feng X, Colina A, Young D, Riabowol K. ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell. 2008;7(6):783–794. doi: 10.1111/j.1474-9726.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- 18.Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45(4):279–285. doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang J-Y, Hung J–J. Overexpression of HDAC1 induces cellular senescence by Sp1/PP2A/pRb pathway. Biochem Biophys Res Commun. 2011;407(3):587–592. doi: 10.1016/j.bbrc.2011.03.068. [DOI] [PubMed] [Google Scholar]
- 20.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(99):1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35(22):7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquard L, Poulsen CB, Gjerdrum LM, de Nully Brown P, Christensen IJ, Jensen PB, Sehested M, Johansen P, Ralfkiaer E. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology. 2009;54(6):688–698. doi: 10.1111/j.1365-2559.2009.03290.x. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, Konishi I, Shiozawa T. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: hDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J Cancer. 2010;127(6):1332–1346. doi: 10.1002/ijc.25151. [DOI] [PubMed] [Google Scholar]
- 24.Klar AJ, Fogel S, Macleod K. MAR1 – a regulator of the HMa and HMalpha loci in Saccharomyces cerevisiae. Genetics. 1979;93(1):37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarente L, Picard F. Calorie restriction – the SIR2 connection. Cell. 2005;120(4):473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11(1):83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 27.Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci U S A. 1997;94(6):2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppikofer M, Kueng S, Martino F, Soeroes S, Hancock SM, Chin JW, Fischle W, Gasser SM. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 2011;30(13):2610–2621. doi: 10.1038/emboj.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273(2):793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 30.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40(51):15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 31.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 Is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10(12):M111.012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finkel T, Deng C-X, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460(7255):587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, Ji J, Wang XW, Park SH, Cha YI, Gius D, Deng CX. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Marcos PJ, Jeninga EH, Cantó C, Harach T, de Boer VC, Andreux P, Moullan N, Pirinen E, Yamamoto H, Houten SM, Schoonjans K, Auwerx J. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep. 2012;2:425. doi: 10.1038/srep00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng H-L, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 37.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23(24):2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen HY. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 41.Cantó C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11(3):213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schenk S, McCurdy CE, Philp A, Chen MZ, Holliday MJ, Bandyopadhyay GK, Osborn O, Baar K, Olefsky JM. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J Clin Invest. 2011;121(11):4281–4288. doi: 10.1172/JCI58554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurd BJ, Yoshida Y, McFarlan JT, Holloway GP, Moyes CD, Heigenhauser GJ, Spriet L, Bonen A. Nuclear SIRT1 activity, but not protein content, regulates mitochondrial biogenesis in rat and human skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2011;301(1):R67–R75. doi: 10.1152/ajpregu.00417.2010. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh HS, McBurney M, Robbins PD. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010;5(2):e9199. doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44(6):851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13(4):225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R-H, Kim H-S, Xiao C, Xu X, Gavrilova O, Deng C-X. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J Clin Invest. 2011;121(11):4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timmers S, Konings E, Bilet L, Houtkooper RH, van de Weijer T, Goossens GH, Hoeks J, van der Krieken S, Ryu D, Kersten S, Moonen-Kornips E, Hesselink MKC, Kunz I, Schrauwen-Hinderling VB, Blaak EE, Auwerx J, Schrauwen P. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14(5):612–622. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong L, Urso AD, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebastián C, Zbm M, Silberman DM, Gymrek MA, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, Mac Donald A, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R. The histone deacetylase SIRT6 is a novel tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9(2):162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 52.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326(5955):948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315(5812):649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. 2006;127(9):741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127(1):48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 58.Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109(4):447–458. doi: 10.1016/S0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- 59.Furuyama T, Banerjee R, Breen TR, Harte PJ. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr Biol. 2004;14(20):1812–1821. doi: 10.1016/j.cub.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 60.Astrom SU, Cline TW, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163(3):931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frankel S, Ziafazeli T, Rogina B. dSir2 and longevity in Drosophila. Exp Gerontol. 2010;46(5):391–396. doi: 10.1016/j.exger.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park S-K, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sasaki T, Maier B, Bartke A, Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell. 2006;5(5):413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 64.Han L, Zhou R, Niu J, McNutt MA, Wang P, Tong T. SIRT1 is regulated by a PPARγ-SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res. 2010;38(21):7458–7471. doi: 10.1093/nar/gkq609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez-Pastor B, Mostoslavsky R. Sirtuins, metabolism, and cancer. Front Pharmacol. 2012;3:22. doi: 10.3389/fphar.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou S, Chen HZ, Wan Yz, Zhang QJ, Wei YS, Huang S, Liu JJ, Lu YB, Zhang ZQ, Yang RF, Zhang R, Cai H, Liu DP, Liang CC. Repression of P66Shc expression by SIRT1 contributes to the prevention of hyperglycemia-induced endothelial dysfunction. Circ Res. 2011;109(6):639–648. doi: 10.1161/CIRCRESAHA.111.243592. [DOI] [PubMed] [Google Scholar]
- 67.Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, Toussaint L, Dequiedt F, Mostoslavsky R, Schmidt MH, Zimmermann B, Brandes RP, Mione M, Westphal CH, Braun T, Zeiher AM, Gerhardt H, Dimmeler S, Potente M. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473(7346):234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiang L, Lin HV, Kim-Muller JY, Welch CL, Gu W, Accili D. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through creb deacetylation. Cell Metab. 2011;14(6):758–767. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9(2):285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 70.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Kim DH, Kim JY, Yu BP, Chung HY. The activation of NF-kappaB through Akt-induced FOXO1 phosphorylation during aging and its modulation by calorie restriction. Biogerontology. 2008;9(1):33–47. doi: 10.1007/s10522-007-9114-6. [DOI] [PubMed] [Google Scholar]
- 72.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, Barnard D, Ward WF, Qi W, Ingram DK, de Cabo R. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489(7415):318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483(7388):218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 74.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun. 2011;2:433–437. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Szerlong HJ, Prenni JE, Nyborg JK, Hansen JC. Activator-dependent p300 acetylation of chromatin in vitro: enhancement of transcription by disruption of repressive nucleosome–nucleosome interactions. J Biol Chem. 2010;285(42):31954–31964. doi: 10.1074/jbc.M110.148718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lerin C, Rodgers JT, Kalume DE, Kim S-h, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 2006;3(6):429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 80.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26(7):1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA Induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42(4):426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002;62(21):6231–6239. [PubMed] [Google Scholar]
- 83.Russell M, Berardi P, Gong W, Riabowol K. Grow-ING, Age-ING and Die-ING: iNG proteins link cancer, senescence and apoptosis. Exp Cell Res. 2006;312(7):951–961. doi: 10.1016/j.yexcr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 84.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406(6792):207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 85.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37(4):469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35(3):352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim S, Ohkuni K, Couplan E, Jazwinski SM. The histone acetyltransferase GCN5 modulates the retrograde response and genome stability determining yeast longevity. Biogerontology. 2004;5(5):305–316. doi: 10.1007/s10522-004-2568-x. [DOI] [PubMed] [Google Scholar]
- 89.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26(37):5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 90.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102(37):13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, Tashiro S, Nakanishi M. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24(4):333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barlow AL, van Drunen CM, Johnson CA, Tweedie S, Bird A, Turner BM. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp Cell Res. 2001;265(1):90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- 93.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simboeck E, Ribeiro JD, Teichmann S, Di Croce L. Epigenetics and senescence: learning from the INK4-ARF locus. Biochem Pharmacol. 2011;82(10):1361–1370. doi: 10.1016/j.bcp.2011.07.084. [DOI] [PubMed] [Google Scholar]
- 95.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 96.Berger JH, Bardeesy N. Modeling INK4/ARF tumor suppression in the mouse. Curr Mol Med. 2007;7(1):63–75. doi: 10.2174/156652407779940477. [DOI] [PubMed] [Google Scholar]
- 97.Wang X, Feng Y, Xu L, Chen Y, Zhang Y, Su D, Ren G, Lu J, Huang B. YY1 restrained cell senescence through repressing the transcription of p16. Biochim Biophys Acta. 2008;1783(10):1876–1883. doi: 10.1016/j.bbamcr.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 98.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15(11):1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang J, Gan Q, Han L, Li J, Zhang H, Sun Y, Zhang Z, Tong T. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS One. 2008;3(3):e1710. doi: 10.1371/journal.pone.0001710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N, Murphy MM, Appella E, Alt FW. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2(1):67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 101.Narala SR, Allsopp RC, Wells TB, Zhang G, Prasad P, Coussens MJ, Rossi DJ, Weissman IL, Vaziri H. SIRT1 acts as a nutrient-sensitive growth suppressor and its loss is associated with increased AMPK and telomerase activity. Mol Biol Cell. 2008;19(3):1210–1219. doi: 10.1091/mbc.E07-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21(10):2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 105.Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40(2):333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585(13):2041–2048. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39(5):724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17(10):1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, Grianti P, Pagani M, Bonaldi T, Ragoussis J, Friedman N, Camilloni G, Bianchi ME, Agresti A. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011;9(6):e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Das C, Tyler JK. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta. 2012;1819(3-4):332–342. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 113.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8(1):19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 114.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 115.Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a) Cell Cycle. 2011;10(3):457–468. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]
- 116.Baird DM. Telomere dynamics in human cells. Biochimie. 2008;90(1):116–121. doi: 10.1016/j.biochi.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 117.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, Weinberg RA, Stewart SA, Hahn WC. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114(2):241–253. doi: 10.1016/S0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 118.Pickett HA, Henson JD, Au AY, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet. 2011;20(23):4684–4692. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- 119.Donate LE, Blasco MA. Telomeres in cancer and ageing. Philos Trans R Soc Lond B Biol Sci. 2010;366(1561):76–84. doi: 10.1098/rstb.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8(4):299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 121.Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 123.Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadinanos J, Horner JW, Maratos-Flier E, Depinho RA. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2010;469(7328):102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103(27):10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103(27):10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shimazu T, Hirschey MD, Huang JY, Ho LT, Verdin E. Acetate metabolism and aging: an emerging connection. Mech Ageing Dev. 2010;131(7–8):511–516. doi: 10.1016/j.mad.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 127.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24(41):6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 129.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8(4):311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 131.Cyr AR, Domann FE. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid Redox Signal. 2011;15(2):551–589. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, Guan JS, Lee BH, Moy LY, Giusti P, Broodie N, Mazitschek R, Delalle I, Haggarty SJ, Neve RL, Lu Y, Tsai LH. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60(5):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kruman II, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, Emokpae R, Jr, Gorospe M, Mattson MP. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004;41(4):549–561. doi: 10.1016/S0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- 134.Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22(24):6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26(13):3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28(45):11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu D, Gharavi R, Pitta M, Gleichmann M, Mattson MP. Nicotinamide prevents NAD+ depletion and protects neurons against excitotoxicity and cerebral ischemia: NAD+ consumption by SIRT1 may endanger energetically compromised neurons. Neuromol Med. 2009;11(1):28–42. doi: 10.1007/s12017-009-8058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dietz KC, Casaccia P. HDAC inhibitors and neurodegeneration: at the edge between protection and damage. Pharmacol Res. 2010;62(1):11–17. doi: 10.1016/j.phrs.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 140.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao G, Cui J, Zhang J-G, Qin Q, Chen Q, Yin T, Deng S-C, Liu Y, Liu L, Wang B, Tian K, Wang G-B, Wang C-Y. SIRT1 RNAi knockdown induces apoptosis and senescence, inhibits invasion and enhances chemosensitivity in pancreatic cancer cells. Gene Ther. 2011;18(9):920–928. doi: 10.1038/gt.2011.81. [DOI] [PubMed] [Google Scholar]
- 142.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10(12):819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li Y, Daniel M, Tollefsbol TO. Epigenetic regulation of caloric restriction in aging. BMC Med. 2011;9(1):98. doi: 10.1186/1741-7015-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chauhan D, Bandi M, Singh AV, Ray A, Raje N, Richardson P, Anderson KC. Preclinical evaluation of a novel SIRT1 modulator SRT1720 in multiple myeloma cells. Br J Haematol. 2011;155(5):588–598. doi: 10.1111/j.1365-2141.2011.08888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10(18):3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leggatt GR, Gabrielli B. Histone deacetylase inhibitors in the generation of the anti-tumour immune response. Immunol Cell Biol. 2012;90(1):33–38. doi: 10.1038/icb.2011.94. [DOI] [PubMed] [Google Scholar]
- 147.Woan KV, Sahakian E, Sotomayor EM, Seto E, Villagra A. Modulation of antigen-presenting cells by HDAC inhibitors: implications in autoimmunity and cancer. Immunol Cell Biol. 2012;90(1):55–65. doi: 10.1038/icb.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16(7):1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]