Abstract
The development of expressed sequence tag-derived simple sequence repeats (EST-SSRs) provided a useful tool for investigating plant genetic diversity. In the present study, 22 polymorphic EST-SSRs from grain soybean were identified and used to assess the genetic diversity in 48 vegetable soybean accessions. Among the 22 EST-SSR loci, tri-nucleotides were the most abundant repeats, accounting for 50.00% of the total motifs. GAA was the most common motif among tri-nucleotide repeats, with a frequency of 18.18%. Polymorphic analysis identified a total of 71 alleles, with an average of 3.23 per locus. The polymorphism information content (PIC) values ranged from 0.144 to 0.630, with a mean of 0.386. Observed heterozygosity (H o) values varied from 0.0196 to 1.0000, with an average of 0.6092, while the expected heterozygosity (H e) values ranged from 0.1502 to 0.6840, with a mean value of 0.4616. Principal coordinate analysis and phylogenetic tree analysis indicated that the accessions could be assigned to different groups based to a large extent on their geographic distribution, and most accessions from China were clustered into the same groups. These results suggest that Chinese vegetable soybean accessions have a narrow genetic base. The results of this study indicate that EST-SSRs from grain soybean have high transferability to vegetable soybean, and that these new markers would be helpful in taxonomy, molecular breeding, and comparative mapping studies of vegetable soybean in the future.
Keywords: Expressed sequence tag (EST), Simple sequence repeat (SSR), Genetic diversity, Microsatellites, Vegetable soybean
1. Introduction
Soybean [Glycine max (L.) Merr.], the world’s most important cultivated legume crop, can be divided into two categories: vegetable soybean, which is harvested between reproductive stages 6 (R6) and 7 (R7) of growth when the seeds have developed to fill 80%‒90% of the pod, and grain soybean, which is harvested at reproductive stage 8 (R8) when the pod has reached full maturity (Yinbo et al., 1997; Young et al., 2000). Grain soybean is primarily used for manufacturing oil and protein products. Vegetable soybean, however, is consumed mainly as a vegetable or snack. Like grain soybean, vegetable soybean is rich in protein, oil, and other nutritious constituents, but there are many differences between them. For example, vegetable soybeans are usually larger (over 30 g/100 seeds) than grain soybeans (less than 25 g/100 seeds), and have many advantages in terms of sensory attributes over grain soybeans, such as green color, soft texture, sweet taste, higher protein utilization, and higher contents of vitamin A, vitamin C, sucrose, and starch (Saldivar et al., 2010; Keatinge et al., 2011).
China is the world center for vegetable soybean production and the history of vegetable soybean cultivation in China can be traced back 1 000 years (Yinbo et al., 1997; Cornelious and Sneller, 2002; Lu et al., 2006). However, because of a lack of local breeding programs, the commercial cultivars used in production have mainly been introduced from Japan and Taiwan of China. To counter this situation, the Chinese government has started a project to accelerate the breeding of vegetable soybean. However, due to various restrictions, no significant breakthrough has yet been achieved (Arikit et al., 2011).
In modern plant breeding, molecular markers have become important and efficient tools. Molecular markers linked to agronomic traits can increase the accuracy and veracity of selection, thereby reducing the field workload, so the selection of suitable markers has become one of the key factors in the success of molecular breeding programs. Many types of molecular markers, such as restriction fragment length polymorphisms (RFLPs), amplified fragment length polymorphisms (AFLPs), random amplified polymorphic DNA (RAPD), simple sequence repeats (SSRs), and single nucleotide polymorphisms (SNPs), have been used to study the genetic diversity and population structure of plants (Wen et al., 2009). Among these markers, SSRs have stood out and are considered to be the most powerful tools because of their high abundance, co-dominant inheritance, multiple alleles, reproducibility, extensive genome coverage, and ease-of-detection by polymerase chain reaction (PCR) (Moe et al., 2010). However, due to the long time and high cost of their development, the wide use of SSRs is often limited.
The rapid development of studies of expressed sequence tags (ESTs) has generated a new source for developing SSRs. Compared with genomic-SSR markers, EST-SSRs have some intrinsic advantages: (1) they are less costly to identify; (2) they are directly associated with transcribed genes; and (3) they have high transferability among related species (Varshney et al., 2005). Recently, grain soybean studies have generated a large number of ESTs in various public databases, and EST-SSR development has been carried out in some studies. For example, Hisano et al. (2007) designed 6 920 primer pairs from 63 676 grain soybean ESTs and using polymorphism analysis, obtained 680 polymorphic EST-SSRs. A further genetic linkage map study indicated that 935 loci detected by these markers were successfully mapped onto 20 linkage groups, covering 2 700.3 cM of the soybean genome. Li et al. (2010) developed 34 EST-SSRs from grain soybean globular embryo ESTs, and the analysis indicated that 11 of them were polymorphic. Liu et al. (2010) developed 37 EST-SSRs from the grain soybean cDNA library, and these markers could be used successfully to distinguish cultivated soybean from wild soybean. In spite of these studies in grain soybean, no EST-SSR studies have yet been reported for vegetable soybean. The objective of this study was to develop EST-SSR markers for vegetable soybean, examine their polymorphism, and evaluate the genetic diversity of vegetable soybean accessions. The results might be useful for marker-assisted selection and breeding of vegetable soybean in the future.
2. Materials and methods
2.1. Materials
Forty-eight vegetable soybean accessions, including 45 spring-sown types and 3 autumn-sown types, were used in the study. Among the 48 accessions, 43 were from China, 3 from Japan, and 2 from the USA (Table 1). DNA was extracted from 20-d old seedlings grown in a glasshouse. A total of 0.2 g of fresh leaves were used for each repeat and DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method following the manufacturer’s instructions. The relative purity and concentration of the extracted DNA were estimated with NanoDrop ND-1000 (NanoDrop Technologies Inc., Wilmington, DE, USA). The final concentration of each DNA sample was adjusted to 20 ng/ml.
Table 1.
List of 48 vegetable soybean accessions used to determine genetic diversity
| No. | Accession name | Origin |
| 1 | LDY081011 | Liaoning, China |
| 2 | LDY081014 | Liaoning, China |
| 3 | LDY081015 | Liaoning, China |
| 4 | LDY081018 | Liaoning, China |
| 5 | LDY081019 | Liaoning, China |
| 6 | LDY081021 | Liaoning, China |
| 7 | LDY081023 | Liaoning, China |
| 8 | LDY081025 | Liaoning, China |
| 9 | ZXN081029 | Zhejiang, China |
| 10 | ZXN081031 | Zhejiang, China |
| 11 | ZXN081035 | Zhejiang, China |
| 12 | ZXN081036 | Zhejiang, China |
| 13 | YZT081045 | Jilin, China |
| 14 | YZT081049 | Jilin, China |
| 15 | YZT081051 | Jilin, China |
| 16 | YZT081058 | Jilin, China |
| 17 | YZT081065 | Jilin, China |
| 18 | YZT081069 | Jilin, China |
| 19 | TW091079 | Kaohsiung, Taiwan, China |
| 20 | TW091080 | Kaohsiung, Taiwan, China |
| 21 | TW091081 | Kaohsiung, Taiwan, China |
| 22 | TW091084 | Kaohsiung, Taiwan, China |
| 23 | TW091091 | Kaohsiung, Taiwan, China |
| 24 | ZAL091110 | Jiangsu, China |
| 25 | ZAL091116 | Jiangsu, China |
| 26 | WJH101125 | Heilongjiang, China |
| 27 | WJH101128 | Heilongjiang, China |
| 28 | WJH101134 | Heilongjiang, China |
| 29 | JAP101136 | Hokkaido, Japan |
| 30 | JAP101139 | Hokkaido, Japan |
| 31 | JAP101141 | Hokkaido, Japan |
| 32 | USA101148 | North Carolina, USA |
| 33 | USA101155 | North Carolina, USA |
| 34 | SWK111158 | Liaoning, China |
| 35 | SWK111175 | Liaoning, China |
| 36 | SWK111176 | Liaoning, China |
| 37 | SWK111177 | Liaoning, China |
| 38 | WGP111178 | Jiangsu, China |
| 39 | WGP111181 | Jiangsu, China |
| 40 | WGP111186 | Jiangsu, China |
| 41 | WGP111187 | Jiangsu, China |
| 42 | YLF111192 | Shanghai, China |
| 43 | YLF111197 | Shanghai, China |
| 44 | YLF111203 | Shanghai, China |
| 45 | ZLH111205 | Fujian, China |
| 46 | ZLH111209 | Fujian, China |
| 47 | ZLH111217 | Fujian, China |
| 48 | ZXH111221 | Guangdong, China |
2.2. Primer selection and prescreening
A total of 172 grain soybean EST-SSRs developed in previous studies (Song et al., 2004; Hisano et al., 2007; Shultz et al., 2007; Xia et al., 2007) were selected and prescreened on five vegetable soybean accessions (one Chinese landrace, one Chinese cultivar, and one cultivar each from Japan, Taiwan of China, and USA) to select suitable primers for further analysis. Based on the results of prescreening, 64 primer pairs were designed and synthesized to assess polymorphism. The forward primers of each pair were labeled with 6-FAM fluorescent dye.
2.3. SSR amplification and PCR product analysis
PCRs were conducted using a Peltier thermo cycler (PTC)-225 thermal cycler from MJ Research Inc. (Waltham, MA, USA). All PCR amplifications were carried out in 20 μl reaction mixtures containing 20 ng genomic DNA template, 1× PCR buffer (containing Mg2+), 0.2 mmol/L dNTPs, 0.2 mmol/L forward and reverse primers, and 1 U of Taq polymerase (TaKaRa, Dalian, China). The PCR program was as follows: 95 °C for 5 min, 36 cycles each at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 60 s, and a final extension at 72 °C for 10 min. The PCR reaction products were diluted and detected on a MegaBACE 1000 DNA analysis system as previously described (Gong et al., 2011). Analysis of amplified fragment size was performed using methods described by Gong et al. (2010a).
2.4. Data and statistics
The variability at each locus was measured in terms of the number of alleles (N a), observed heterozygosity (H o), expected heterozygosity (H e), polymorphic information content (PIC), Nie’s gene diversity (D A) and a Chi-square test for Hardy-Weinberg equilibrium (P HW). The N a, H o, and H e were calculated using POPGENE software, Version 1.3 (Choudhary et al., 2009). The PIC value was calculated using the formula developed by Anderson et al. (1993). D A was determined using NTSYSpc Version 2.10 (NTSYS-PC 2.10, Applied Biostatistics, Setauket, NY, USA). A dendrogram was constructed using unweighted pair group method with arithmetic means (UPGMA).
3. Results
3.1. Characteristics of EST-SSR markers in vegetable soybean
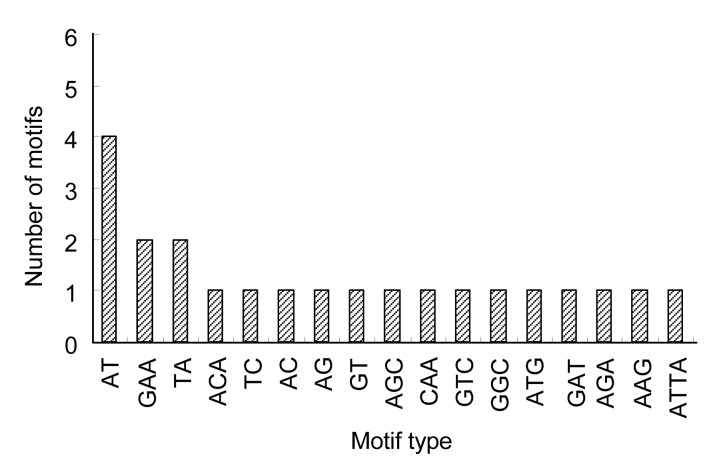
Based on the prescreening results, 64 primer pairs with clean amplification products were designed and explored over 48 vegetable soybean accessions. Among these primers, 34.38% (22) were polymorphic, which was higher than previous results of 9.83% and 32.35% in grain soybean (Hisano et al., 2007; Li et al., 2010). The number of repeats ranged from 8 to 22, with an average of 11.59. Out of all the repeat motifs, the length of SSRs ranged from 22 to 44 bp, with an average of 28.21 bp (Table 2). Among the 22 SSR loci, tri-nucleotides were the most abundant repeats, accounting for 50.00%, followed by di-nucleotides (45.45%) and tetra-nucleotides (4.55%). GAA was the most common motif among the tri-nucleotide repeats, with a frequency of 18.18%, followed by AGC, CAA, GTC, GGC, ATG, GAT, AGA, and AAG, each with a frequency of 9.09%. The dominant repeat motif of the di-nucleotides was AT with a frequency of 40.00%, followed by TA (20.00%), and TC, AC, AG, and GT, each accounting for 10.00% (Table 3, Fig. 1). More details about the different repeat motifs are listed in Table 2.
Table 2.
Characteristics of 22 polymorphic vegetable soybean EST-SSR markers
| Locus name | Corresponding ID from references | Primer sequence (5′–3′) | Motif | Annealing Temp. (°C) | Expected size (bp) | Practical size (bp) | Number of alleles | Reference or information source |
| Gmp-009 | AW620774 | Forward: TCACCCACCCAATACACAAA | (GAA)9 | 56 | 174 | 166–181 | 2 | Song et al., 2004 |
| Reverse: TTCGCAACTTGTTACAACGG | ||||||||
| Gmp-017 | CG819919.1 | Forward: ACCTCTTCCCCCATTCAGTT | (AT)12 | 55 | 236 | 198–236 | 3 | Shultz et al., 2007 |
| Reverse: ACCTCTTCCCCCATTCAGTT | ||||||||
| Gmp-045 | CSSR381 | Forward: AACACCGTTGGTTTTCATGC | (AGC)8 | 55 | 167 | 156–169 | 2 | Xia et al., 2007 |
| Reverse: TTGCAGTTTGGGTTTGAACA | ||||||||
| Gmp-046 | CSSR385 | Forward: AACCCTTCTTCCACTTCCGT | (CAA)9 | 55 | 201 | 197–211 | 3 | Xia et al., 2007 |
| Reverse: AACCCTTCTTCCACTTCCGT | ||||||||
| Gmp-048 | CSSR391 | Forward: CCGCCGAAGTACGAAGTAGA | (GTC)9 | 54 | 260 | 247–263 | 4 | Xia et al., 2007 |
| Reverse: CCGCCGAAGTACGAAGTAGA | ||||||||
| Gmp-049 | CSSR400 | Forward: CTTCTCTCAGCACCCTCCAC | (TC)18 | 54 | 269 | 250–283 | 3 | Xia et al., 2007 |
| Reverse: AACCCTTCTTCCACTTCCGT | ||||||||
| Gmp-050 | CSSR405 | Forward: AACAACAACAGCCACCACAA | (CAA)8 | 54 | 219 | 197–238 | 6 | Xia et al., 2007 |
| Reverse: CTGGCATTGACACTGTTGCT | ||||||||
| Gmp-058 | CSSR443 | Forward: GTATGGATTTCATGGGTGGG | (GGC)8 | 54 | 124 | 110–160 | 6 | Xia et al., 2007 |
| Reverse: GTTCCTCGGAGACAGCAAAG | ||||||||
| Gmp-059 | CSSR449 | Forward: GAAATGACAATAATGCCGGG | (AT)21 | 54 | 183 | 177–182 | 3 | Xia et al., 2007 |
| Reverse: TTCCATTCAAAGCAGAAGCA | ||||||||
| Gmp-066 | CSSR472 | Forward: GGTTACGGCACTTCCTACCA | (AAC)9 | 55 | 225 | 202–235 | 2 | Xia et al., 2007 |
| Reverse: AATTTTTGCGTTGTTGAGGG | ||||||||
| Gmp-072 | CSSR498 | Forward: GAGATTCATGAGAAGGGCCA | (ATG)9 | 56 | 160 | 144–165 | 4 | Xia et al., 2007 |
| Reverse: CTCCCCGTGTTAGGTGTTGT | ||||||||
| Gmp-088 | CSSR540 | Forward: GAGGTTGGTGCCTGGAGATA | (GAT)9 | 56 | 211 | 197–235 | 3 | Xia et al., 2007 |
| Reverse: TGGCGAGTTACGAGGCTATT | ||||||||
| Gmp-112 | GMES0255 | Forward: TCACCCACCCAATACACAAA | (AT)11 | 56 | 154 | 140–156 | 4 | Hisano et al., 2007 |
| Reverse: TTCGCAACTTGTTACAACGG | ||||||||
| Gmp-116 | GMES0633 | Forward: GCCTGTGGTTGGTCTTCATT | (AT)22 | 54 | 189 | 172–192 | 3 | Hisano et al., 2007 |
| Reverse: AAAACCATATGCTTGCGGAC | ||||||||
| Gmp-122 | GMES0644 | Forward: AGATTGGAAGAGCCATCCCT | (AGA)12 | 54 | 294 | 294–308 | 4 | Hisano et al., 2007 |
| Reverse: ACTTCTCGCCCTCGTTCTTT | ||||||||
| Gmp-128 | GMES0687 | Forward: CATCCTAACCGGTCAAAAACA | (ATTA)7 | 55 | 152 | 138–154 | 4 | Hisano et al., 2007 |
| Reverse: GGAACTTCTCCCTTGGGTTC | ||||||||
| Gmp-133 | GMES0709 | Forward: ACAGGTTGTGGGACGGTAAA | (ACA)9 | 55 | 217 | 197–221 | 3 | Hisano et al., 2007 |
| Reverse: ACCAAATAGCTGGAATCCCC | ||||||||
| Gmp-141 | GMES0799 | Forward: CCCTTCCCTCTTCTCCTGTT | (TA)11 | 54 | 133 | 107–128 | 2 | Hisano et al., 2007 |
| Reverse: ATGCAAACAAGAATCCTGGC | ||||||||
| Gmp-149 | GMES1455a | Forward: ACCAGCACCGCGTAATAAAT | (AG)12 | 54 | 180 | 197–210 | 2 | Hisano et al., 2007 |
| Reverse: AACCAGGACAACCGAGTCAC | ||||||||
| Gmp-166 | GMES3937 | Forward: GGAGAAGGCCTCATCAACAG | (AAG)9 | 56 | 186 | 183–186 | 2 | Hisano et al., 2007 |
| Reverse: AGGTACTGGACACTCGGTGG | ||||||||
| Gmp-197 | GMES4774 | Forward: AGGATCACATACCAGGCACC | (TA)18 | 56 | 277 | 253–285 | 4 | Hisano et al., 2007 |
| Reverse: AGGATCACATACCAGGCACC | ||||||||
| Gmp-199 | GMES6145 | Forward: CTCCGATGCTTATCCCAAAA | (GT)15 | 56 | 184 | 168–182 | 2 | Hisano et al., 2007 |
| Reverse: AAAACCCTCCAAAATCAGGG |
Table 3.
Occurrence of different types of SSRs in vegetable soybean
| SSR motif | Motif type | Occurrence |
||||||||||||||
| Number of repeats |
Total | |||||||||||||||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | >18 | |||
| Di- | AT | 1 | 1 | 2 | 4 | |||||||||||
| TC | 1 | 1 | ||||||||||||||
| AC | 1 | 1 | ||||||||||||||
| TA | 1 | 1 | 2 | |||||||||||||
| AG | 1 | 1 | ||||||||||||||
| GT | 1 | 1 | ||||||||||||||
| Total | 2 | 3 | 1 | 2 | 2 | 10 | ||||||||||
| Tri- | GAA | 1 | 1 | 2 | ||||||||||||
| AGC | 1 | 1 | ||||||||||||||
| CAA | 1 | 1 | ||||||||||||||
| GTC | 1 | 1 | ||||||||||||||
| GGC | 1 | 1 | ||||||||||||||
| ACA | 1 | 1 | ||||||||||||||
| ATG | 1 | 1 | ||||||||||||||
| GAT | 1 | 1 | ||||||||||||||
| AGA | 1 | 1 | ||||||||||||||
| AAG | 1 | 1 | ||||||||||||||
| Total | 3 | 7 | 1 | 11 | ||||||||||||
| Tetra- | ATTA | 1 | 1 | |||||||||||||
|
| ||||||||||||||||
| Total | 0 | 1 | 3 | 7 | 0 | 3 | 3 | 0 | 0 | 1 | 0 | 0 | 2 | 2 | 22 | |
Fig. 1.
Distribution of different motifs in vegetable soybean EST-SSRs
3.2. Polymorphism of EST-SSRs
The 22 EST-SSRs detected a total of 71 alleles in 48 vegetable soybean accessions (Table 1). For each locus, the number of alleles ranged from 2 (Gmp-009, Gmp-045, Gmp-066, Gmp-141, Gmp-149, Gmp-166, and Gmp-199) to 6 (Gmp-050 and Gmp-058), with an average of 3.23 (Table 4). The most frequent number of alleles per locus was 2. This result was slightly lower than the value for wild soybean (3.80) (Liu et al., 2010). PIC values varied from a high value of 0.630 (Gmp-050) to a low value of 0.144 (Gmp-046), with a mean value of 0.386, which was similar to the average value of 0.400 in grain soybean (Hisano et al., 2007). As a strictly self-fertilizating crop, vegetable soybean was expected to have lower heterozygosity than hybrid crops. However, H o values varied widely among these markers, ranging from 0.0196 to 1.0000, with an average of 0.6092. Also, H e values varied significantly, ranging from 0.1502 to 0.6840, with a mean of 0.4616, which was significantly higher than in grain soybean (0.0140) and wild soybean (0.0690) (Li et al., 2008; Liu et al., 2010). In addition, 16 EST-SSRs (72.73%) had significantly higher levels of H o than did H e. All loci except Gmp-116, Gmp-149, and Gmp-166, showed significant deviations from the Hardly-Weinberg equilibrium (P<0.01). Considering that the accessions used in this study included cultivars and landraces from China, Japan, and USA, these results were not surprising.
Table 4.
Characterization of the 22 microsatellite loci for vegetable soybean, including observed heterozygosity (H o), expected heterozygosity (H e), polymorphism information content (PIC), and Chi-square test for Hardy-Weinberg equilibrium (P HW)
| Locus name | H o | H e | PIC | P HW |
| Gmp-009 | 0.1923 | 0.3107 | 0.262 | ** |
| Gmp-017 | 0.7000 | 0.5850 | 0.495 | ** |
| Gmp-045 | 1.0000 | 0.5000 | 0.375 | ** |
| Gmp-046 | 0.0400 | 0.1502 | 0.144 | ** |
| Gmp-048 | 0.9038 | 0.6433 | 0.570 | ** |
| Gmp-049 | 0.3409 | 0.5338 | 0.473 | ** |
| Gmp-050 | 0.3529 | 0.6840 | 0.630 | ** |
| Gmp-058 | 0.8654 | 0.5775 | 0.486 | ** |
| Gmp-059 | 0.1429 | 0.4638 | 0.399 | ** |
| Gmp-066 | 0.9804 | 0.5438 | 0.441 | ** |
| Gmp-072 | 0.8431 | 0.4931 | 0.372 | ** |
| Gmp-088 | 0.7885 | 0.4776 | 0.364 | ** |
| Gmp-112 | 0.6905 | 0.5558 | 0.456 | ** |
| Gmp-116 | 0.3462 | 0.2862 | 0.245 | |
| Gmp-122 | 0.0196 | 0.1624 | 0.152 | ** |
| Gmp-128 | 1.0000 | 0.6248 | 0.554 | ** |
| Gmp-133 | 1.0000 | 0.5185 | 0.403 | ** |
| Gmp-141 | 0.9615 | 0.4993 | 0.375 | ** |
| Gmp-149 | 0.2500 | 0.2188 | 0.195 | |
| Gmp-166 | 0.2400 | 0.2112 | 0.189 | |
| Gmp-197 | 1.0000 | 0.5000 | 0.375 | ** |
| Gmp-199 | 0.7451 | 0.6146 | 0.533 | ** |
denotes significant deviation from Hardy-Weinberg equilibrium (P<0.01)
3.3. Assessment of genetic diversity in 48 vegetable soybean accessions
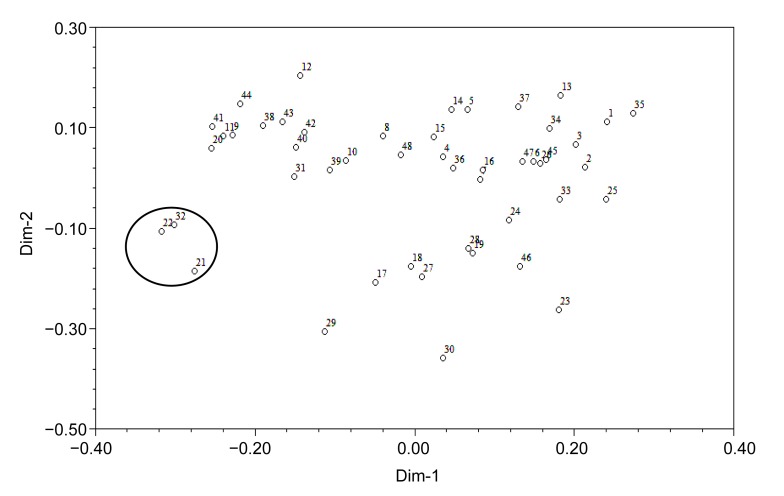
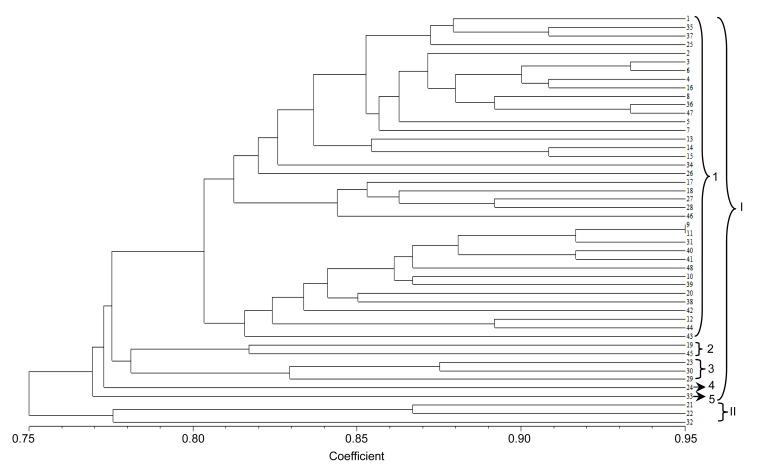
To evaluate the usefulness of these EST-SSRs, a genetic variation study was carried out in the 48 accessions. A cluster graph produced by two-dimensional principle coordinate analysis indicated that cultivars 21, 22, and 32 (within the circle in Fig. 2), which were autumn-sown types, were clustered in the lower-left side and separated from other accessions. Consistent with this result, phylogenetic tree analysis showed that at a coefficient of 0.76, all accessions were clustered into two main groups: spring-sown type (I) and autumn-sown type (II) (Fig. 3). At a coefficient of 0.80, accessions of the spring-sown type were clustered into five groups. Most Chinese cultivars and landraces fell into Group 1. Group 2 was comprised of two cultivars, one from Kaohsiung, Taiwan, China (No. 19) and the other from Fujian, China (No. 45). Group 3 contained three cultivars, two from Hokkaido, Japan (Nos. 29 and 30) and the other from Kaohsiung, Taiwan, China (No. 23). Groups 4 and 5 each had only one accession, from Jiangsu, China (No. 24) and North Carolina, USA (No. 33), respectively. Group 1 was further divided into two subgroups at a coefficient of 0.81: one group comprised 24 accessions mainly from the northern regions of China, and the other group included 14 accessions mainly from the southern regions of China. These results indicated that the average genetic similarity among Chinese accessions was high, meaning that, although some accessions are far from each other in terms of geographic distance, they have a similar genetic background. However, the genetic relationships among Chinese, Japanese, and USA accessions are distant.
Fig. 2.
Two-dimension principlal coordinate analysis of 48 vegetable soybean accessions
The numbers represent different accessions as listed in Table 1
Fig. 3.
Dendrogram of 48 vegetable soybean accessions based on 22 EST-SSR data using the UPGMA clustering method
The numbers represent different accessions as listed in Table 1
4. Discussion
In this study, a total of 172 grain soybean EST-SSR markers were selected and screened for useful markers for vegetable soybean. Finally, we obtained 64 primer pairs with clean amplification products. Among these primers, 22 loci were polymorphic with a polymorphism percentage of 34.38%, which was higher than previous results of 9.83% and 32.35% in grain soybean (Hisano et al., 2007; Li et al., 2010). These results indicated that grain soybean EST-SSRs have high transferability to vegetable soybean and could be a potential resource for the development of vegetable soybean EST-SSR markers. Further analysis showed that tri-nucleotides were the most abundant motif in vegetable soybean (Table 3). It has been suggested that the abundance of tri-nucleotide SSRs in plants might be attributed to the absence of frameshift mutations, due to the variety of tri-nucleotide repeats (Metzgar et al., 2000). Also, the predominance of tri-nucleotide repeats in EST-SSRs is due to the suppression of non-trinucleotide SSRs in coding regions, which could reduce the occurrence of frameshift mutations within transcribed genes. Morgante et al. (2002) reported that the high frequency of tri-nucleotide repeats in plant ESTs is due to mutation pressure and putative selection for specific amino acid stretches. Previous studies indicated that tri-nucleotides were the most dominant repeat motif in legume crops, such as grain soybean, wild soybean, pea and faba bean (Kuroda et al., 2009; Gong et al., 2010a; 2010b; Liu et al., 2010). Our results support those findings. GAA was found to be the most abundant of the tri-nucleotide repeats, consistent with previous results for grain soybean (Roy et al., 2004). SSRs with long motifs are usually ignored during generation of markers because of their scarcity in plant genomes (Wang et al., 2010). However, one EST-SSR with a tetra-nucleotide repeat motif was found to be polymorphic in our study, which suggested that long motif SSRs may also be useful for generating informative markers.
Generally, N a, PIC, H o, and H e are important indexes of the polymorphism of SSRs. In this study, a total of 71 alleles were detected in 22 loci, with an average of 3.23 per locus, equivalent to about 18% of genomic SSRs in grain soybean (Li et al., 2008). The differences in the number of alleles per locus between EST-SSRs and genomic SSRs are due mainly to the origin of the sequences, which are more conserved in coding regions than in non-coding regions. Thus, the N a values of EST-SSRs are usually lower than those of genomic SSRs (Temnykh et al., 2000). Liu et al. (2010) reported that 37 EST-SSR markers from grain soybean detected a total of 142 alleles in wild soybean, with a mean value of 3.8, which is slightly higher than the value found in our results. The PIC value is the most important index of molecular marker polymorphism. In our study, PIC values among the 22 loci ranged from 0.144 to 0.630, with an average of 0.386, similar to the mean value (0.400) of grain soybean (Hisano et al., 2007). As a strictly self-fertilizating crop, vegetable soybean is expected to have low heterozygosity. However, in this study, the mean value of 48 vegetable soybean accessions was 0.6092, which was significantly higher that the average values of grain soybean (0.0140) and wild soybean (0.0690) (Li et al., 2008; Liu et al., 2010). In addition, 19 (86.36%) loci exhibited significant deviations from Hardy-Weinberg equilibrium corrected for multiple comparisons (P<0.01), which is similar to the value for wild soybean (Liu et al., 2010). These results indicate that these markers are highly informative and could be used to differentiate genotypes and cluster them for genetic diversity analysis.
Assessment of genetic diversity within a population is essential to characterize germplasm and provides insights into evolutionary aspects, conservation, utilization, and establishment of breeding programs (Li et al., 2011). In this study, genetic variation in 48 vegetable soybean accessions was analyzed using principal coordinate analysis and phylogenetic tree. Both results (Figs. 2 and 3) showed that cultivars with the same origin tended to be clustered. These results indicated that the genetic relationship among vegetable soybean accessions was related to a great extent to their geographic distribution, which was similar to the results for wheat (Li et al., 2008). In China, southeastern coastal provinces, including Zhejiang, Jiangsu, Shanghai, and Fujian, are the main cultivation regions for vegetable soybean, while the northeastern provinces, such as Liaoning, Jilin, and Heilongjiang, are the main cultivation regions for grain soybean. The ecological and geographical conditions differ greatly between these areas, and accessions originating from those areas should show high genetic differentiation. However, the results of the cluster analysis in our study (Fig. 3) suggested that most accessions from China were clustered into the same groups. These results suggest that genetic variation among Chinese populations is low and that vegetable soybean in China has a narrow genetic base, forming the major constraint to vegetable soybean breeding programs. However, genetic diversity among Chinese, Japanese, and American accessions was high, indicating that they have a distant relationship. Zhou et al. (2002) suggested that grain soybean collections from China, Japan, and USA have different genetic bases, and that they could be used to increase the diversity in Chinese breeding programs. Mimura et al. (2007) reported that the genetic background of vegetable soybean accessions from Japan and China was different, and the same result was found in grain soybean. Our results were consistent with these previous studies. So, more foreign germplasm needs to be introduced into Chinese breeding programs to broaden the genetic base of vegetable soybean. This will benefit vegetable soybean breeding and variety improvement in the future.
In summary, this paper is the first report concerning the development of EST-SSRs in vegetable soybean. Twenty-two EST-SSR markers from grain soybean showed high polymorphism in vegetable soybean and were successfully used to investigate the genetic diversity among 48 vegetable soybean accessions from China, Japan, and the USA. These results suggest that EST-SSRs from grain soybean have high transferability to vegetable soybean and that these new EST-SSR markers would be useful in vegetable soybean germplasm conservation, cultivar identification, molecular mapping, and marker-assisted selection in the future.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31101538, 31000942, and 31000676), the Grand Science and Technology Special Project of Zhejiang Province (Nos. 2010 C02006, 2012C12903-4-1, and 2012C12903-6-3), the Zhejiang Provincial Natural Science Foundation of China (No. LY12C15004), the Public Welfare Project of Zhejiang Province (No. 2011C22011), and the Shaoxing Important Science and Technology Projects (No. 2012A22008), China
Compliance with ethics guidelines: Gu-wen ZHANG, Sheng-chun XU, Wei-hua MAO, Qi-zan HU, and Ya-ming GONG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing parental selection for genetic linkage maps. Genome. 1993;36(1):181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- 2.Arikit S, Yoshihashi T, Wanchana S, Tanya P, Juwattanasomran R, Srinives P, Vanavichit A. A PCR-based marker for a locus conferring aroma in vegetable soybean (Glycine max L.) Theor Appl Genet. 2011;122(2):311–316. doi: 10.1007/s00122-010-1446-y. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary S, Sethy NK, Shokeen B, Bhatia S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor Appl Genet. 2009;118(3):591–608. doi: 10.1007/s00122-008-0923-z. [DOI] [PubMed] [Google Scholar]
- 4.Cornelious BK, Sneller CH. Yield and molecular diversity of soybean lines derived from crosses of northern and southern elite parents. Crop Sci. 2002;42(2):642–647. doi: 10.2135/cropsci2002.0642. [DOI] [Google Scholar]
- 5.Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li YD. Developing new SSR markers from ESTs of pea (Pisum sativum L.) J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2010;11(9):702–707. doi: 10.1631/jzus.B1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li ZY. Generation and characterization of 11 novel EST derived microsatellites from Vicia faba (Fabaceae) Am J Bot. 2010;97(7):69–71. doi: 10.3732/ajb.1000166. [DOI] [PubMed] [Google Scholar]
- 7.Gong YM, Xu SH, Mao WH, Li ZY, Hu QZ, Zhang GW, Ding J. Genetic diversity analysis of faba Bean (Vicia faba L.) based on EST-SSR markers. Agric Sci China. 2011;10(6):838–844. doi: 10.1016/S1671-2927(11)60069-2. [DOI] [Google Scholar]
- 8.Hisano H, Sato S, Isobe S, Sasamoto S, Wada T, Matsuno A, Fujishiro T, Yamada M, Nakayama S, Nakamura Y, et al. Characterization of the soybean genome using EST-derived microsatellite markers. DNA Res. 2007;14(6):271–281. doi: 10.1093/dnares/dsm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keatinge JDH, Easdown WJ, Yang RY, Chadha ML, Shanmugasundaram S. Overcoming chronic malnutrition in a future warming world: the key importance of mungbean and vegetable soybean. Euphytica. 2011;180(1):129–141. doi: 10.1007/s10681-011-0401-6. [DOI] [Google Scholar]
- 10.Kuroda Y, Tomooka N, Kaga A, Wanigadeva SMSW, Vaughan DA. Genetic diversity of wild soybean (Glycine soja Sieb. et Zucc.) and Japanese cultivated soybeans [G. max (L.) Merr.] based on microsatellite (SSR) analysis and the selection of a core collection. Genet Resour Crop Evol. 2009;56(8):1045–1055. doi: 10.1007/s10722-009-9425-3. [DOI] [Google Scholar]
- 11.Li AQ, Zhao CZ, Wang XJ, Liu ZJ, Zhang LF, Song GQ, Yin J, Li CS, Xia H, Bi YP. Identification of SSR markers using soybean (Glycine max) ESTs from globular stageembryos. Electron J Biotechnol. 2010;13(5):1–11. doi: 10.2225/vol13-issue5-fulltext-5. [DOI] [Google Scholar]
- 12.Li G, Ra WH, Park JW, Kwon SW, Lee JH, Park CB, Park YJ. Developing EST-SSR markers to study molecular diversity in Liriope and Ophiopogon . Biochem Syst Ecol. 2011;39(4-6):241–252. doi: 10.1016/j.bse.2011.08.012. [DOI] [Google Scholar]
- 13.Li YH, Guan RX, Liu ZX, Ma YS, Wang LX, Li LH, Lin FY, Luan WJ, Chen PY, Yan Z, et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet. 2008;117(6):857–871. doi: 10.1007/s00122-008-0825-0. [DOI] [PubMed] [Google Scholar]
- 14.Liu YL, Li YH, Zhou GA, Uzokwe N, Chang RZ, Chen SY, Qiu LJ. Development of soybean EST-SSR markers and their use to assess genetic diversity in the Subgenus soja . Agric Sci China. 2010;9(10):1423–1429. doi: 10.1016/S1671-2927(09)60233-9. [DOI] [Google Scholar]
- 15.Lu YB, Yang YW, Wu PD. Separation of phosphatidylcholine from soybean phospholipids by simulated moving bed. J Zhejiang Univ-Sci B. 2006;7(7):559–564. doi: 10.1631/jzus.2006.B0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10(1):72–80. [PMC free article] [PubMed] [Google Scholar]
- 17.Mimura M, Coyne CJ, Bambuck MW, Lumpkin TA. SSR diversity of vegetable soybean [Glycine max (L.) Merr.] Genet Resour Crop Evol. 2007;54(3):497–508. doi: 10.1007/s10722-006-0006-4. [DOI] [Google Scholar]
- 18.Moe KT, Zhao WG, Song HS, Kim YH, Chung JW, Cho YI, Park PH, Park HS, Chae SC, Park YJ. Development of SSR markers to study diversity in the genus Cymbidium . Biochem Syst Ecol. 2010;38(4):585–594. doi: 10.1016/j.bse.2010.07.004. [DOI] [Google Scholar]
- 19.Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with non-repetitive DNA in plant genomes. Nat Genet. 2002;30(2):194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- 20.Roy JK, Lakshmikumaran MS, Balyan HS, Gupta PK. AFLP-based genetic diversity and its comparison with diversity based on SSR, SAMPL, and phenotypic traits in bread wheat. Biochem Genet. 2004;42(1/2):43–59. doi: 10.1023/B:BIGI.0000012143.48298.71. [DOI] [PubMed] [Google Scholar]
- 21.Saldivar X, Wang YJ, Chen P, Mauromoustakos A. Effects of blanching and storage conditions on soluble sugar contents in vegetable soybean. LWT Food Sci Technol. 2010;43(9):1368–1372. doi: 10.1016/j.lwt.2010.04.017. [DOI] [Google Scholar]
- 22.Shultz JL, Kazi S, Bashir R, Afzal JA, Lightfoot DA. The development of BAC-end sequence-based microsatellite markers and placement in the physical and genetic maps of soybean. Theor Appl Genet. 2007;114(6):1081–1090. doi: 10.1007/s00122-007-0501-9. [DOI] [PubMed] [Google Scholar]
- 23.Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109(1):122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 24.Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.) Theor Appl Genet. 2000;100(5):697–712. doi: 10.1007/s001220051342. [DOI] [Google Scholar]
- 25.Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23(1):48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Wang XB, Mulock B, Guus B, McCallum BT. Development of EST-derived simple sequence repeat markers for wheat leaf rust fungus, Puccinia triticina Eriks. Can J Plant Pathol. 2010;32(1):98–107. doi: 10.1080/07060661003594133. [DOI] [Google Scholar]
- 27.Wen ZX, Ding YL, Zhao TJ, Gai JY. Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor Appl Genet. 2009;119(2):371–381. doi: 10.1007/s00122-009-1045-y. [DOI] [PubMed] [Google Scholar]
- 28.Xia ZJ, Tsubokura Y, Hoshi M, Hanawa M, Yano C, Okamura K, Ahmed T, Anai T, Watanabe S, Hayashi M, et al. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 2007;14(6):257–269. doi: 10.1093/dnares/dsm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yinbo G, Peoples MB, Rerkasem B. The effect of N fertilizer strategy on N2 fixation, growth and yield of vegetable soybean. Field Crop Res. 1997;51(3):221–229. doi: 10.1016/S0378-4290(96)03464-8. [DOI] [Google Scholar]
- 30.Young G, Mebrahtu T, Johnson J. Acceptability of green soybeans as a vegetable entity. Plant Food Hum Nutr. 2000;55(4):323–333. doi: 10.1023/A:1008164925103. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, Carter JTE, Cui Z, Miyazaki S, Burton JW. Genetic diversity patterns in Japanese soybean cultivars based on coefficient of parentage. Crop Sci. 2002;42(4):1331–1342. doi: 10.2135/cropsci2002.1331. [DOI] [Google Scholar]