Abstract
Genome-wide association studies (GWASs) have transformed the field of human genetics and have led to the discovery of hundreds of genes that are implicated in human disease. The technological advances that drove this revolution are now poised to transform genetic studies in model organisms, including mice. However, the design of GWASs in mouse strains is fundamentally different from the design of human GWASs, creating new challenges and opportunities. This Review gives an overview of the novel study designs for mouse GWASs, which dramatically improve both the statistical power and resolution compared to classical gene-mapping approaches.
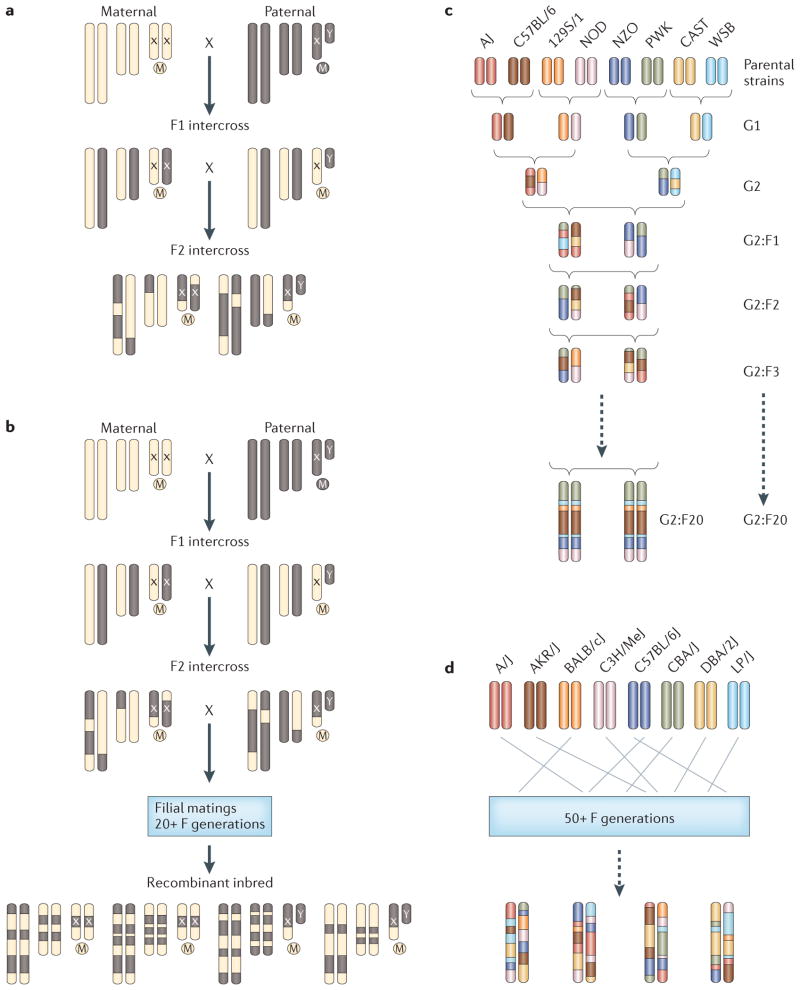
Until recently, the genetic cross was the most common design for identifying the sequence variants that contribute to phenotypic variation in mice. In a cross, two inbred strains are mated, and their offspring are either mated to each other (an intercross design) or to a progenitor strain (a backcross design) (FIG. 1a). Second-generation offspring are then phenotyped and genotyped, and linkage analysis is carried out to identify a region that is associated with the trait1.
Figure 1. Breeding schemes for mouse genome-wide association study populations.
a | In the classic F2 generation cross, two parental strains are mated to generate F1 strains. The F1 strains are then either mated to each other (intercross) or to one of the parental strains (backcross; not shown) to generate F2 offspring. These offspring are then genotyped and phenotyped. b | Recombinant inbred strains are generated by sibling mating F2 intercross animals until the resulting progeny, at least 20 generations later, is fully inbred. These inbred lines are maintained in breeding colonies and can be purchased from commercial vendors. Strategies that use recombinant inbred strains do not require genotyping as the genotypes of each animal in such a strain are identical and available in public databases. c | The Collaborative Cross is a large-scale project for generating recombinant inbred strains from eight parental strains using a breeding scheme that leads to inbred strains with, on average, equal genome content from each parental strain. Because the Collaborative Cross strains are inbred, strategies that use the strains do not require genotyping of the animals as the genotypes are available from public databases. d | Heterogeneous stock animals are the outbred offspring of eight parental strains. The breeding scheme generates animal offspring with, on average, equal genome content from each parental strain. Unlike inbred strains, these animals are genetically unique, and studies that use heterogeneous stock animals require genotyping of each animal included in the study. Panels a–c are reproduced, with permission, from REF. 84 © (2012) Macmillan Publishers Ltd. All rights reserved.
This approach has led to the identification of thousands of quantitative trait loci (QTLs) for various phenotypes and diseases. However, each QTL region is large, often tens of megabases, and contains hundreds of genes. The process of identifying the causal variant and the gene involved is therefore difficult and costly. Of the thousands of QTLs identified, only a small fraction of genes has been identified.
These results compare poorly to the recent success of human genome-wide association studies (GWASs), which have discovered hundreds of genes that are involved in dozens of common diseases2. However, the mouse studies also reveal that in a genetic cross only a few hundred animals are required to identify loci that together explain 50% or more of the phenotypic variance for a particular trait3. This finding is particularly striking compared to human studies, in which typically tens of thousands of individuals are required to identify loci that are involved in traits, and in which the loci identified typically explain only a small fraction of phenotypic variance4. These observations bode well for developing new ways of exploiting the genetic variation in mice to identify the basis of disease and traits related to those being mapped in humans. Furthermore, as discussed below, mouse studies have several advantages over human studies, including the accessibility of disease-relevant tissues, a greater ability to control environmental factors and an advanced genetic toolkit for the functional characterization of candidate genes5,6.
Indeed, alternative strategies to the genetic cross are now being developed in mice and are being fuelled by the same transformative technologies that have revolutionized human genetics. These strategies are taking advantage of advances in microarray7,8 and sequencing technologies9, which are yielding an almost complete map of genetic variation in various laboratory strains. For example, in the past year, the Sanger Institute has sequenced 17 mouse genomes and discovered 71 million SNPs10. Meanwhile, The Jackson Laboratory, in conjunction with the University of North Carolina, has developed a mouse-genotyping microarray — the Mouse Diversity Array, which measures 600,000 genotypes — and applied it to several hundred laboratory strains11. The result of these efforts is an almost complete picture of genetic variation and insights into the origin12–14 of the laboratory mouse.
In a separate effort, large-scale mouse-breeding programmes have generated inbred strains with advantageous genetic properties for carrying out genetic studies. These include efforts to expand existing populations of recombinant inbred strains (FIG. 1b) as well as to develop new strains, such as those developed by the Collaborative Cross project15. In addition, recent efforts to characterize commercially available outbred mouse resources provide gene-level resolution for mapping complex traits.
In this Review, we first discuss the general considerations for GWAS approaches, then describe how recently developed mouse GWAS strategies have been made possible by new mouse-breeding efforts and tools for large-scale sequencing, genotyping and phenotyping. These strategies differ in the genetic background of the mice that are included in the study, the genetic information that is collected and how the phenotypes are measured. They have been applied to identify loci that are involved in dozens of complex traits, including lipid and triglyceride levels16,17, bone density18, fear conditioning19, albumin-to-creatinine ratios in urine17, blood pressure17, exercise behaviour and metabolism20, and susceptibility to infectious diseases20.
We also discuss how the most appropriate GWAS strategy greatly depends on the goals of the study. The design of these strategies, as well as the analysis of data obtained, raises novel challenges and opportunities. However, all strategies have a common feature: they implicate much smaller regions than those that are typically found in classical genetic crosses, thus facilitating the process of identifying the underlying genes involved. Because so many models of human disease have been developed in mice, the developments provide a powerful and complementary approach towards understanding the genetics of human disease.
Mouse genetic studies: design considerations
There are various factors to bear in mind when designing a GWAS in any species. We begin by describing properties of GWAS designs and proceed to a discussion of the various advantages and disadvantages of each of the novel mouse GWAS strategies.
Statistical power
The most important property defining the expectations of a genetic mapping study is the anticipated or predicted statistical power. The statistical power measures the likelihood of detecting a genetic effect of a certain size given the number and type of animals included in the study. Informally, the statistical power can be thought of as the probability that the study will discover a locus of a certain effect on the trait. For example, a study of 90% power to detect a QTL that explains 10% of the variation will find a QTL of this magnitude, or larger, with a probability of 0.9, assuming that such a QTL exists in the population studied.
Mapping resolution
Mapping resolution measures the size of the interval implicated by the study. The resolution affects the number of genes that will be identified as candidates for harbouring the variant affecting the trait. A low resolution means that more genes and variants will have to be tested to confirm or to exclude their role in contributing to phenotypic variation; a high resolution means that fewer genes and variants need be tested.
Cost
The cost of a study includes the cost of obtaining and breeding the animals, maintaining them while phenotypes are scored and obtaining genetic information. For some animals, such as inbred strains, genetic information is publicly available, whereas for other animals the extent of genetic variation must be experimentally determined.
A genetic cross entails substantial mouse breeding and genotyping costs. For example, if the parental strains are obtained at breeding age, the total time until F2 generation mice are ready to be phenotyped is approximately 5 to 6 months. If a study consists of 200 F2 mice, the total number of animals involved in the study, including those generated by breeding, is typically about 300 animals. Each F2 mouse must be genotyped because it has a unique mixture of parental chromosomes.
Coverage
The coverage of a study refers to the extent to which the genome is polymorphic between the animals being analysed. Strategies with a higher coverage have the potential to discover more variants that affect traits. For example, a standard F2 genetic cross only covers the genetic variation in the two parental strains. Thus, if a genetic variant that affects the trait is present in a third strain but not in the parental strains, this variant cannot be discovered in the cross.
Reproducibility
This property refers to how easily the study can be replicated. Studies that use inbred strains, such as the recombinant inbred lines, can be fully replicated because the animals in a replication study are genetically identical to the animals in the original study. By contrast, animals from an intercross or a backcross are genetically unique, so although it is possible to replicate the cross, the specific genotypes of the resultant animals will be different each time.
Novel strategies for genetic studies in mice
Modern strategies for mouse association studies differ in the genetic structure of the mice included in the studies and will therefore vary with respect to the principles outlined above. As our analysis below shows, each strategy involves trade-offs, and exactly which strategy is best for a specific study greatly depends on the goals of the study. The following sections focus on the most widely applied mapping strategies, which are summarized in TABLE 1. These are the classical inbred strain association, the Hybrid Mouse Diversity Panel (HMDP), the Collaborative Cross, heterogeneous stocks and commercial outbred stocks. A general overview of mouse GWASs is shown in FIG. 2.
Table 1.
A summary of the different strategies for mouse genome-wide association studies
| Strategy | Requires genotyping? | Requires breeding? | Genetic diversity? | Genome- wide power | Resolution | Refs* |
|---|---|---|---|---|---|---|
| Classic cross | Yes | Yes | No | Yes | Low | 50 |
| Inbred strain association | No | No | No‡ | No | High | 22 |
| Hybrid Mouse Diversity Panel | No | No | No | Yes | High | 16 |
| Collaborative Cross | No | No | Yes | Yes | Medium | 15 |
| Heterogeneous stock | Yes | No | No | Yes | High | 38 |
| Commercial outbred stock | Yes | No | No | Yes | Very high | 40 |
A representative reference for each study type. For a more comprehensive list of mouse genome-wide association studies, please see Supplementary information S1 (table).
Inbred strain association strategies have a low genetic diversity if they include only classical inbred strains, but they have a higher genetic diversity if they include wild-derived inbred strains.
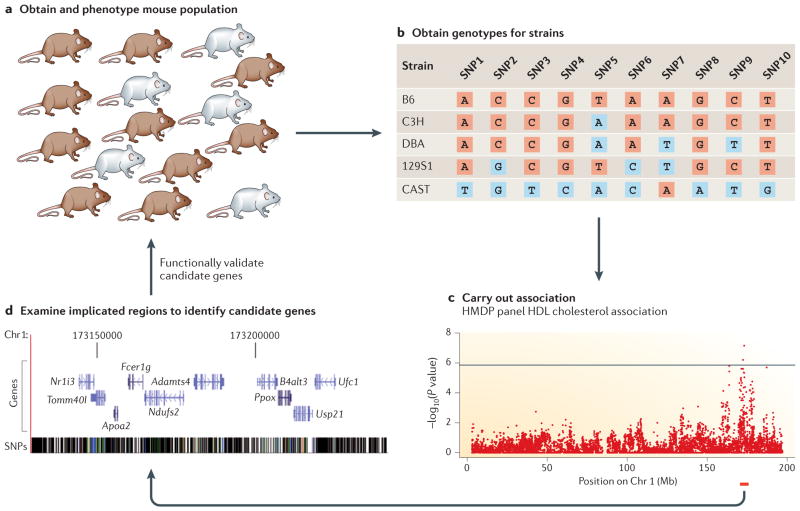
Figure 2. Overview of mouse GWASs.
Mouse genome-wide association studies (GWASs) follow a common general approach. a | Mice in the study population are phenotyped for the traits of interest. Deciding which mice and their corresponding genetic structure to include in the study population is a key design consideration in a mouse GWAS. b | Genotypes for each mouse are then obtained either by direct genotyping for outbred animals or by sourcing them from publicly available SNP maps for inbred strains. c | Association testing is then carried out, typically using a statistical method for correcting for population structure such as efficient mixed-model association (EMMA)31 or resample model averaging (RMA)79. d | Implicated regions are then examined for candidate genes, which are then functionally validated. Apoa2, apolipoprotein A2; Chr, chromosome; HDL, high-density lipoprotein; HMDP, Hybrid Mouse Diversity Panel. The data in part c are derived from REF. 16.
Classical inbred strain association
The first strategy to use the tools of association studies in mouse strains applied an association methodology to a set of commonly used laboratory strains21–24. Pletcher et al.22 described the first mouse GWAS, which led to 11 distinct loci being reported for high-density lipoprotein (HDL) cholesterol levels. In their pioneering work, the authors collected genotypes at 10,990 SNPs in 48 inbred mouse strains. A GWAS was then carried out by measuring HDL cholesterol in 25 of these strains and correlating the collected phenotypes and genotypes.
The inbred strains association approach gives a much higher mapping resolution than a typical genetic cross, because the inbred mouse strains are separated from their founders by more generations. These more distant relationships include many more recombination events between the genomes of the founding strains, thereby increasing the mapping resolution down to approximately 2 Mb, as reported in the initial studies. The approach had several other advantages compared to genetic crosses. First, classical inbred strain GWASs do not require any breeding steps as the inbred strains required for phenotyping can be directly purchased from a vendor such as The Jackson Laboratory, reducing the time and cost of the study compared to a genetic cross. Second, classical inbred strain GWASs sample a larger amount of variation, because they include many more strains than just the two parental strains of a cross. Third, they are completely reproducible as the exact strains can be used in a replication study, so that the genetic structure of each of the animals in the original and replication study are identical. Finally, inbred strains are homozygous at each locus, thereby increasing the power of the association approach, particularly for recessive loci.
The first inbred strain association studies fuelled an interest in applying association techniques in larger cohorts of inbred strains. This interest drove the development of the Mouse Phenome Project25–27, which is a project at The Jackson Laboratory that aims to create a large catalogue of phenotypes for each of the inbred strains. Most strains have now been genotyped or sequenced, and their genotypes are available in public databases10–14,28–30, so investigators do not need to obtain genotypes experimentally.
At the time of the first inbred strain studies, it appeared that the strategy had adequate power to identify genetic variation involved in traits. However, it is now understood that many of the identified associations were driven by population structure (BOX 1); the differing degrees of relatedness among the strains gave rise to false-positive associations16,31,32. After correcting for population structure, studies using this design were shown to have limited statistical power to identify genes30: most of the reported associations were false positives32,33.
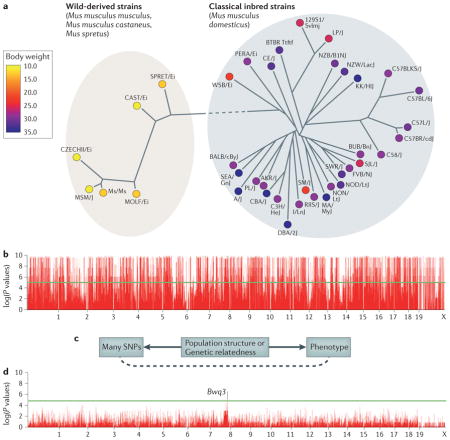
Box 1. Population structure.
A challenge in mouse genome-wide association studies (GWASs) is the complex genetic relationships between strains included in the study. Some of these differences stem from the distinct ancestral origins of the mice, such as the differences between wild-derived strains and classical inbred strains, which are primarily descended from domesticated mice12–14. Additionally, among strains, there is variability in the degree to which particular genomic regions are shared owing to the complex breeding history.
Traditional association statistical tests make the assumption that the phenotypes of individuals in an association are independent. However, owing to the complex genetic relationships, this assumption is violated for mouse GWASs. Closely related strains will have more similar phenotype values than more distant strains. This phenomenon, which is termed population structure, causes spurious associations in GWASs. Recently, statistical methods have been developed to address this problem, including efficient mixed-model association (EMMA)31 and resample model averaging (RMA)80, which are widely used in mouse GWASs, and EIGENSTRAT81 and EMMAX82, which are widely used in human studies. The figure demonstrates this problem for mouse GWASs.
Panel a shows body-weight data for 38 inbred strains from the Mouse Phenome Database as analysed in REF. 31. A phylogeny of the strains is shown, demonstrating a clear genetic distinction between the wild-derived strains and the classical inbred strains. Note that all wild-derived strains have a lower body weight than classical inbred strains. Panel b shows a Manhattan plot with the association results for 140,000 SNPs30 and body weight. Almost every locus appears to be associated with body weight as each of the many SNPs that differentiate the wild-derived and classical inbred strains appears to be associated with body weight. A visualization of the cause of the spurious associations is shown panel c. Many SNPs and the phenotype are both correlated with the genetic relatedness or population structure among the strains. Statistical techniques can take into account the genetic relationships between the strains to correct for population structure, thus minimizing spurious associations. In this example, EMMA was applied to the data (panel d). The highest peak, although not genome-wide significant, occurs on chromosome 8 and is near the logarithm of the odds (lod) peak of a previously known body weight quantitative trait locus Bwq3 (REF. 83). Panels b and d are reproduced, with permission, from REF. 31 © (2008) Genetics Society of America.
Hybrid Mouse Diversity Panel
Whereas classical association studies had advantages in terms of resolution, cost, coverage and reproducibility, their main weakness was a lack of power for genome-wide association and a failure to deal with population structure. An extension of the classical inbred strain design is the HMDP16, which increases the statistical power of the classical association studies by including a set of 70 recombinant inbred strains in the mapping panel. In this design, approximately 100 strains are phenotyped (30 classical inbred strains and 70 recombinant inbred strains), and association is carried out after correcting for population structure using efficient mixed-model association (EMMA) (BOX 1). The mapping resolution of an association study using recombinant inbred strains is actually lower than in a design that uses classical inbred strains. However, using the combined population included in the HMDP provides a high statistical power (from the recombinant inbred strains) and a high resolution (from the classical inbred strains). The set of strains included in the HMDP was determined by carrying out power simulations31. HMDP studies have a comparable resolution to the classical inbred strain strategy of approximately 2 Mb interval sizes. Because these strains are all inbred, they retain the advantages of the classical inbred strain design, such as cost, coverage and reproducibility.
A limitation of the HMDP is the number of available inbred strains, particularly recombinant inbred strains. It is likely that the maximum number of strains that can be used in such a study is between 200 and 300, even if all currently available strains are used. This results in an upper limit on the statistical power of the HMDP. The currently used 100 strain HMDP panel with 8–10 animals per strain has about 80% power to detect loci that account for approximately 5% of the variance of the trait.
The HMDP approach has been applied to a host of phenotypes, including lipid levels16, bone density18 and fear conditioning19. These studies highlight the advantage of increased resolution, allowing identification of genes that underlie implicated loci, such as the previously known apolipoprotein A2 (Apoa2) in HDL cholesterol16 and the novel association of additional sex combs-like 2 (Asxl2) with bone mineral density18.
The Collaborative Cross
The Collaborative Cross is a large-scale effort to create a set of recombinant inbred strains that are specifically designed for mapping traits and, more generally, for applying a systems-biology approach to mouse genetics15,20,34. Unlike the HMDP, which consists of currently available strains, the Collaborative Cross has generated new inbred strains using a specific breeding scheme: recombinant inbred strains are being derived from eight founder strains with the goal of producing animals that have on average an equal combination of ancestry from each founder (FIG. 1c). Five of the founders are classical inbred strains and three are wild-derived strains. The classical inbred strains lack variants in many genes, so using wild-derived strains introduces a substantial amount of genetic diversity into the Collaborative Cross and thus provides the advantage of covering more variation compared with other strategies. In particular, there are likely to be more variants in genes that perturb traits of interest. A second advantage of the Collaborative Cross is that there is less population structure compared to other strategies. Whereas techniques such as EMMA are available to correct for population structure, the presence of population structure still has a negative effect on statistical power.
There has recently been a tremendous amount of activity related to the Collaborative Cross as the first strains are nearing completion35,36. Although the final Collaborative Cross strains are not complete, studies are being undertaken using ‘pre-Collaborative Cross’ strains, which are incompletely inbred versions of the Collaborative Cross strains. A recent study successfully mapped several haematological phenotypes37.
Heterogeneous stock
The limited number of inbred strains that is either currently available or that can be generated by efforts such as the Collaborative Cross poses a fundamental limit on the statistical power that can be attained in a mouse genetic study that uses inbred strains. An alternative strategy is to use out-bred mice. These include heterogeneous stock mice38 (for which animals are descended from eight classical inbred founder strains (FIG. 1d) and the Diversity Outbred mice39 (which comprises animals descended from the eight Collaborative Cross founder strains). Both heterogeneous stock and Diversity Outbred animals are similar to F2 animals generated from a cross, but they have ancestry from eight founder strains instead of only two, and the population is bred for more generations. The main advantage of heterogeneous stock strategies is that they can be used to generate an almost limitless number of animals, enabling large studies to be carried out that can find weak genetic effects. In addition, owing to their breeding history, animals have undergone many more recombination events with mapping resolution of approximately 3 Mb for a typical QTL. However, unlike inbred strains, each heterogeneous stock mouse is unique and does not have the phenotyping and reproducibility advantages offered by inbred strains nor the availability of genotypes. Furthermore, the population structure in the heterogeneous stock designs complicates the analysis and reduces power. A very large study consisting of more than 2,000 outbred mice used an extensive high-throughput phenotyping strategy38. A total of 101 phenotypes were measured, including behavioural traits, common-disease-related traits, biochemistry, haematology and immunology-related traits. The study successfully implicated loci in almost all of the phenotypes38.
Commercially available outbred stock
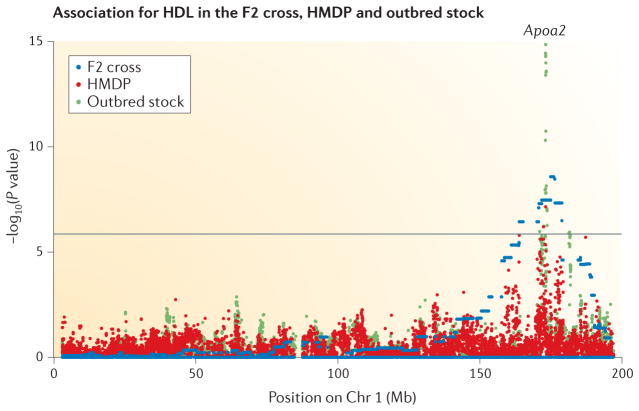
An alternative source of outbred mice is available from commercial vendors. These animals have a genetic structure that differs from the carefully constructed heterogeneous stock approach described above. Many such vendors have large colonies that they have maintained for many generations, although they are descended from a small number of classical inbred strains. A recent characterization of these populations has shown that some stocks have desirable properties for high-resolution mapping owing to the large number of generations since the founding of the stock. In fact, some stocks can achieve a resolution of interval sizes of less than 100 kb40. FIGURE 3 shows a comparison of the resolution of studies using a genetic cross, the HMDP and commercial outbred stocks for the association for HDL cholesterol on chromosome 1.
Figure 3. Comparison of mouse GWASs for HDL cholesterol.
The figure highlights differences in resolution of genome-wide association studies (GWASs) of high-density lipoprotein (HDL) between the strategies of the F2 generation genetic cross85, the Hybrid Mouse Diversity Panel (HMDP)16 and commercial outbred stocks17. The results are shown for chromosome 1, in which a known gene, apolipoprotein A2 (Apoa2), has been previously linked to HDL levels86 and is likely to drive the association signal in all three studies. All three strategies successfully identify an association in the region. However, as shown in the figure, the genetic cross study implicates a very broad region covering a substantial fraction of the chromosome owing to the limited number of recombinations between the parental strains of the cross. The HMDP study narrows down the association signal to a much smaller region owing to the larger number of generations that separate the inbred strains. The outbred stock study localizes the association even further owing to the large number of generations since the founding of the stocks. Chr, chromosome. The data sets presented are from REFS 16,17,85.
Large differences in allele frequencies between populations mean that no single commercial stock outbred population is ideal, but they also mean that if mapping in one population fails, mapping in another population may succeed because of differences in the linkage disequilibrium patterns between populations. The populations differ owing to the unique population history of each stock, yet for all of these stocks, the vast majority of the variation present in the animals is derived from the genetic differences present in the founding inbred strains. This insight makes it possible to obtain accurate genotype information for each animal by collecting only several hundred markers and applying imputation41–43. Commercial out-bred mice were recently used to map variants associated with HDL cholesterol, systolic blood pressure, triglyceride levels, glucose and urinary albumin-to-creatinine ratios17, and simulations using the panel of mice used in the mapping showed that most of the peak associations are within 500 kb of the causal variant.
Choosing a strategy for mouse GWASs
Which approach works best for a given study depends on many factors, including the genetic architecture of the trait of interest and the goals of the study. We discuss these issues in terms of the design choices that underlie the mouse GWAS strategies.
Inbred or outbred?
A key differentiator between the novel mouse GWASs is that some of the strategies use inbred strains, such as those from the HMDP or the Collaborative Cross, whereas other GWASs use outbred animals, such as heterogeneous stock and commercial outbred mice. The inbred strains have the advantage of genomic homozygosity and complete reproducibility of phenotype measurements. In fact, if a phenotype has a low heritability or if the assay to measure it has a large amount of noise, multiple genetically identical animals can be measured to obtain accurate estimates of the phenotype in each strain. These measurements can then be combined with any other measurements on the same set of strains, resulting in a much richer set of phenotypes. However, the number of available inbred strains is limited. If the variants to be identified have sufficiently small effect sizes that even using all available inbred strains will have only limited power, the only practical approach is to use a large number of outbred animals.
Genetic diversity: more or less?
Another key differentiating factor is whether to include animals that are descended from wild-derived strains in addition to animals that are descended from classical inbred strains. The Collaborative Cross includes several wild-derived founder strains, and for this reason it includes an order of magnitude larger amount of diversity compared to the other strategies34. However, a high proportion of alleles is derived from just one of the wild-derived strains. Linkage disequilibrium decay for these private variants solely depends on the number of recombination events that have accumulated during the creation of the population; this reduces the resolution compared to strategies based on inbred strains. Furthermore, as the amount of genetic variation increases in a study, the power to detect variants is reduced for two reasons. First, there are likely to be many more variants involved in the trait, reducing the relative effect size of any given variant. Second, there are many more polymorphic variants, decreasing the amount of linkage disequilibrium and increasing the multiple testing penalty when carrying out GWASs. However, increased diversity has several advantages, including that there are more variants with genetic effects that can be found. In addition, if the goal of a study is to follow up specific human GWAS results, greater diversity increases the chance that variants in the gene of interest that affect the trait are present in the chosen mouse population.
Breeding scheme or opportunism?
The final consideration is whether it is preferable to use populations that are specially designed for mapping studies (such as the Collaborative Cross or heterogeneous stock) or whether opportunistically to use currently available mouse resources that were not designed for mapping (such as the HMDP or commercially available outbred stock). In general, the specially designed mouse populations each have some distinct advantages, such as lower amounts of population structure and more control over the level of genetic diversity. However, resolution is bounded by the number of recombination events that can be limited for specially designed mouse populations that were only recently generated.
From implicated loci to genes
Three common ways to prove the involvement of a gene or gene variant in a complex trait are as follows. The first way is reciprocal hemizygosity, which was developed in yeast and in which a pair of F1 strains is generated that harbour a mutant allele of a gene of interest on one chromosome and a deletion of that gene on the homologous chromosome44 (to allow the phenotypic effect of the allele of interest to be seen). The mutant alleles differ between the two F1 strains, and a difference in phenotype between these F1 strains implicates the gene in the phenotype. The second technique is quantitative complementation, which was developed in Drosophila melanogaster45,46. When this method is applied to mice, a pair of strains that carry different alleles at the locus is taken from the mouse GWAS. This pair of strains is then bred with a knockout strain for the gene and with the background strain for the knockout. Four F1 strains are generated, and their phenotypes are analysed to test for an interaction between a null allele and the QTL (rather than for a main effect of either); a significant interaction indicates that the allelic differences in the gene are (at least partly) responsible for the QTL. The third technique is finding an enrichment of rare or low-frequency variants in candidate genes in cases relative to controls, as determined by resequencing candidate genes. This strategy was developed for human studies and has successfully identified human genetic variants that are associated with type 1 diabetes47 or Crohn’s disease48.
To date, these three methods have not been widely used in mouse genetics. Reciprocal hemizygosity requires such complex genome manipulation that it is almost impossible to implement in mice. Quantitative complementation is technically possible and has been implemented49,50,51, but it requires co-isogenic wild-type strains, which can be difficult to obtain in mice. Most mouse knockouts are still created in a 129 strain and are back-crossed onto a different strain (typically C57BL/6) so that often no pure co-isogenic wild-type strain is available. Finally, resequencing candidate genes requires access to thousands of unrelated individuals, and this is not an option in mouse genetics.
Perhaps not surprisingly, therefore, mouse geneticists have resorted to applying less-stringent tests for establishing the causality of candidate genes at QTLs. One simple test is whether a knockout of the candidate gene has an abnormal phenotype52. It should be noted that this does not prove that the QTL acts either at or through the gene. A knockout allele is rarely the same as the variant allele at a QTL, so finding that the knockout has an effect on the phenotype does not prove the candidacy of the gene. Almost all variants that act at QTLs do so in subtle ways, perhaps by increasing or decreasing expression of a transcript; moreover, contrasting phenotypes can be attributed to different alleles at the same gene53. Therefore, it would be wrong to expect the complete removal of a transcript to model the QTL effect. Some phenotypes, such as height or weight, are influenced by so many genes that more than one-third of all knockouts may show a phenotype54. In fact, relying on knockouts to implicate a gene at a QTL can give rise to both false-negative and false-positive results. False positives can occur if the knocked-out gene is not the gene that harbours the variant that causes the association signal observed at the QTL but nonetheless has an effect on the phenotype. False negatives can occur if the knocked-out gene harbours the causal variant, but the knockout does not share the same phenotype.
An alternative approach towards identifying the relevant gene is the labour-intensive process of generating congenic strains55–58, which are specially constructed strains that are designed to fine-map a region. However, these approaches have yielded only a few gene associations and are successful only when the effect of the gene on the trait is very large. The promise of the recent developments in mouse genetics has been to provide alternative strategies for identifying QTLs with orders of magnitude greater resolution than for traditional linkage analyses, thus in principle facilitating the identification of causal genes or variants. However, even with these new strategies, the resolution of mouse genetic studies is still poorer than the resolution of human genetic studies.
Supplementary information S1 (table) provides a summary of candidate genes identified since 2005 and lists the ways in which this has been achieved. The table clearly shows that the use of gene expression data is a common and successful method for prioritizing candidate genes. In most cases, investigators look for an association between differential gene expression levels and the trait of interest as a way to implicate a gene. However, more complex methods are possible: for example, by comparing a statistical model — in which genetic variation contributes to transcript variation and that in turn contributes to phenotypic variation — with alternative non-causal models59. This approach can be extended so that an entire network of transcripts is associated with a phenotype60.
In most cases, investigators use a composite method to find genes — often referred to as a ‘bioinformatics tool-box’ — in which data from sequence, expression and the published literature are synthesized to increase the likelihood that the causal gene has been found61,62. The adequacy of this method has, however, yet to face rigorous testing. One assessment of the efficacy of incorporating expression data for isolating genes at QTLs concluded, after reviewing 37 studies, that the method had limited success: although it reduces the numbers of potential candidates (just 1.9% of candidate genes showed differential expression), meta-analysis showed that this filtering of candidates resulted in no significant over-representation of genes in QTL regions in 70% of studies63.
Supplementary information S1 (table) also shows the continuing importance of the intercross for identifying QTLs. Despite the availability of advanced intercrosses, heterogeneous stocks, outbred stocks and now the Collaborative Cross, the workhorse of genetic mapping remains the intercross. Overall, this table demonstrates that the field of mouse complex trait genetics has yet to exploit fully the new resources and technologies that are now available. Moving from genetic mapping towards identifying the causal variant, and identifying the genes that the variant affects, remains a major challenge.
Mouse or human genetic studies?
The ultimate goal of mouse genetic studies is to understand the mechanism of human disease. There are many trade-offs between mouse and human studies. The most fundamental is that mouse studies are limited to disease models that are only approximations of human disease.
The genetic structure of mouse and human studies differs greatly. The process of inbreeding a mouse strain can be thought of as obtaining a single haplotype from a wild population. When two inbred strains are crossed, the resulting offspring will have allele frequencies close to 50% for all polymorphic variants, independent of their effect sizes. This breaks the inverse relationship between allele frequency and effect size that is observed in humans19 and other wild populations. Thus, variants with large effect sizes, which were correspondingly rare in wild mouse populations, might be artificially increased in frequency in a mouse study. This leads to associations being discovered in only a few hundred animals, and together these associations explain a large fraction of the phenotype3. This compares favourably with the results of human studies that involve hundreds of thousands of individuals64,65 and that identify loci with only small effects that account for a small fraction of the variance66. Because they require many fewer samples, mouse GWASs are typically much cheaper than human studies, particularly for strategies that do not require any genotyping. In addition, human geneticists working with disease phenotypes have no choice but to use a case–control design, comparing individuals with and without the disease. By contrast, mouse phenotypes are not so constrained. The mouse models of disease almost always contain a quantitative measure of the phenotype, and this increases the power to map loci that affect the phenotype.
However, even with the novel mapping strategies, the resolution of a human GWAS is still superior to a mouse mapping study. This is because human population history has resulted in more extensive levels of linkage disequilibrium decay; therefore, QTLs identified in human studies are usually shorter and contain fewer genes than regions implicated by mouse studies. Furthermore, GWASs of humans typically use larger numbers of individuals than GWASs of mice, and this also increases mapping resolution.
Despite these disadvantages, mouse genetic studies have several major advantages relative to human studies for dissecting the genetic mechanism of disease. Whereas human studies can access only a few tissue types, mouse studies can easily measure traits in any tissue, including the tissue that is most relevant to the disease, which is often inaccessible in human studies. Similarly, unlike in human studies, the environment in mouse studies can be carefully controlled, facilitating studies that examine gene-by-environment interactions. Mouse studies also facilitate the functional characterization of candidate causal genes or variants by using, for example, gene knockouts and allele swaps, which are impossible in human studies.
In summary, although identifying the gene that underlies a QTL can be more difficult in mouse models, following up this gene to understand the mechanism is easier than in human studies.
Future directions
The strategies discussed in this Review are only a starting point for improving the ability of mouse genetic studies to identify genes that underlie human-disease-related traits. Many other strategies are currently being proposed3,67.
Exploiting the genetic diversity of wild mice
Wild mice, which should not be confused with wild-derived inbred strains, are substantially more outbred than laboratory strains: they contain more genetic diversity and have undergone a larger number of recombination events between the founder genomes, leading to a higher mapping resolution68. Each wild-derived inbred strain can be thought of as capturing a single haplotype from the population of wild mice. Genetic studies using wild mice will probably have a resolution that is similar to human studies, but they are also likely to require thousands of individuals: similar in number to the individuals involved in a human GWAS. One difficulty posed by this strategy is that wild mice need to be sequenced to obtain their genetic variation. Genotyping microarrays such as the Mouse Diversity Array may perform poorly on these animals because the genetic variation that is polymorphic in wild mice may not be captured in the microarray, as it represents only a small number of wild-derived laboratory strains. A further difficulty is that wild mice are infected with many diseases that prohibit their introduction into animal breeding facilities.
Advantages of using F1 animals
A strategy that uses laboratory mice directly involves including F1 strains in mouse association studies in addition to inbred strains, making more strains available for a study. F1 strains are the offspring of two inbred strains and can be generated from any pair of inbred strains, including either classical inbred strains or the Collaborative Cross strains. In principle, the added value of such F1 mice depends on the genetic inheritance model of the trait. If the mode of inheritance is assumed to be additive, then the use of F1 animals will add little to the statistical power of the study. Conversely, if the genetic model assumes dominant or recessive effects, then the F1 strains will enhance statistical power owing to the many heterozygous loci in the F1, but not the inbred, mice.
F1 strains can also be used to identify genes that modify a dominant genetic variant69. In this approach, a strain that is homozygous for a dominant variant is bred to many inbred strains, generating F1 mice with one parent transmitting the dominant variant and the second parent transmitting a chromosome without the variant. Each F1 mouse is phenotyped, and association is carried out between the genotypes of the alternate parent and the phenotype. The results of this association are loci that modify the dominant variant effect.
Cell-based strategies
Cell lines that have been generated from mouse strains can also be used for genetic studies70,71. The idea behind these studies is that cell lines are generated from each mouse strain and that phenotyping is carried out on these cell lines. Although the phenotyping strategies greatly differ from whole-animal studies, the phenotypes can be mapped in the same way. Advantages of cell-based strategies include their low cost and the ease of both measuring phenotypes and carrying out manipulations (such as exposure to oxidative stress72). However, a disadvantage is that the measured cell-based phenotypes may not be as relevant to human disease as they would be in whole-animal studies.
Promoting recombination events
There has been great progress in understanding recombination with the discovery of recombination hotspots and the molecular mechanisms that promote recombination73,74. Potentially, these discoveries can be leveraged either by implanting hotspots into regions of the mouse genome in which we would like to observe more recombination events in breeding or by increasing the total number of recombinations per generation.
Exploring quantitative genetics
Finally, the human GWAS results — characterized by the small effect sizes of implicated variants that collectively explain only a small portion of the genetic variance of a trait — have led to great debate on the cause for this so-called ‘missing heritability’66. Many candidates have been proposed, including epistatic interactions75, gene-by-environment interactions76, rare variants77 and structural variants66,78. The ability to manipulate the genomes and environment of mouse studies provides exciting opportunities to explore these issues in an attempt to understand the causes of missing heritability in human GWASs79. In addition, although the focus of this Review has been on medical traits, mouse GWASs can be used as tools to analyse fundamental biological processes in mammals and to provide insights into general principles of development, physiology and evolution in mammals.
Overall, having been driven by the same advances that have transformed human genetics in the past several years, recent developments in mouse studies have led to a proliferation of methods for increasing the mapping resolution of GWASs compared to genetic crosses. Using these strategies, many groups are rapidly identifying regions in the mouse genome that have been implicated in complex traits that are relevant to human disease, leading to the discovery of additional genes involved in human disease. Moreover, the genetic tractability of mice provides great potential for functional characterization of the implicated genes, thus contributing to the elucidation of human disease mechanisms.
Supplementary Material
Acknowledgments
J.F. is supported by the Wellcome Trust. E.E. is supported by US National Science Foundation grants 0513612, 0731455, 0729049, 0916676 and 1065276, and US National Institutes of Health grants K25-HL080079, U01-DA024417, P01-HL30568 and PO1-HL28481. This research was supported in part by the University of California, Los Angeles, subcontract of contract N01-ES-45530 from the National Toxicology Program and the National Institute of Environmental Helath Sciences to Perlegen Sciences.
Glossary
- Inbred strains
Mouse strains that have been sibling-mated for at least 20 generations to the point that both alleles at each locus are expected to be identical
- Linkage analysis
A statistical method for identifying a region of the genome that is implicated in a trait by observing which region is inherited from the parental strain carrying the trait in offspring that carry the trait
- Quantitative trait loci
(QTLs). Regions of the genome that are implicated in a quantitative trait
- Recombinant inbred strains
Inbred strains that are generated by sibling-mating the offspring of a genetic cross until the progenies are inbred
- Collaborative Cross project
A large panel of inbred mouse strains that are currently being developed through a community effort. The strains are derived from an eight-way cross using a set of founder strains that include three wild-derived strains
- Population structure
Differences in levels of genetic similarity between individuals in the study population. Population structure can cause spurious associations in genetic studies
- Imputation
A statistical procedure to predict the values of genetic variation which was not collected using observed genetic variation and genetic reference data sets
- Heritability
A measure of the genetic component of phenotypic variance of a trait
- Linkage disequilibrium decay
The decrease in the correlation between genetic variants as the distance between the variants increases
- Private variants
Genetic variants that are confined to single individuals, families or populations
- Multiple testing
A statistical problem that arises from carrying out many (in the order of thousands) hypothesis tests together. The significance threshold must be appropriately corrected to avoid false positives; for example, by using the Bonferroni correction
- F1 strains
Mouse strains that are generated by breeding two inbred strains together. An F1 mouse has one chromosome from each of the parental strains
- Co-isogenic wild-type strain
A strain that differs from the wild-type strain at only a single locus through a mutation occurring in the wild-type strain
- Congenic strains
Strains that are produced by a breeding strategy in which recombinants between two inbred strains are backcrossed to produce a strain that carries a single genomic segment from one strain on the genetic background of the other
- Additive
In the context of a genetic effect, the linear relationship between the replacement of an allele and its effect on the phenotype
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Jonathan Flint’s homepage: http://www.well.ox.ac.uk/flint-2
Eleazar Eskin’s homepage: http://www.cs.ucla.edu/~eeskin
GeneNetwork: http://www.genenetwork.org
The Jackson Laboratory: http://www.jax.org
MGI — Mouse Genome Informatics: http://www.informatics.jax.org
Mouse Genome Variation Identifier: http://www.berndtlab.pitt.edu
Mouse Phenome Database: http://phenome.jax.org
Nature Reviews Genetics Series on Genome-wide association studies: http://www.nature.com/nrg/series/gwas/index.html
Nature Reviews Genetics Series on Study designs: http://www.nature.com/nrg/series/studydesigns/index.html
UCLA Mouse Genetics and Genomics Software and Tools: http://mouse.cs.ucla.edu
References
- 1.Silver LM. Mouse Genetics: Concepts and Applications. Oxford Univ. Press; 1995. This book is the classic resource on mouse genetics. [Google Scholar]
- 2.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke DT, et al. Dissection of complex adult traits in a mouse synthetic population. Genome Res. 2012;22:1549–1557. doi: 10.1101/gr.135582.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ringwald M, et al. The IKMC web portal: a central point of entry to data and resources from the international knockout mouse consortium. Nucleic Acids Res. 2011;39:D849–D855. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nature Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nature Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 9.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. This paper describes an almost complete map of genetic variation in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, et al. A customized and versatile high-density genotyping array for the mouse. Nature Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frazer KA, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nature Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nature Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Churchill GA, et al. The collaborative cross, a community resource for the genetic analysis of complex traits. Nature Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. This paper describes the motivation and rationale for the development of the Collaborative Cross. [DOI] [PubMed] [Google Scholar]
- 16.Bennett BJ, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. This paper describes the HMDP mouse GWAS strategy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, et al. Genome-wide association mapping of quantitative traits in outbred mice. G3. 2012;2:167–174. doi: 10.1534/g3.111.001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farber CR, et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 2011;7:e1002038. doi: 10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CC, et al. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. doi: 10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aylor DL, et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grupe A, et al. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- 22.Pletcher MT, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cervino AC, Darvasi A, Fallahi M, Mader CC, Tsinoremas NF. An integrated in silico gene mapping strategy in inbred mice. Genetics. 2007;175:321–333. doi: 10.1534/genetics.106.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClurg P, et al. Genomewide association analysis in diverse inbred mice: power and population structure. Genetics. 2007;176:675–683. doi: 10.1534/genetics.106.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogue MA, Grubb SC, Maddatu TP, Bult CJ. Mouse phenome database (MPD) Nucleic Acids Res. 2007;35:D643–D649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Mouse Phenotype Database Integration Consortium. Integration of mouse phenome data resources. Mamm Genome. 2007;18:157–163. doi: 10.1007/s00335-007-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2009;37:D720–D730. doi: 10.1093/nar/gkn778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beck JA, et al. Genealogies of mouse inbred strains. Nature Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 29.Wade CM, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 30.Kirby A, et al. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. This paper describes the EMMA approach, which is widely applied in mouse GWASs and is used for the correction of population structure in association studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manenti G, et al. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive loci. PLoS Genet. 2009;5:e1000331. doi: 10.1371/journal.pgen.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Payseur BA, Place M. Prospects for association mapping in classical inbred mouse strains. Genetics. 2007;175:1999–2008. doi: 10.1534/genetics.106.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philip VM, et al. Genetic analysis in the collaborative cross breeding population. Genome Res. 2011;21:1223–1238. doi: 10.1101/gr.113886.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Threadgill DW, Churchill GA. Ten years of the collaborative cross. Genetics. 2012;190:291–294. doi: 10.1534/genetics.111.138032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collaborative Cross Consortium. The genome architecture of the collaborative cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. This paper provides a description of genetic architecture of the generated Collaborative Cross strains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelada SN, et al. Genetic analysis of hematological parameters in incipient lines of the Collaborative Cross. G3. 2012;2:157–165. doi: 10.1534/g3.111.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdar W, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nature Genet. 2006;38:879–887. doi: 10.1038/ng1840. This paper provides a description of the heterogeneous stock strategy for mouse GWASs. [DOI] [PubMed] [Google Scholar]
- 39.Svenson KL, et al. High-resolution genetic mapping using the mouse diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yalcin B, et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 2010;6:e1001085. doi: 10.1371/journal.pgen.1001085. This paper describes the commercially available outbred stock mouse GWAS strategy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Mach: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JR, et al. Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics. 2012;190:449–458. doi: 10.1534/genetics.111.132381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinmetz LM, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 45.Long AD, Mullaney SL, Mackay TF, Langley CH. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting bristle number in Drosophila melanogaster. Genetics. 1996;144:1497–1510. doi: 10.1093/genetics/144.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackay TF. Quantitative trait loci in Drosophila. Nature Rev Genet. 2001;2:11–20. doi: 10.1038/35047544. [DOI] [PubMed] [Google Scholar]
- 47.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yalcin B, et al. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nature Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- 50.Su Z, et al. Four additional mouse crosses improve the lipid QTL landscape and identify Lipg as a QTL gene. J Lipid Res. 2009;50:2083–2094. doi: 10.1194/jlr.M900076-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nature Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackay TF. Complementing complexity. Nature Genet. 2004;36:1145–1147. doi: 10.1038/ng1104-1145. [DOI] [PubMed] [Google Scholar]
- 53.Wilkie AO. Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 2005;16:187–203. doi: 10.1016/j.cytogfr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- 55.Wakeland E, Morel L, Achey K, Yui M, Longmate J. Speed congenics: a classic technique in the fast lane (relatively speaking) Immunol Today. 1997;18:472–477. doi: 10.1016/s0167-5699(97)01126-2. [DOI] [PubMed] [Google Scholar]
- 56.Markel P, et al. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nature Genet. 1997;17:280–284. doi: 10.1038/ng1197-280. [DOI] [PubMed] [Google Scholar]
- 57.Shao H, et al. Analyzing complex traits with congenic strains. Mamm Genome. 2010;21:276–286. doi: 10.1007/s00335-010-9267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis RC, et al. A genome-wide set of congenic mouse strains derived from CAST/ei on a C57BL/6 background. Genomics. 2007;90:306–313. doi: 10.1016/j.ygeno.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nature Rev Genet. 2012;13:523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- 62.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nature Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 63.Verdugo RA, Farber CR, Warden CH, Medrano JF. Serious limitations of the QTL/microarray approach for QTL gene discovery. BMC Biol. 2010;8:96. doi: 10.1186/1741-7007-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manolio TA, Collins FS. The HapMap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443–456. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan YF, et al. Parallel selection mapping using artificially selected mice reveals body weight control loci. Curr Biol. 2012;22:794–800. doi: 10.1016/j.cub.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 68.Phifer-Rixey M, et al. Adaptive evolution and effective population size in wild house mice. Mol Biol Evol. 2012 Apr 3; doi: 10.1093/molbev/mss105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bennett BJ, et al. High-resolution association mapping of atherosclerosis loci in mice. Arterioscler Thromb Vasc Biol. 2012;32:1790–1798. doi: 10.1161/ATVBAHA.112.253864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhasin JM, et al. Sex specific gene regulation and expression QTLs in mouse macrophages from a strain intercross. PLoS ONE. 2008;3:e1435. doi: 10.1371/journal.pone.0001435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith JD, et al. Transcriptome profile of macrophages from atherosclerosis-sensitive and atherosclerosis-resistant mice. Mamm Genome. 2006;17:220–229. doi: 10.1007/s00335-005-0099-7. [DOI] [PubMed] [Google Scholar]
- 72.Romanoski CE, et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nature Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 74.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nature Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas D. Gene-environment-wide association studies: emerging approaches. Nature Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nature Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 78.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker CC, Palmer AA. Dark matter: are mice the solution to missing heritability? Front Genet. 2011;2:32. doi: 10.3389/fgene.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 82.Valdar W, Holmes CC, Mott R, Flint J. Mapping in structured populations by resample model averaging. Genetics. 2009;182:1263–1277. doi: 10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anunciado RV, et al. Quantitative trait loci for body weight in the intercross between SM/J and A/J mice. Exp Anim. 2001;50:319–324. doi: 10.1538/expanim.50.319. [DOI] [PubMed] [Google Scholar]
- 84.Hunter KW. Mouse models of cancer: does the strain matter? Nature Rev Cancer. 2012;12:144–149. doi: 10.1038/nrc3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Nas A, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Warden CH, Hedrick CC, Qiao JH, Castellani LW, Lusis AJ. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.