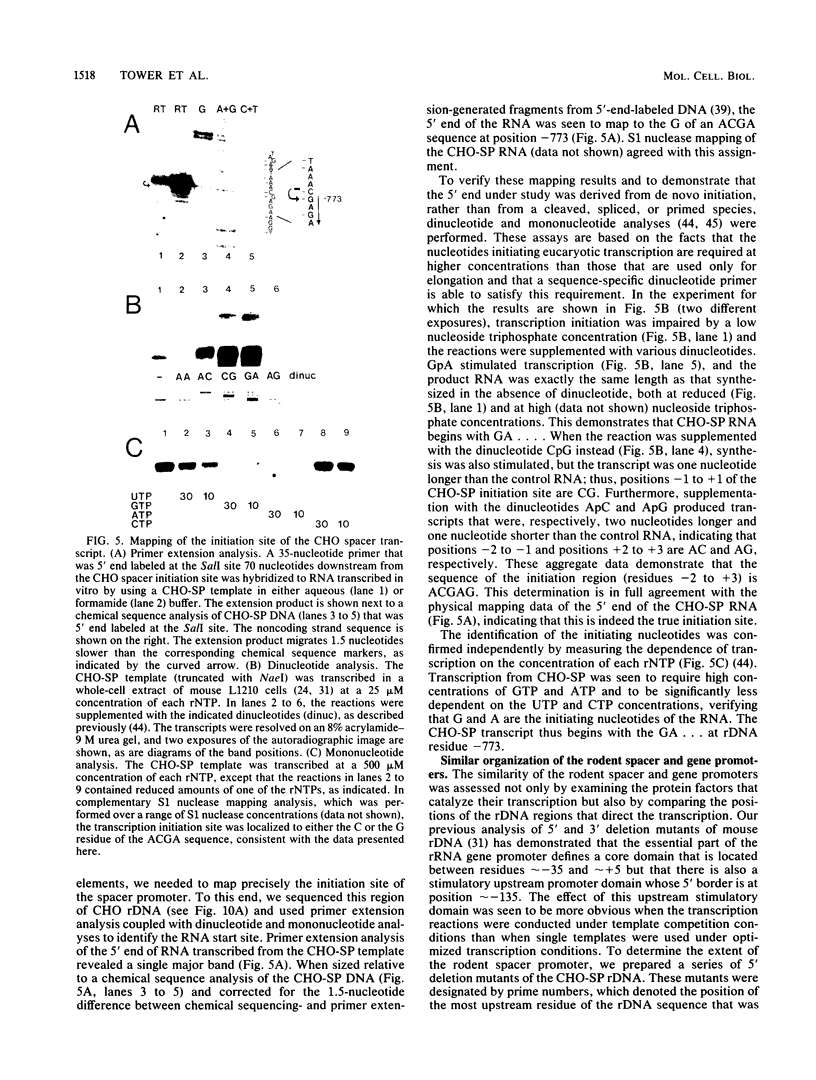
Abstract
We report results of experiments in which we demonstrated the existence of a polymerase I promoter within the ribosomal DNA spacer upstream from the rRNA initiation site in Chinese hamsters and mice. Transcription of the CHO spacer promoter was achieved by the same protein factors, C and D, that catalyzed transcription of the gene promoter, and these factors bound stably to the CHO spacer promoter in a preinitiation complex, just as they did to the gene promoter. In contrast to the CHO spacer promoter, which was transcribed in vitro nearly as efficiently as the gene promoter, the mouse spacer promoter was far less active; this low activity was attributable to the fact that the mouse spacer promoter bound factor D inefficiently. It is striking that the active CHO spacer promoter violated the otherwise universal rule that metazoan RNA polymerase I promoters all have a G residue at position -16. Sequence comparisons also revealed a great similarity between the CHO and mouse spacer promoter regions, yet there was much less similarity between the flanking sequences. There was also only limited homology between the spacer and gene promoter regions, but despite this the two kinds of initiation regions were organized similarly, both consisting of an essential core promoter domain and a stimulatory domain that extended upstream to approximately residue -135. Evolutionary considerations argue strongly that the presence of ribosomal DNA spacer promoters offers a significant selective advantage.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach R., Allet B., Crippa M. Sequence organization of the spacer in the ribosomal genes of Xenopus clivii and Xenopus borealis. Nucleic Acids Res. 1981 Oct 24;9(20):5311–5330. doi: 10.1093/nar/9.20.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy B. G., Yang-Yen H. F., Rothblum L. I. Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol. 1987 Jul;7(7):2388–2396. doi: 10.1128/mcb.7.7.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cizewski V., Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983 Oct 25;11(20):7043–7056. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta V. C., Wilkinson J. K., Sollner-Webb B. Mouse and frog violate the paradigm of species-specific transcription of ribosomal RNA genes. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7498–7502. doi: 10.1073/pnas.84.21.7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. A complex array of sequences enhances ribosomal transcription in Xenopus laevis. J Mol Biol. 1987 Aug 20;196(4):813–827. doi: 10.1016/0022-2836(87)90407-4. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell. 1986 Jan 31;44(2):313–318. doi: 10.1016/0092-8674(86)90765-8. [DOI] [PubMed] [Google Scholar]
- Dumenco V. M., Wejksnora P. J. Characterization of the region around the start point of transcription of ribosomal RNA in the Chinese hamster. Gene. 1986;46(2-3):227–235. doi: 10.1016/0378-1119(86)90407-5. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil I., Gallego M. E., Renart J., Cruces J. Identification of the transcriptional initiation site of ribosomal RNA genes in the crustacean Artemia. Nucleic Acids Res. 1987 Aug 11;15(15):6007–6016. doi: 10.1093/nar/15.15.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Harrington C. A., Chikaraishi D. M. Transcription of spacer sequences flanking the rat 45S ribosomal DNA gene. Mol Cell Biol. 1987 Jan;7(1):314–325. doi: 10.1128/mcb.7.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. L., Ryan K., Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989 Feb;3(2):212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Kishimoto T., Nagamine M., Sasaki T., Takakusa N., Miwa T., Kominami R., Muramatsu M. Presence of a limited number of essential nucleotides in the promoter region of mouse ribosomal RNA gene. Nucleic Acids Res. 1985 May 24;13(10):3515–3532. doi: 10.1093/nar/13.10.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Nontranscribed spacer sequences promote in vitro transcription of Drosophila ribosomal DNA. Nucleic Acids Res. 1982 Nov 11;10(21):6879–6886. doi: 10.1093/nar/10.21.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller H. T., Frondorf K. A., Maschner P. D., Vaughn J. C. In vivo transcription from multiple spacer rRNA gene promoters during early development and evolution of the intergenic spacer in the brine shrimp Artemia. Nucleic Acids Res. 1987 Jul 10;15(13):5391–5411. doi: 10.1093/nar/15.13.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M., Arnheim N. Nucleotide sequence of the genetically labile repeated elements 5' to the origin of mouse rRNA transcription. Nucleic Acids Res. 1983 Jan 11;11(1):211–224. doi: 10.1093/nar/11.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987 Nov;6(11):3487–3492. doi: 10.1002/j.1460-2075.1987.tb02673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- McStay B., Bird A. The origin of the rRNA precursor from Xenopus borealis, analysed in vivo and in vitro. Nucleic Acids Res. 1983 Dec 10;11(23):8167–8181. doi: 10.1093/nar/11.23.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Miller J. R., Hayward D. C., Glover D. M. Transcription of the 'non-transcribed' spacer of Drosophila melanogaster rDNA. Nucleic Acids Res. 1983 Jan 11;11(1):11–19. doi: 10.1093/nar/11.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan G. T., Reeder R. H., Bakken A. H. Transcription in cloned spacers of Xenopus laevis ribosomal DNA. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6490–6494. doi: 10.1073/pnas.80.21.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Murtif V. L., Rae P. M. A transcriptional function for repetitive ribosomal spacers in Xenopus? Nature. 1983 Aug 11;304(5926):561–562. doi: 10.1038/304561c0. [DOI] [PubMed] [Google Scholar]
- Murtif V. L., Rae P. M. In vivo transcription of rDNA spacers in Drosophila. Nucleic Acids Res. 1985 May 10;13(9):3221–3239. doi: 10.1093/nar/13.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Scheer U., Trendelenburg M. F., Krohne G., Franke W. W. Lengths and patterns of transcriptional units in the amplified nucleoli of oocytes of Xenopus laevis. Chromosoma. 1977 Mar 16;60(2):147–167. doi: 10.1007/BF00288462. [DOI] [PubMed] [Google Scholar]
- Skinner J. A., Ohrlein A., Grummt I. In vitro mutagenesis and transcriptional analysis of a mouse ribosomal promoter element. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2137–2141. doi: 10.1073/pnas.81.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Tower J., Culotta V. C., Sollner-Webb B. Factors and nucleotide sequences that direct ribosomal DNA transcription and their relationship to the stable transcription complex. Mol Cell Biol. 1986 Oct;6(10):3451–3462. doi: 10.1128/mcb.6.10.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Sollner-Webb B. Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell. 1987 Sep 11;50(6):873–883. doi: 10.1016/0092-8674(87)90514-9. [DOI] [PubMed] [Google Scholar]
- Wilkinson J. A., Miller K. G., Sollner-Webb B. Dinucleotide primers facilitate convenient identification of the mouse ribosomal DNA transcription initiation site. A general method for analysis of transcription by RNA polymerases I and III. J Biol Chem. 1983 Nov 25;258(22):13919–13928. [PubMed] [Google Scholar]
- Wilkinson J. K., Sollner-Webb B. Transcription of Xenopus ribosomal RNA genes by RNA polymerase I in vitro. J Biol Chem. 1982 Dec 10;257(23):14375–14383. [PubMed] [Google Scholar]
- Wood K. M., Bowman L. H., Thompson E. A., Jr Transcription of the mouse ribosomal spacer region. Mol Cell Biol. 1984 May;4(5):822–828. doi: 10.1128/mcb.4.5.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Yen H. F., Rothblum L. I. Partial nucleotide sequence of a 3.4 kb fragment from the rat ribosomal DNA nontranscribed spacer. Nucleic Acids Res. 1986 Jul 11;14(13):5557–5557. doi: 10.1093/nar/14.13.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]