Summary
Circadian rhythms in mammals are generated by a transcriptional negative feedback loop that is driven primarily by oscillations of PER and CRY, which inhibit their own transcriptional activators, CLOCK and BMAL1. Current models posit that CRY is the dominant repressor while PER may play an accessory role. In this study, however, constitutive expression of PER, and not CRY1, severely disrupted the clock in fibroblasts and liver. Furthermore, constitutive expression of PER2 in the brain and SCN of transgenic mice caused a complete loss of behavioral circadian rhythms in a conditional and reversible manner. These results demonstrate that rhythmic levels of PER2, rather than CRY1, are critical for circadian oscillations in cells and in the intact organism. Biochemical evidence supports an elegant mechanism for the disparity: PER2 directly and rhythmically binds to CLOCK:BMAL1, while CRY only interacts indirectly; PER2 bridges CRY and CLOCK:BMAL1 to drive the circadian negative feedback loop.
Introduction
In mammals, daily physiological processes such as sleep/wake cycles, hormone production, and metabolism are governed by endogenous circadian clocks (Allada et al., 2001; Hastings et al., 2003; Lowrey and Takahashi, 2004; Panda et al., 2002; Reppert and Weaver, 2002). In the suprachiasmatic nuclei (SCN) of the anterior hypothalamus resides a master clock, which coordinates synchronization of other clocks in the brain as well as clocks in the liver, kidney, and other peripheral tissues (Yamazaki et al., 2000; Yoo et al., 2004). These peripheral tissues also contain self-sustained circadian oscillators that are believed to have a similar molecular composition and operational mechanism as the SCN (Liu et al., 2007; Yagita et al., 2001).
Forward and reverse genetic studies have successfully revealed molecular components of the circadian clock (Bae et al., 2001; Bunger et al., 2000; King et al., 1997; Preitner et al., 2002; Sun et al., 1997; Takahashi, 2004; Tei et al., 1997; van der Horst et al., 1999; Zheng et al., 2001; Zheng et al., 1999). Such molecules have been shown to form a network of transcriptional/translational feedback loops. However, much remains to be discovered in order to delineate the precise biochemical interactions through which these molecules generate a circadian timekeeper.
It is generally acknowledged that at the core of the clock mechanism, molecular rhythms are generated by a circadian autoregulatory feedback loop that contains both positive and negative transcriptional elements (Allada et al., 2001; Lowrey and Takahashi, 2004; Reppert and Weaver, 2002; Sato et al., 2006; Schibler, 2005). Three basic-helix-loop-helix (bHLH)/PAS-containing transcription factors, CLOCK, NPAS2 and BMAL1, constitute the positive (activator) elements, as CLOCK:BMAL1 (hereafter used as shorthand for the heterodimer of CLOCK [or NPAS2] and BMAL1) activates the transcription of Period (Per) and Cryptochrome (Cry) genes, which constitute the negative (repressor) elements. As the levels/activities of the negative elements rise, they progressively repress their own transcription by interfering with the function of the positive elements. This inhibition is gradually relieved as the negative elements are degraded by the proteasome pathway through specific E3 ubiquitin ligases (Busino et al., 2007; Eide et al., 2005; Godinho et al., 2007; Reischl et al., 2007; Shirogane et al., 2005; Siepka et al., 2007), thus creating oscillating transcription with a period of ~24 hours.
Expression of the positive elements, Bmal1 and Clock, is regulated in a circadian manner by a second transcriptional feedback loop involving the two nuclear receptors, REV-ERBα and RORα (Liu et al., 2008; Preitner et al., 2002; Sato et al., 2004). However, oscillations in Bmal1 and Clock mRNA levels are not required for circadian rhythm generation; although these rhythms are abolished in Rev-erbα deficient mice, behavioral and molecular rhythms remain largely intact (Preitner et al., 2002). Furthermore, constitutive expression of Bmal1 in Bmal1-deficient mice can restore circadian rhythmicity in the arrhythmic mutant mice (McDearmon et al., 2006). A clock must have oscillating components; because oscillations in Clock and Bmal1 are not required, oscillations in Per and/or Cry genes are likely to be essential in order to complete a negative feedback loop. This general question (which oscillations are essential for clock function?) has been addressed in model organisms such as Neurospora, Drosophila and cyanobacteria (Aronson et al., 1994; Kitayama et al., 2008; Yang and Sehgal, 2001), but no conclusive experiments have been performed in mammalian circadian systems thus far. Although a few studies implicated Per or Cry oscillations as important for rhythm generation (Numano et al., 2006; Ueda et al., 2005; Yamamoto et al., 2005), their results have been contradicted by other studies (Fan et al., 2007; Fujimoto et al., 2006; Yamanaka et al., 2007). Furthermore these studies focused on clock outputs and did not provide mechanistic insight into the underlying molecular clock.
To address more definitively whether oscillations of PER, CRY or both are required for circadian rhythm generation, and to determine how constitutive expression of the proteins can affect the molecular clock, we abolished PER or CRY oscillations in fibroblasts and intact mice, and assessed both rhythmic outputs and the molecular clock mechanism. We report here that PER, and not CRY, is a critical oscillating component in the mammalian clock mechanism. Biochemical analysis reveals that PER globally regulates the molecular oscillator through both transcriptional and post-transcriptional mechanisms. Importantly, our findings suggest that CRY cannot inhibit CLOCK:BMAL1 directly, but rather relies on PER to bridge it to the CLOCK:BMAL1 complex. These results argue that rhythms in PER, rather than CRY, define a critical rhythmic nodal point for the generation of circadian rhythms and highlight the conserved role of PER among animals (e.g., Drosophila and mammals) in the generation of circadian rhythms.
Results
Constitutive overexpression of PER1 and 2, but not CRY1, abolishes the circadian rhythmicity in luciferase activity in MEFs
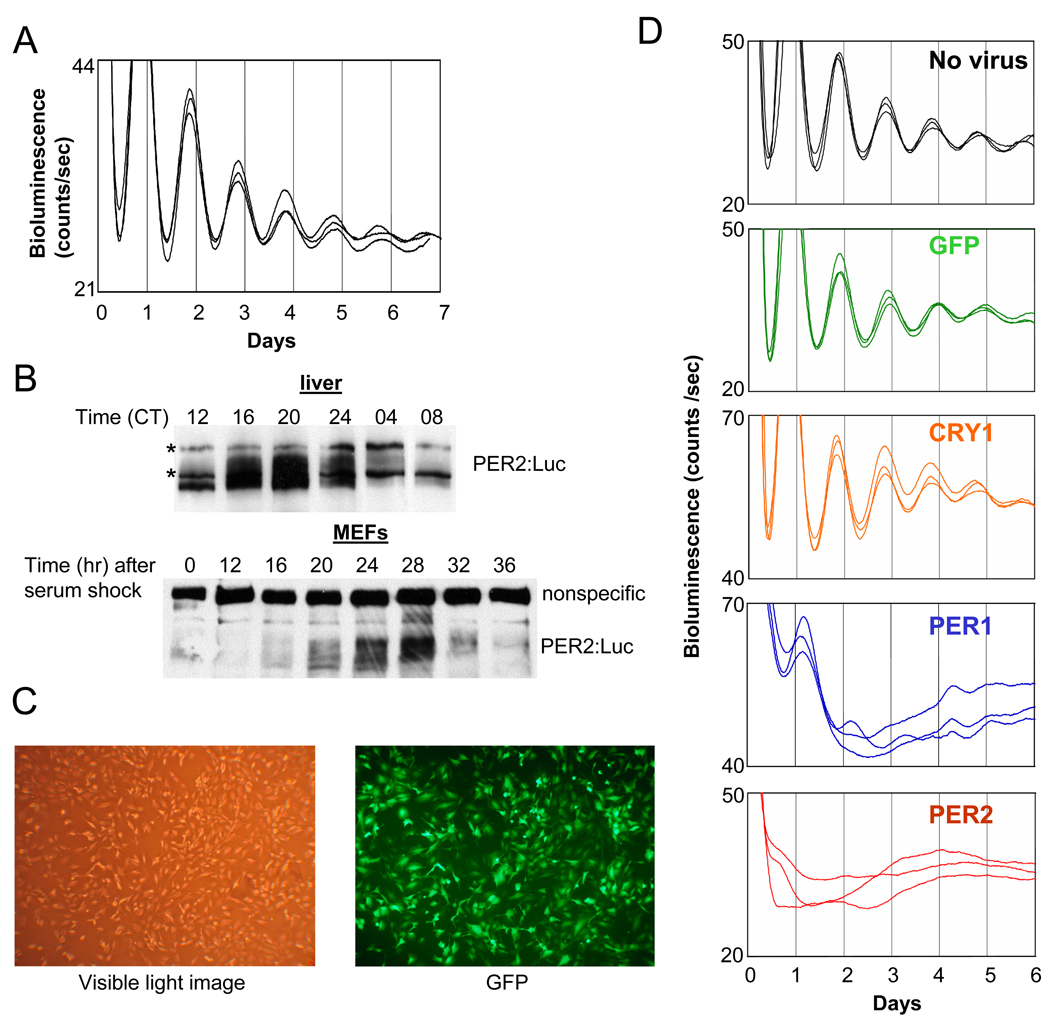
We used mouse embryonic fibroblasts (MEFs) derived from the Per2Luc mouse as a model for in vivo circadian clocks, since the monitoring of circadian rhythms in living cells and tissues derived from this reporter-knockin mouse has been well established (e.g., (Liu et al., 2007; Yoo et al., 2004)]). We confirmed that the MEFs exhibited robust rhythms in luciferase activity over 7 days (Fig 1A), as has been reported for various tissues derived from the knockin mice (Yoo et al., 2004). In addition, the PER2:Luciferase fusion protein oscillated in abundance and phosphorylation in MEFs, much as it did in liver (Fig 1B).
Fig. 1.
Bioluminescence rhythms in MEFs are abolished by constitutively expressed PER, but not by GFP or CRY1. (A) Per2Luc MEFs exhibit circadian bioluminescence rhythms over 7 days after serum shock. (B) PER2:Luciferase fusion protein shows circadian oscillations in abundance and phosphorylation in both liver and MEFs. * indicates nonspecific bands. (C) The adenoviral vector efficiently infects MEFs. MEFs were infected with GFP-adenovirus and fixed 24 hr later to take a visible light image and a fluorescence (GFP) image. (D) MEFs were infected with adenoviral vector expressing GFP, CRY1, PER1 or PER2 or mock infected with medium only (no virus) for 2 hours and then serum shocked for 2 hours before measurement of bioluminescence.
We then assessed the circadian clock in MEFs after abolishing oscillations in CRY1, PER1, or PER2 by constitutively expressing these genes from the cytomegalovirus (CMV) promoter in an adenoviral vector (Fig 1C). Adenoviral infection and expression of a non-specific protein such as Green Fluorescent Protein (GFP) did not significantly affect the overt luciferase rhythms compared to the no-virus control (Fig 1D). Constitutive expression of CRY1 was no more effective than GFP at affecting the endogenous clock: luciferase rhythms of CRY1-overexpressing MEF cells were robust and normal (Fig 1D). However, when PER1 or 2 was constitutively expressed, circadian bioluminescence rhythms were completely abolished (Fig 1D). The suppression of the luciferase rhythms by PER2 was maintained even when the titer of PER2 virus was 1/6 that of the others (Suppl. Fig 1).
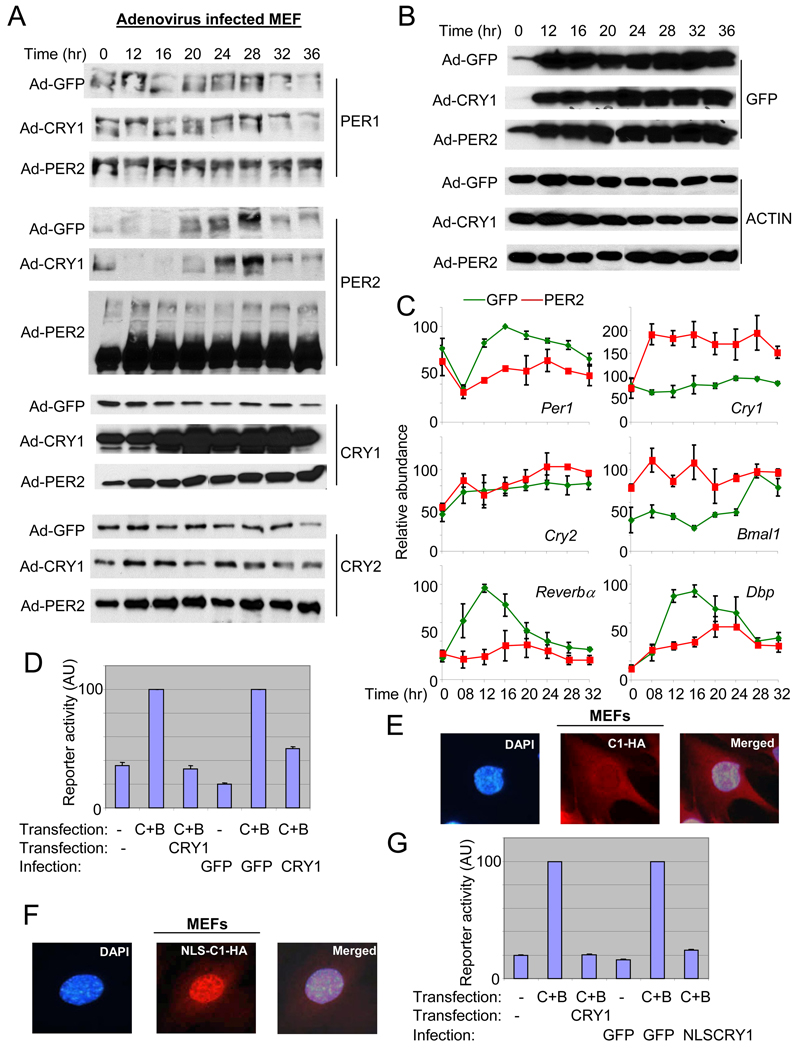
The ineffectiveness of CRY1 did not result from inadequate expression of the target protein, since both PER2 and CRY1 were more than 10-fold overexpressed relative to their endogenous counterparts, as measured in GFP-MEFs (Fig 2A). GFP and ACTIN blots demonstrate that similar titers of adenovirus and similar amounts of total protein were used for all three cases (Fig 2B). Consistent with our luciferase data, rhythms (in abundance and phosphorylation) of endogenous PER1 and PER2 were intact in GFP- and CRY1-overexpressing MEFs, while those rhythms were completely abolished in PER2-overexpressing MEFs (Fig 2A). PER2 overexpression was associated with other interesting changes as well, including a dramatic increase in the levels of endogenous CRY1 and 2 (Fig 2A). Cry2 mRNA levels were not significantly altered (Fig 2C), indicating that the increased CRY2 levels were due to altered posttranscriptional regulation of Cry2. However, both altered transcriptional and posttranscriptional regulation of Cry1 seemed to contribute to the increased levels of CRY1 in PER2-overexpressing MEFs (Fig 2C and see below): transcriptional because mRNA levels increased, but also posttranscriptional because protein levels increased even more (~4 fold increase for protein vs. ~2 fold increase for mRNA). Bmal1 mRNA levels were elevated in PER2-MEFs, whereas mRNA levels of Per1 and clock-controlled genes, Dbp and Rev-erbα, were suppressed compared to GFP-MEFs (Fig 2C).
Fig. 2.
Constitutive expression of PER2, and not CRY1, disrupts PER protein rhythms and mRNA rhythms of clock and clock-controlled genes. (A) (B) MEFs were infected and serum shocked as described in Fig 1. Both endogenous (the fusion protein; top band) and exogenous (bottom band) PER2 are shown for PER2 immunoblot of PER2-MEFs. The adenoviral vector has dual CMV promoters, one default CMV promoter for GFP and the other one for the gene of interest (He et al., 1998), in our case PER2 or CRY1. Thus, similar intensities of GFP signal indicate similar viral titers among the three groups. (C) Quantitation of mRNA levels of clock and clock-controlled genes in Adenovirus GFP (green)- and Adenovirus PER2 (red)-MEFs. mRNA levels were measured by quantitative real time PCR. Data are shown as mean+/−SEM of three experiments. For some data points, error bars (if less than 3) are obscured by the square symbols. (D) The viral CRY1 can inhibit CLOCK:BMAL1-driven transcription in reporter assays. The inhibitory activity between plasmid (pcDNA3.1) and adenoviral CRY1 on CLOCK:BMAL1-driven transcription was compared in transient transfection assays. The first three reactions (bars) were done only with plasmid transfection but the other three reactions were performed with transfection (CLOCK and BMAL1) and viral infection (CRY). Results are shown as mean+/−SEM of triplicate samples and are representative of three independent experiments. (E) (F) The viral CRY1-HA was both cytoplasmic and nuclear (E), but the viral NLS-CRY1-HA was predominantly nuclear (F). The overexpressed viral CRY1-HA was visualized with immunofluorescence using anti-HA antibody. These single cells correspond to the cells in white boxes in Suppl Fig 2B–C, magnified as representatives. (G) The inhibitory activity of plasmid (pcDNA3.1)-expressed CRY and adenoviral NLS-CRY1 on CLOCK:BMAL1-driven transcription was compared in COS7 cells, as in (D).
CRY proteins are the major inhibitors of CLOCK:BMAL1 driven transcription in reporter gene assays (Griffin et al., 1999; Kume et al., 1999). Why, then, does the overexpression of CRY1 fail to affect the overt rhythm in our system? The protein analysis discussed above (Fig 2A) demonstrates that inadequate expression levels are not a concern. We also tested the potency of virally expressed CRY1 in inhibiting CLOCK:BMAL1-activated transcription and found that viral CRY1 is comparable to plasmid-expressed CRY1 in potency for inhibiting CLOCK:BMAL1 when normalized for expression level (Fig 2D). We also assessed subcellular localization of the viral CRY1, and found that it is mostly nuclear (Suppl Fig 2A) as is pcDNA Cry1 in COS7 cells (Kume et al., 1999). However, in MEFs we found that the exogenous viral CRY1 was distributed between nucleus and cytoplasm (Fig 2E and Suppl Fig 2B), reminiscent of endogenous CRY proteins in vivo (Lee et al., 2001; Preitner et al., 2002). To rule out that viral CRY1 failed to disrupt circadian rhythms when overexpressed in MEFs because it did not adequately accumulate in the nucleus, a strong nuclear localization signal (NLS) derived from the simian virus 40 large T antigen was added to the N-terminus of CRY1-HA. This NLS increased nuclear localization of CRY1 in our MEFs (Fig 2F and Suppl Fig 2C) and enhanced CRY1’s inhibitory activity on CLOCK:BMAL1 in the reporter assay in COS7 cells (Fig 2G), but still failed to disrupt luciferase rhythms in MEFs (Suppl Fig 2D). Thus, we can conclude that the circadian clock in MEFs is not perturbed by overexpression of CRY, even though the overexpressed CRY can potently inhibit CLOCK:BMAL1 transcription in reporter gene assays and has significant nuclear localization.
Constitutive overexpression of PER2, but not CRY1, disrupts circadian rhythms in liver
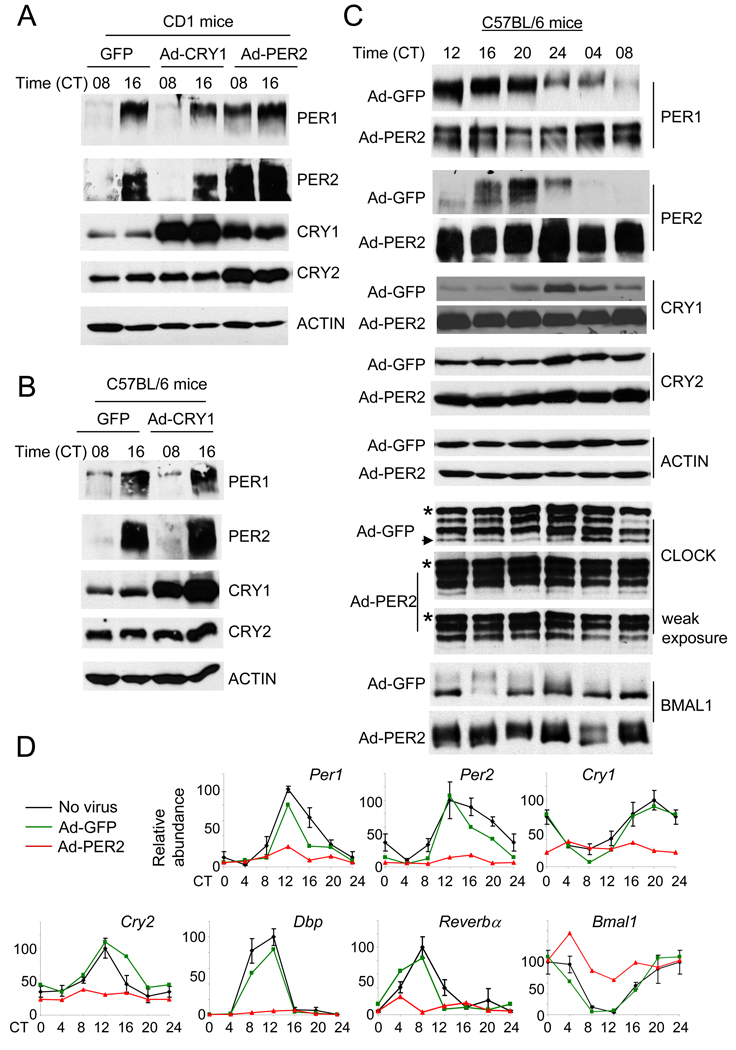
Our results showing that CRY rhythms and proper levels are not required for clock function are surprising given that the current working model of clock function involves direct and rhythmic binding of CRY to the CLOCK:BMAL complex (Reppert and Weaver, 2002). We therefore tested whether our findings are limited to the clock system in fibroblast cells or whether similar results could be reproduced in intact mice. For this purpose, we generated tissue (liver)-specific transient transgenic mice by systemically delivering the recombinant adenoviruses into mice through tail vein injection (Herzig et al., 2001). In our hands, we routinely observed more than an 80% infection rate in liver as judged by the number of GFP-expressing cells (Suppl. Fig 3). We injected GFP, CRY1 or PER2 adenovirus into an outbred CD1 strain and an inbred C57BL/6 strain to demonstrate that our results are not strain specific (Fig 3). In both strains of mice, the adenoviral-overexpressed PER2 and CRY1 levels were several fold higher than peak levels of endogenous PER2 and CRY1 in GFP expressing mice (see Suppl. Fig 4 for quantitative comparison). The extent and range of phosphorylation of overexpressed PER2 were similar to endogenous PER2 (Suppl. Fig 4), suggesting that the level of overexpression did not saturate endogenous phosphorylation machinery.
Fig. 3.
Constitutive expression of GFP, CRY1 or PER2 in mouse liver produced similar results as in MEFs. (A) (B) (C) CD-1 (A) and C57BL/6 (B and C) strains were injected with adenovirus expressing GFP, CRY1 or PER2 and sacrificed at the indicated circadian times. Liver extracts were subjected to immunoblotting. Results are representative of at least 2 mice per time point. * indicates a nonspecific band in the CLOCK immunoblot. The arrow indicates a hypophosphorylated isoform, which is much weaker in PER2-liver. Note that the top isoform is much more enriched in the PER2-liver. (D) Quantification of mRNA levels of clock and clock-controlled genes in control (no virus, black), GFP (green)-, and PER2 (red)-liver. mRNA levels were measured by quantitative real-time PCR. Results are average+/−SEM of three mice for controls, and representative of two mice per time point for GFP and PER2-adenovirus injected mice. The data for the second set of the GFP and PER2 mice are shown in Suppl Fig 6. Levels were normalized so that the highest levels of the control mice equaled 100 arbitrary units. The CT 0 data were plotted twice as CT 0 and 24.
In CRY1- and GFP-overexpressing liver, PER1 and PER2 were rhythmic in both CD1 (Fig 3A) and C57BL/6 (Fig 3B) strains of mice. By contrast, in PER2-overexpressing liver, circadian rhythms were severely disrupted (Fig 3A and C). As seen in MEFs (Fig 2A), CRY protein levels were dramatically increased in PER2 liver. To examine more carefully how overexpression of PER2 disrupted the circadian transcription/translation feedback loop, we measured levels of clock components at four-hour intervals over one circadian cycle in C57BL/6 mice (Fig 3C and D). As before, PER2 overexpression severely disrupted PER1 rhythms and dramatically increased CRY levels (Fig 3C). CLOCK levels were slightly elevated (<2-fold) and BMAL1 levels were more strongly elevated (2- to 3-fold) compared to those in GFP liver. Phosphorylation patterns also differed with PER2 overexpression: for CLOCK, hyperphosphorylated isoforms were predominant and constitutively present, while for BMAL1, both hypo- and hyperphosphorylated isoforms were present in the PER2 liver (see Suppl Fig 5 for side by side comparison for BMAL1). The increase in BMAL1 levels were apparently due to both transcriptional and posttranscriptional regulation since Bmal1 mRNA levels were elevated, but less than 2-fold (Fig 3D and Suppl Fig 6). The elevation of Bmal1 mRNA levels is reminiscent of previous data in Rev-erbα deficient mice (Preitner et al., 2002), suggesting that it could be due to suppression of Rev-erbα expression.
Constitutive overexpression of PER2 decreased mRNA levels of all clock genes except for Bmal1 (Fig 3D and Suppl Fig 6), consistent with an increase in PER:CRY-mediated transcriptional repression; viral infection and GFP expression in and of themselves did not significantly affect clock gene mRNA levels. Although the mRNA levels for most clock and clock-controlled genes were similarly modulated between MEFs and liver by overexpressed PER2, we observed different effects on Cry mRNA. In MEFs, Cry1 mRNA levels were elevated and those for Cry2 were neither increased nor decreased; in liver, Cry1 and 2 mRNA levels were both decreased. This may be due to a possible difference in clock mechanisms between peripheral tissues or the presence of systemic signals in intact mice that are absent when studying cultured MEFs.
Constitutive overexpression of PER2 in liver results in increased and constitutive interaction between CRY and CLOCK:BMAL1, and this interaction is missing in Per1/2 mutant mice
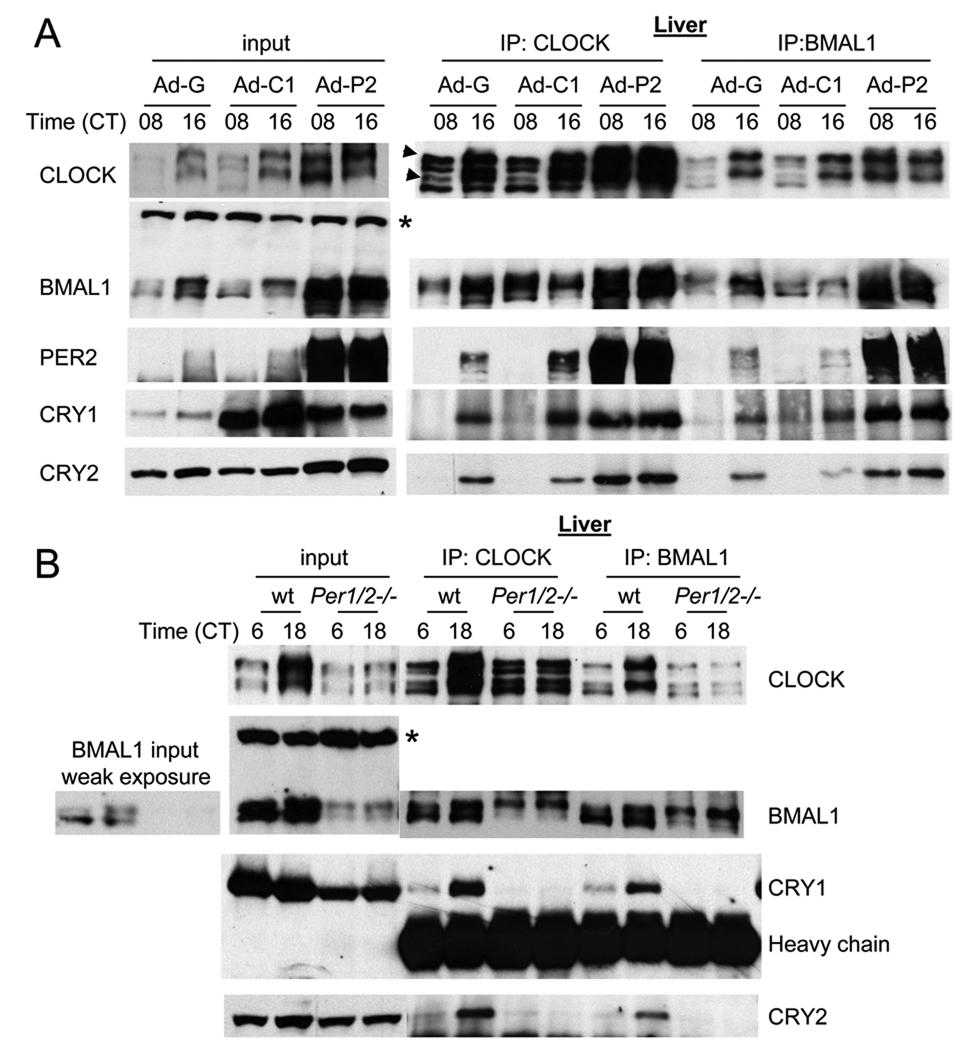
Although CRY1 overexpression did not disrupt rhythms, previous work has argued that CRYs play a major role in the inhibition of CLOCK:BMAL1 (Griffin et al., 1999; Kume et al., 1999; Lee et al., 2001). If the interaction of CRY with CLOCK:BMAL1 remains tightly regulated despite CRY overexpression, then that could explain why the clock is not sensitive to overall CRY levels. If CRY binding to CLOCK:BMAL1 is regulated by PER, then that would explain why PER2-overexpression disrupts the clock and results in an apparent excess of CLOCK:BMAL1 inhibition, suggested by the suppression of CLOCK:BMAL1 target gene mRNA levels (Fig 3D). If these hypotheses are correct, then CLOCK:BMAL1 would be rhythmically bound by CRY1/2 despite constitutive overexpression of CRY1, and constitutively bound by CRY1/2 under conditions of PER2 overexpression. We assessed CRY binding to CLOCK:BMAL1 using coimmunoprecipitation experiments.
In GFP- or CRY1-overexpressing livers, much higher levels of CRY1 and 2 were copurified with CLOCK and BMAL1 at CT16 than at CT08 (Fig 4A), in accordance with the normal protein and mRNA abundance rhythms in these livers (Fig 3) (Lee et al., 2001). The exogenously expressed CRY1 interacted with endogenous PER2, CLOCK and BMAL1, indicating that the exogenous CRY1 can interact with endogenous clock proteins normally (Suppl Fig 7A). Although the interaction remained rhythmic in CRY1-overexpressing liver, the levels of copurified CRY1 were slightly increased compared to GFP-liver. However, in PER2-overexpressing liver, this rhythmic interaction was abolished and levels of copurified CRY were dramatically increased at both times compared to CRY1- or GFP-overexpressing livers at CT16 (Fig 4A). Thus, in liver, the patterns of CRY interaction with CLOCK:BMAL1 match our prediction that rhythmic CRY interaction with CLOCK:BMAL1 persists in the face of constitutive overexpression of CRY1, a finding that is difficult to reconcile with the current working model of the clock, which posits direct interaction of CRY with CLOCK:BMAL1. Coimmunoprecipitation assays in MEFs produced similar results (Suppl. Fig 7B).
Fig. 4.
In GFP- or CRY-overexpressing liver, rhythmic interaction between CLOCK:BMAL1 and PER/CRY is maintained, but the interaction is constitutively elevated in PER2-overexpressing liver and almost absent in Per1/2ldc mutant mice. (A) Liver samples at CT 08 and 16 in Fig 4A (CD1 mice) were subjected to IP for CLOCK and BMAL1 and the resulting immunocomplexes were immunoblotted for clock proteins. Two arrow heads indicate two hyperphosphorylated isoforms of CLOCK compared to the other two isoforms as has been shown previously (Lee et al., 2001). The asterisk denotes a nonspecific band in the BMAL1 immunoblot ensuring equal loading of total protein. (B) IP for CLOCK and BMAL1 was performed using liver samples at CT06 and 18 from Per1/2ldc mutant and matching wt mice. To compensate for lower amounts of clock proteins in the mutant mice, twice as much starting material was used for the IP experiments. Note that CLOCK is hypophosphorylated, but BMAL1 is hyperphosphorylated in the mutant mice.
These results suggest that endogenous PER2 is a limiting factor for CRY interaction with CLOCK:BMAL1. This idea is further supported by the reduced levels of CRY2 that are copurified with CLOCK:BMAL1 in CRY1-overexpressing liver and MEFs compared to GFP controls (Fig 4A and Suppl Fig 7B). This reduction is presumably due to overexpressed CRY1 binding endogenous PER2 (which is limiting), making less PER2 available for facilitating CRY2 binding with CLOCK:BMAL1. When PER2 is constitutively overexpressed, on the other hand, CRY1 and 2 interact with CLOCK:BMAL1 in a greatly increased constitutive manner, indicating that the rhythm in CRY:CLOCK:BMAL1 interaction is driven by temporal abundance of PER2, not CRY1. The PER-mediated interaction is further supported by the finding that although CLOCK and BMAL1 were readily copurified with each other, neither of them significantly interacted with CRY in Per1/2ldc mutant mice (Fig 4B) (Lee et al., 2004). In contrast to the results in PER2-overexpressing liver, CLOCK was hypophosphorylated at both CT06 and 18, and its levels were slightly reduced. Likewise, BMAL1 levels were greatly reduced and it was hyperphosphorylated at both times. Since Bmal1 mRNA levels are comparable between wt and the mutant mice (Suppl Fig 7C), the increased BMAL1 and CLOCK are due at least in part to posttranscriptional mechanisms in the PER2-overexpressing liver.
In PER2-overexpressing liver, more CLOCK and BMAL1 are present than in GFP- or CRY1-overexpressing liver, and more CLOCK was copurified by IP for BMAL1 and vice versa (Fig 4A). These data indicate that interaction between CLOCK and BMAL1 in PER2-overexpressing liver is proportionally increased with the increased levels of the proteins in the starting extracts, suggesting that the increased interaction between negative (PER;CRY) and positive (CLOCK:BMAL1) elements does not disrupt the CLOCK:BMAL1 complex (Lee et al., 1999; Lee et al., 2001).
The negative complex, PER:CRY, interacts with the positive complex (CLOCK:BMAL1) through PER, and formation of the PER:CRY complex is essential for the clock
Our data above suggests that PER is the interface or scaffolding molecule that brings the positive and negative components together, and that CRY may not bind directly to CLOCK:BMAL1 in its normal rhythmic interaction in vivo. Since these results contradict the current model of the clock mechanism, we sought to test this hypothesis critically by carefully measuring and comparing the interactions of PER and CRY for CLOCK:BMAL1 in vivo and in vitro.
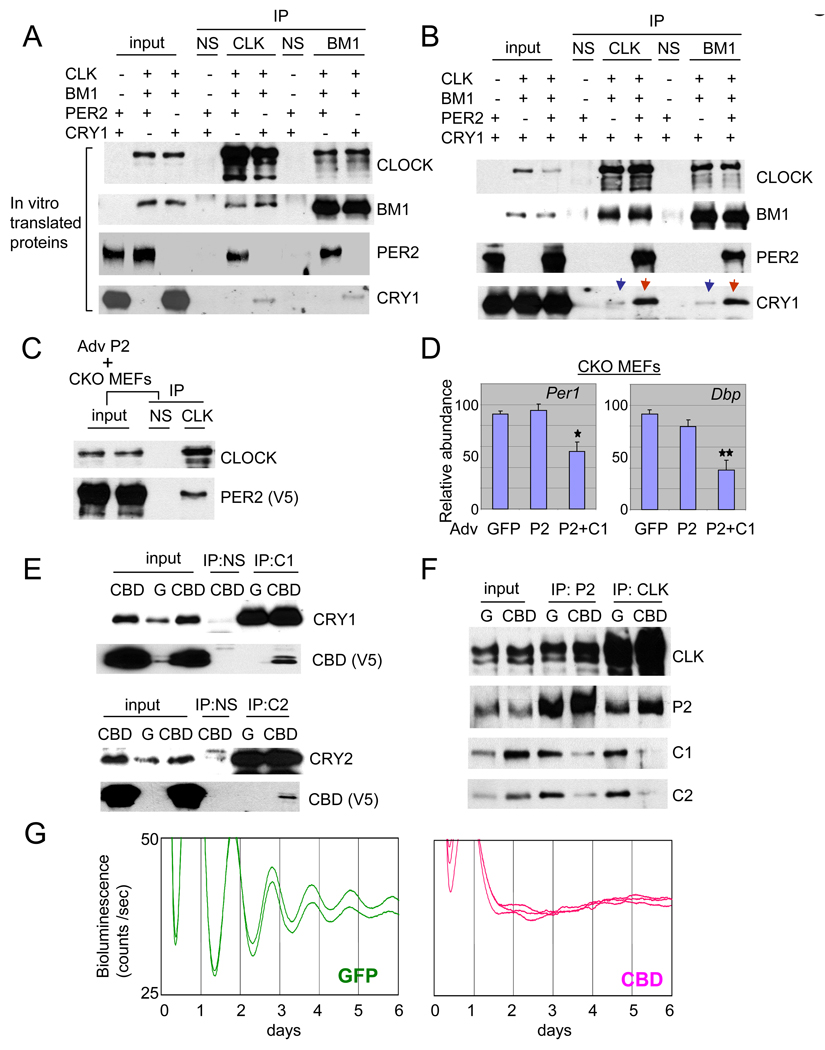
First, to reduce the possibility that interactions could be due to unknown intervening molecules in our binding assay, we employed in vitro translated clock proteins (Fig 5A). We found that IP for CLOCK or BMAL1 co-purified robust quantities of PER2, whereas CRY1, while detectable, was barely above background level. Similar results were reproduced when CLOCK and BMAL1 were incubated separately with PER2 or CRY1 in vitro (Suppl Fig 8A) and when the clock proteins were co-expressed in HEK 293 cells (Suppl Fig 8B and C). The transiently expressed CRY1, CLOCK and BMAL1 were predominantly nuclear in HEK 293 cells (Suppl Fig 9). Importantly, copurified CRY1 with CLOCK or BMAL1 was dramatically increased when PER2 and CRY1, instead of CRY1 alone, were incubated with CLOCK and BMAL1 in the in vitro experiments (Fig 5B). The interaction between CLOCK:BMAL1 and PER2 did not require CRY, since PER2 can be copurified with CLOCK in Cry double knockout MEFs (Fig 5C). Interestingly, however, overexpression of PER2 alone did not suppress mRNA levels of endogenous Per1 and Dbp in the mutant MEFs (Fig 5D), whereas overexpression of both PER2 and CRY1 reduced mRNA levels of those genes (Fig 5D). Thus, our data support the conventional model in that CRY is required for inhibition, but differs in suggesting that CRY may do so indirectly, with PER bridging CRY interaction with CLOCK:BMAL1.
Fig. 5.
PER is the scaffolding molecule between the activator complex (CLOCK:BMAL1) and the negative complex (PER:CRY). (A) PER2 has a much higher affinity than CRY1 for binding to CLOCK:BMAL1. Equal moles of in vitro translated proteins were mixed in different combinations and subjected to IP for CLOCK or BMAL1. (B) CRY1 copurified with CLOCK and BMAL1 is dramatically increased in the presence of PER2 in vitro. Equimolar amounts of the in vitro translated proteins were mixed as indicated and subjected to IP for CLOCK or BMAL1. Note that copurified CRY1 levels are dramatically different when PER2 is absent (the blue arrows) and present (the red arrows). (C) PER2 does not require CRY for binding CLOCK. PER2-V5 adenovirus was infected into primary Cry double-mutant MEFs to test if PER2 can interact with CLOCK:BMAL1 in the absence of CRYs. (D) Adenoviral overexpression of PER2 in the Cry double-knockout (CKO) MEFs does not suppress endogenous mRNA expression of Per1 and Dbp, but co-overexpression of PER2 and CRY1 does. Average+/−SEM of three experiments is shown. GFP and PER2+CRY1 samples differed significantly (Student t-test; one star: p<0.05; two stars: p<0.01). (E) CRY-binding domain of PER2 (CBD) interacts with endogenous CRY1 and 2. MEFs were infected with GFP or CBD (CBD-V5) adenovirus and subjected to IP for CRY1 or CRY2. (F) CBD disrupts CRY interaction with clock protein complexes. Results from IP for BMAL1 are shown in Suppl. Fig 11. (G) PER2:CRY interaction is essential for circadian rhythm generation. When the interaction is disrupted by CBD, overt rhythms were completely abolished.
Coimmunoprecipitation studies with serial deletion mutants of PER2 demonstrated that the PAS domain in PER2 can interact with both CLOCK and BMAL1 (Suppl. Fig 10). Given that all three proteins have a PAS domain, and PAS domains are frequently involved in protein-protein interactions (Huang et al., 1993), this result is not surprising. However, the interaction assay revealed that at least one more region downstream of PAS in PER2 can interact with BMAL1 almost as strongly as the PAS domain (Suppl. Fig 10C). We call this the BMAL1-binding domain (BBD). Thus, there are at least two domains on PER2 that can interact directly with CLOCK:BMAL1.
Finally, we disrupted the in vivo interaction between PER and CRY by overexpressing the CRY-Binding Domain (CBD) (Miyazaki et al., 2001; Yagita et al., 2002) of PER2 in MEFs using the adenoviral vector to determine which molecule is the interface in interacting with CLOCK:BMAL1. Since the CBD consists of only 101 amino acids on the C-terminus of PER2, it does not bind CLOCK:BMAL1 when transiently expressed in cells (data not shown). We assume that for a CLOCK:BMAL1/PER2/CRY complex, the CBD is unlikely to disrupt the interface between the CLOCK:BMAL1 dimer and PER2 or CRY; instead, the CBD would be expected to disrupt only the interaction between PER2 and CRY. Thus, if CRY binds directly to CLOCK and/or BMAL1, the CBD would only reduce the amount of PER2 copurified by IP for CLOCK (or BMAL1), since the CBD would not disrupt the interaction between CLOCK:BMAL1 and CRY. However, if PER binds the complex, the IP would produce only less copurified CRY and would not disrupt the CLOCK:BMAL1:PER2 complex. We first confirmed that the CBD readily interacted with in vivo CRY1/2 (Fig 5E). When the CBD was overexpressed in MEFs, IP for PER2 produced less copurified CRYs compared to those in GFP MEFs, demonstrating that the CBD indeed disrupted the interaction between PER2 and CRYs (Fig 5F). This occurred even in the face of increased levels of CRYs. We observed that CRY levels were increased by only overexpressing the CBD of PER2 (compare inputs between GFP and CBD MEFs), but hyperphosphorylation of CLOCK was not induced (compare between Fig 5F and Suppl. Fig 7B). These data suggest that increased levels of CRY in PER2-overexpressing cells are a direct effect of CRY interaction with PER2. Copurified CRY1 and 2 by IP for CLOCK or BMAL1 were greatly reduced by the CBD (Fig 5F and Suppl. Fig 11), consistent with our hypothesis that CRY interaction with the CLOCK:BMAL1 complex is mediated by PER2. The importance of PER2:CRY interaction is demonstrated by complete disruption of bioluminescence rhythms in MEFs when the interaction is disrupted by the CBD (Fig 5G), further suggesting that both PER and CRY are integral components of the negative complex.
Oscillations of PER play a dominant role in circadian rhythm generation over those of CRY and BMAL1
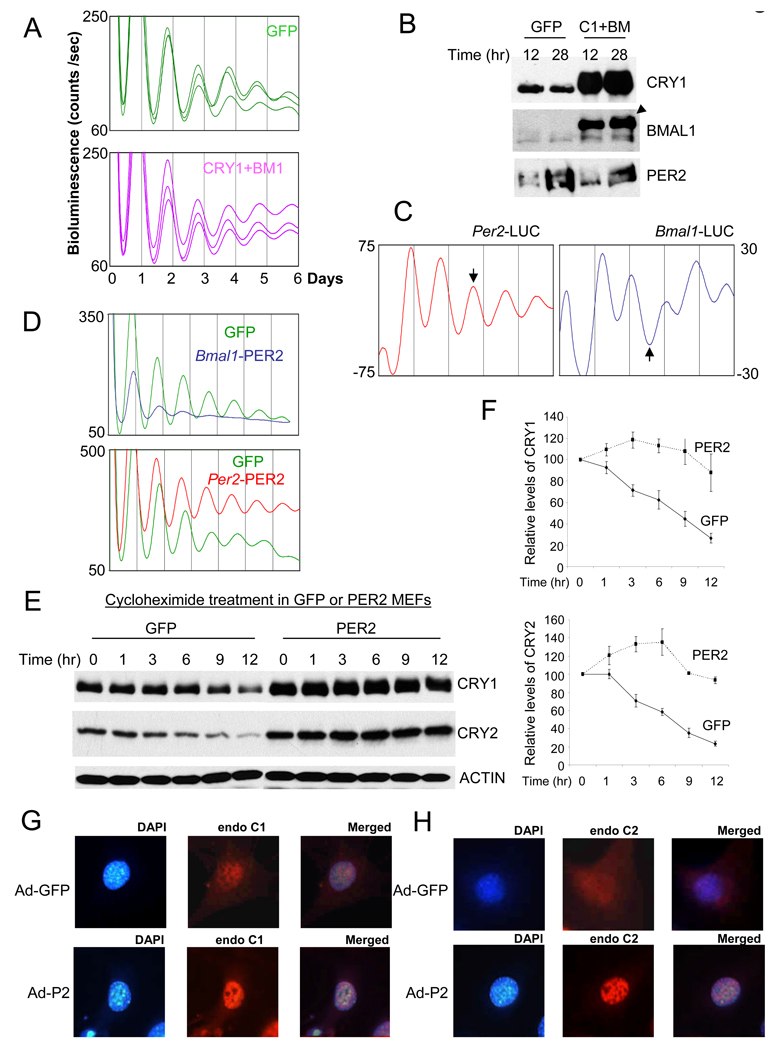
Because the Bmal1 feedback loop involving REV-ERBα and RORα represents a second feedback loop within the clock mechanism, it is possible that circadian rhythms could persist in CRY1-overexpressing cells as a consequence of this second feedback loop. To test this possibility, we disrupted the normal oscillations/levels of both Bmal1 and Cry1 by constitutively overexpressing them in the MEFs. We found that circadian bioluminescence rhythms were still robustly rhythmic in BMAL1 and CRY1-overexpressing MEFs, similar to GFP-MEFs (Fig 6A). The expression levels of BMAL1 and CRY1 in the co-infected cells were more than 10-fold higher than endogenous levels of the proteins in GFP-MEFs (Fig 6B). Thus, we can rule out a role for BMAL1 oscillations underlying the persistence of rhythms under conditions of constitutive CRY1 overexpression.
Fig. 6.
Per oscillation plays a dominant role in circadian rhythm generation. (A) Overexpression of both CRY1 and BMAL1 does not affect the circadian rhythms. MEFs were co-infected with CRY1 and BMAL1 adenoviruses. (B) PER2 remains rhythmic in MEFs overexpressing CRY1 and BMAL1. MEFs were harvested 12 and 28 hr after GFP or CRY1/BMAL1 adenovirus infection and serum shock. The arrow head indicates the exogenous BMAL1 (2XHA-BMAL1). (C) Per2 and Bmal1 promoters drive antiphasic oscillations in MEFs. Adenovirus expressing Luciferase under Per2 or Bmal1 promoter was infected into wt MEFs and bioluminescence rhythms were measured. The arrows indicate the antiphasic oscillations by two promoters. (D) PER2 expression from the Bmal1 promoter severely compromised the circadian rhythms in Per2Luc MEFs, whereas that from Per2 promoter did not disrupt the circadian rhythms. (E) (F) Extended half-lives of endogenous CRYs in PER2-MEFs. To measure half-lives of endogenous CRYs, cycloheximide was added 12 hrs after GFP or PER2 adenovirus infection and serum shock. (F) shows mean +/−SEM of three experiments. (G) (H) Nuclear staining of endogenous CRYs is more pronounced in PER2-MEFs than in GFP-MEFs.
Because the overexpression levels of PER2 in MEFs were more than 10 times higher than peak endogenous levels of PER2, we tested whether the clock would still be functional, albeit at a weak level, if the constitutive levels were closer to the endogenous levels. To effectively disrupt the endogenous oscillations of PER2 near the peak endogenous levels, Bmal1 promoter was used to drive expression of Per2 in antiphase to endogenous Per2. First, by driving expression of Luciferase under control of the Bmal1 and Per2 promoters in wildtype MEFs, we confirmed that oscillations driven by these two promoters occur in antiphase (Fig 6C). When PER2 was exogenously expressed anti-phase by the Bmal1 promoter, the levels were ~4 times higher than the peak level of endogenous PER2 in GFP control MEFs (Suppl Fig 12A). At this relatively modest level of PER2 overexpression, bioluminescence rhythms were still severely compromised in Per2Luc MEFs (Fig 6D). However, when PER2 was expressed at similarly high levels but in-phase with the endogenous PER2 under the Per2 promoter (Suppl Fig 12B), the circadian rhythms were still robustly rhythmic (Fig 6D), suggesting that a rhythm of mPER in the correct phase is far more critical than mPER’s absolute abundance levels. Interestingly, the period of the rhythms was significantly shorter (~ 1 hr) than that of GFP control MEFs, consistent with studies in Drosophila showing that increased transcription of dper from the endogenous promoter or increased dper gene dose does not disrupt rhythms but shortens the period (Baylies et al., 1987; Hao et al., 1999; Kadener et al., 2008; Rutila et al., 1992). Exogenous antiphasic-oscillating expression of dClock does not disrupt circadian rhythms in Drosophila (Kim et al., 2002), further supporting the critical role of PER oscillation.
Our earlier experiments demonstrate that PER affects CRY stability as well as CRY binding with CLOCK:BMAL1. These results suggest that temporal PER abundance controls critical circadian parameters such as timing and duration of the negative feedback inhibition through posttranslational regulation of CRY. To analyze the effects of PER2 on CRY stability more quantitatively, we examined the stability of CRY1 and 2 in the presence of overexpressed GFP or PER2. While GFP expression or viral infection of MEFs did not significantly affect the stability of CRY (data not shown), PER2 overexpression greatly extended CRY’s half life (Fig 6E and F). Immunocytochemistry experiments showed that nuclear staining of endogenous CRY was more pronounced after PER2 overexpression, indicating that PER2 affects nuclear accumulation as well as the stability of CRY (Fig 6G and H), consistent with previous work (Lee et al., 2001). These results are likely relevant to the circadian clock, and not an artifact due to PER2 overexpression, because we observed the opposite results in Per1/2 double mutant mice. CRY1/and 2 levels in the mutant mice were much lower than those in wild type mice, even though the mRNA levels were comparable (Suppl. Fig 13).
Transgenic mice expressing PER2 under control of brain specific promoters exhibit loss of rhythmicity in a conditional manner
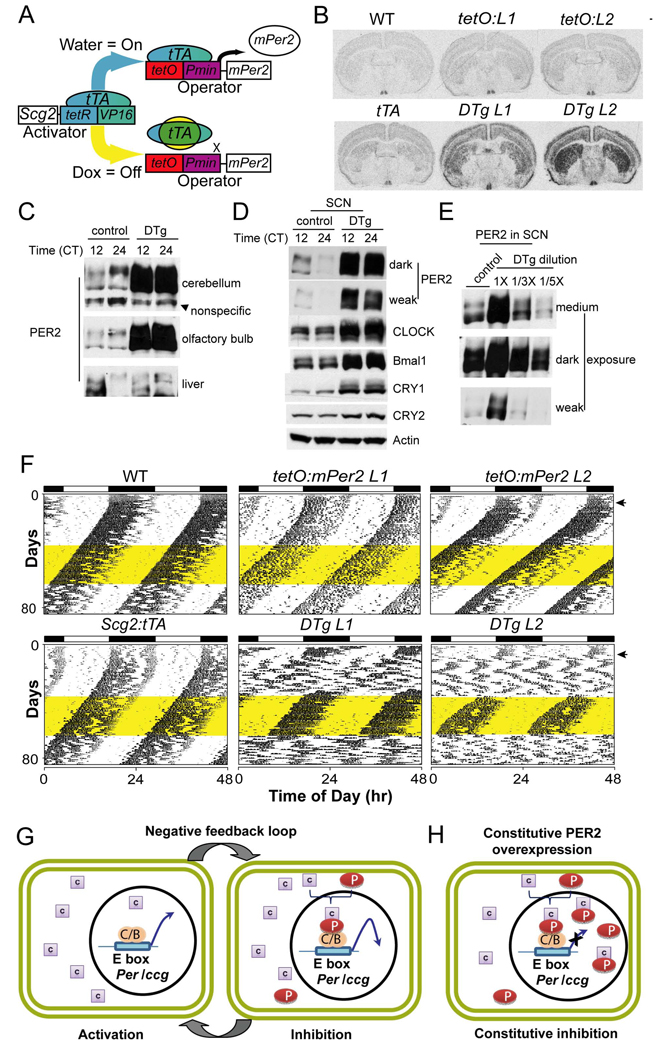
Our results thus far consistently showed that PER rhythms are critical for clock function in two distinct peripheral tissues and two mouse strains. To test whether the same is true for the master circadian pacemaker in the SCN, behavioral rhythms were assessed in conditional transgenic mice constitutively overexpressing Per2 under a brain-specific promoter (Scg2) in a tetracycline-dependent manner (Hong et al., 2007) (Fig 7A). Two independent lines of tetO:Per2 transgenic mice were established and the overexpression of Per2 was validated by in situ hybridization (Fig 7B and Suppl Fig 14A), immunoblots (Fig 7C, D, E) and quantitative PCR (Suppl Fig 14B) in various brain tissues including the SCN. Double-transgenic animals exhibited levels of PER2 in the SCN that were 3–5 fold greater than levels seen in the single transgenic controls, when the levels were measured by X-ray film and quantitative chemiluminescence imaging with serially diluted samples (Fig 7E and Suppl Fig 15).
Fig. 7.
PER2 overexpression in the brain, including SCN, abolished locomotor activity rhythms in constant darkness (DD). (A) Conditional Per2 expression. The system works by driving the tetracycline transactivator (tTA) expression from a tissue-specific promoter (Scg2), which binds the tetracycline operator (tetO) and drives the transcription of Per2 through a minimal promoter sequence (Pmin). However, doxycycline (Dox) potently inhibits tTA, turning off Per2 transgene expression. (B) In situ hybridization for Per2 confirms exogenous expression throughout the brain with enriched expression in the SCN. Representative sections encompassing the SCN are shown for six genotypes: wild type (WT); tetO:Per2 Line1 Only (tetO:L1); tetO:Per2 Line 2 Only (tetO:L2); Scg2:tTA Only (tTA); tetO:Per2 Line 1/Scg2:tTA (DTg L1); tetO:Per2 Line 2/Scg2:tTA (DTg L2). Levels of Per2 mRNA expression in the SCN of double transgenic (DTg) mice were 2–3 fold higher than those in the SCN of WT and other control mice (Suppl. Fig 14A). (C) (D) PER2 levels in DTg were higher in olfactory bulbs (OB), cerebellum and SCN than those in control mice. The single transgenic (tetO:L2) mouse was used as the control. The tissues from control and DTg mice were collected at CT12 and 24. Unlike OB and cerebellum, there was no sign of overexpression in liver. Note that the phase of PER2 rhythm in control mice is different between SCN and the other brain areas. (E) The serial dilution samples of the DTg SCN extract at CT12 indicate that the levels of PER2 overexpression were 3–5 fold higher than those of PER2 in the control mice at CT12. Quantitative chemiluminescence imaging is shown in Suppl Fig 15. (F) Disrupted circadian rhythms of PER2-overexpressing double-transgenic mice. Representative actograms for the two double transgenic lines show a loss of rhythmicity in DD when compared to WT and single transgenic controls. Locomotor behavior returns to a WT state in the double transgenic animals with a treatment of 10μg/ml dox. This treatment can then be reversed with a water washout converting the double transgenic animals back to a state with no circadian rhythmicity. The yellow shading denotes the time of dox administration in DD. (G) A revised mammalian circadian clock model. When PER is absent, CRY is predominantly cytoplasmic and transcriptional activation by CLOCK:BMAL1 is permitted (activation phase). However, when PER abundance reaches a certain level, PER:CRY complexes enter or accumulate in the nucleus. In the nucleus, PER directly binds the CLOCK:BMAL1 complex to mediate the negative feedback effects of the PER:CRY complex (inhibition phase of the negative feedback loop). (H) In the presence of constitutively overexpressed PER, excess PER:CRY complexes accumulate in the nucleus and constitutively inhibit CLOCK:BMAL1 complex, leading to constitutive inhibition of the negative feedback loop.
As seen in PER-overexpressing MEFs and liver, higher levels of CRY, CLOCK and BMAL1 were measured in the SCN of double transgenic mice (Fig 7D). Moreover, circadian rhythms in locomotor activity were abolished in both double-transgenic lines overexpressing Per2 (Fig 7F). Loss of circadian rhythmicity was quantified by the power of the Fast Fourier Transform (FFT) in the circadian period range in constant darkness (Suppl Fig 16). This loss of circadian rhythmicity caused by the overexpression of Per2 was conditional; treatment with 10 µg/ml doxycycline (Dox) silenced the transgene, and the circadian rhythms of double transgenic animals returned to a wild type state. This change was reflected in the return of circadian rhythmicity with an appropriate free running period (Fig 7F) as well as in the increased amplitude of the FFT in the circadian range (Suppl Fig 16). These phenotypic effects were seen in two independent tetO:Per2 transgenic lines, consistent with the idea that the phenotypes are not due to position or deleterious effects of the transgenes. To further support these claims, the tetO:Per2 Line 2 was crossed with an independent brain specific tTA (CamK2:tTA) line with enriched SCN expression (Mayford et al., 1996). These animals also showed no circadian rhythm in constant darkness and this deficit was reversed by treatment with10 µg/ml of dox (Suppl. Fig 17). Taken together these results demonstrate that circadian rhythms can be completely disrupted by constitutive expression of Per2 in the brain, as seen in MEFs and liver, and that PER oscillations are critical for not only peripheral clocks but also for the master circadian pacemaker in SCN driving locomotor activity rhythms.
Discussion
Although it has been postulated that oscillations of key clock components play a central role in circadian rhythm generation, it has not been previously demonstrated in vivo that oscillation of a particular clock component is critical for the mammalian circadian clock. In the work presented here, contrary to the current models of the clock mechanism in mammals (Lowrey and Takahashi, 2004; Panda et al., 2002; Reppert and Weaver, 2002), constitutive overexpression of CRY1 did not significantly affect the circadian oscillator in MEFs and intact mice. The molecular oscillator can function normally even without oscillations and proper levels of both Cry1 and Bmal1. In contrast, we have shown that oscillations of PER are critical for the circadian rhythm in cultured MEFs as well as in mouse liver and brain in vivo.
We have also described a mechanism explaining the critical role of PER rhythms. The rhythmic interaction between the positive complex (CLOCK:BMAL1) and the negative complex (PER:CRY) is mediated by the rate-limiting PER, and this rhythmic inhibition drives the negative feedback loop. Our results suggest that not only is PER the limiting factor in the formation of the PER:CRY inhibitor complex, but that PER also initiates negative feedback regulation by bringing CRY into contact with CLOCK:BMAL1.
How do we reconcile this mechanism (based on our in vivo data) with models based on numerous transient transfection studies suggesting the direct inhibition of CLOCK:BMAL1 by CRY (Griffin et al., 1999; Kume et al., 1999; Shearman et al., 2000b)? One possible explanation would be that endogenous PER in mammalian cells or some unknown Drosophila protein (possibly dPER) in S2 cells may have complexed with the transfected CRY for inhibition of CLOCK:BMAL1. Another scenario (more likely) is that the transient transfection assays do not reflect the true in vivo mechanism for inhibition of CLOCK:BMAL1 by PER/CRY. Unlike artificial promoters used in those assays, native promoters would contain more endogenous regulatory elements and normal chromatin structure, whose regulation by epigenetic factors in a circadian manner may play essential roles in the clock mechanism (Doi et al., 2006; Etchegaray et al., 2003; Ripperger and Schibler, 2006). As discussed in the study by Fan et al. (Fan et al., 2007), the discrepancy could be due to the artificial promoters missing essential regulatory elements other than the E-Box. Alternatively, considering the complexity of endogenous chromatin structure and regulation, the lack of proper chromatin structure in the transfected promoters may account for the discrepancy. Indeed, numerous studies have shown that transfected DNA templates cannot form the same high order chromatin structure as their endogenous counterpart genes (Cervoni and Szyf, 2001; Fryer and Archer, 1998). As a result, transfected promoters are differentially regulated from endogenous or stably integrated promoters by the same transcription factors in some cases. For example, the transcription factor adenovirus E1A inhibits glucocorticoid-dependent transcription from the murine mammary tumor virus promoter in a chromatin context-dependent manner; E1A inhibits the transcription from stably integrated promoter, but not from transfected promoter (Burkhart et al., 2005; Soeth et al., 2002). Taken together, these findings suggest that simplified and artificial reporter assays based on transient transfection may not reflect the true in vivo mechanisms for gene expression and regulation. If this is the case, then in transient transfection assays CRY may well inhibit CLOCK:BMAL1 through a direct mechanism, but in vivo CRY depends on PER to interact with and inhibit CLOCK:BMAL1.
Our model of the clock’s negative feedback mechanism is supported not only by the elimination of rhythms by PER2 overexpression and the failure of CRY overexpression to do the same, but also by a deeper analysis of biochemical mechanisms. Our stoichiometric binding assay using in vitro translated proteins showed that PER2 has much greater binding capacity than CRY1 for CLOCK and BMAL1, supporting the idea that PER brings CRY close to CLOCK:BMAL1 by directly binding to the complex, not the other way around. This is further supported by in vivo data showing that disruption of PER:CRY interaction by CRY-Binding Domain (CBD) mainly removes CRY from the large clock protein complex (CLOCK:BMAL1:PER:CRY) indicating that PER is at the interface for interaction between CRY and CLOCK:BMAL1. Our data suggest that the scaffolding role of PER may be conserved between Drosophila and mammals, since recent studies showed that dPER may be an indirect inhibitor as a molecular bridge between positive and negative components in Drosophila (Kim et al., 2007; Nawathean et al., 2007). Although both PER and CRY can interact with CLOCK:BMAL1 independent of each other in artificial systems, our data suggest that a PER:CLOCK:BMAL1 interaction predominates in vivo. Thus, direct CRY:CLOCK:BMAL1 interactions, if present in vivo, may be stoichiometrically or functionally insignificant for the clock mechanism.
The overexpression levels of PER2 in MEFs and transgenic mice were at least 3–5 fold higher than those of its endogenous counterpart, and thus may exceed the physiological range. One could argue that the overexpressed PER2 could disrupt the molecular clock by a non-physiological mechanism unavailable to overexpressed CRY1 and BMAL1. However, our data showing that overexpression itself does not disrupt circadian rhythms strongly argue against this possibility. When PER2 was similarly overexpressed using either the Bmal1 or Per2 promoter in MEFs, circadian rhythms were eliminated only in Bmal1-PER2 MEFs. Per2-PER2 MEFs exhibited rhythms as robust as in control cells, suggesting that a PER2 rhythm in the correct phase is far more critical than mere PER2 levels, and that 3–5 fold overexpression does not cause any deleterious effects on the molecular oscillator.
Our data clearly show that PER2 rhythms are far more critical than those of CRY for clock function, and therefore PER is a preeminent point of clock regulation. This work has focused on the effects that PER has on CRYs, but PER’s roles extend to other clock components as well. Phosphorylation and possibly abundance of CLOCK and BMAL1 are greatly affected by PER levels, suggesting that PER may regulate transcriptional activity of the transcription factors through posttranslational modifications such as phosphorylation, as has been shown in Drosophila (Kim et al., 2007; Yu et al., 2006). Although the functions of these phosphorylation changes have not yet been determined, prior studies have shown that hyperphosphorylation of dCLOCK induced by dPER is associated with rhythmic transcription of Drosophila clock genes and dCLOCK stability (Kim et al., 2007; Yu et al., 2006). These functions may be conserved in the mammalian system. In terms of clock regulation, robust temporal oscillations in PER levels are vital to the mammalian clock; thus it will be important in future work to focus attention on how PER levels are regulated via transcriptional and posttranscriptional mechanisms.
Experimental Procedures
Animals, Cells and Antibodies
All animals were maintained in a climate-controlled room and used according to the FSU and Northwestern University Animal Care and Use Committee’s guidelines. All experiments involving animals were performed according to Committee-approved protocols. CD1 and C57BL/6 mice were purchased from Charles River laboratory or The Jackson Laboratory. The Per1/2 mutant mice were described previously (Bae et al., 2001). The Cry1/2 double mutant mouse was kindly provided by Dr. Aziz Sancar (Univ. of North Carolina at Chapel Hill). Wild type MEFs and tail fibroblasts were isolated from the Per2Luc mice and immortalized by retroviral introduction of a dominant-negative mutant p53 (GSE56) (Ossovskaya et al., 1996). MEFs and tail fibroblasts exhibited similar results in experiments in Fig 1. Results from MEFs are shown in Fig 1. Primary MEFs from the Cry double mutant mice were used in Fig 5D. COS7 and HEK 293 cells were obtained from ATCC. Antibodies to clock proteins were described previously (Lee et al., 2001; Lee et al., 2004). PER1-1-R, PER2-1-R, CLK-1-GP, BM1-2-GP, C1-GP and C2-GP antibodies were used. Monoclonal anti-GFP antibody was purchased from Boehringer Mannheim. Rabbit anti-ACTIN antibody was purchased from Sigma. Anti-HA (3F10) and V5 were from Roche (Germany) and Invitrogen (Carlsbad, CA), respectively.
Immunoblotting, Immunoprecipitation (IP)
Immunoblotting and IP were performed as described previously (Lee et al., 2001; Shearman et al., 2000a). In Suppl Fig 18, we show that our extraction condition can extract the majority (>90%) of CLOCK and BMAL1 proteins.
Supplementary Material
Acknowledgements
We thank Ailing Zheng and Jiangqin Lai for excellent technical assistance during the project. We thank Dennis Chang for assistance with manuscript revisions. This work was supported by NIH grant NS-053616 awarded to Choogon Lee and by a Silvio O. Conte Center NIH Grant P50 MH074924 to J.S. Takahashi. J.S.T. is an Investigator, S.-H. Yoo is an Associate and V. Kumar was an Associate with the Howard Hughes Medical Institute.
Footnotes
See the supplementary data for other experimental procedures.
References
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart BA, Hebbar PB, Trotter KW, Archer TK. Chromatin-dependent E1A activity modulates NF-kappaB RelA-mediated repression of glucocorticoid receptor-dependent transcription. J Biol Chem. 2005;280:6349–6358. doi: 10.1074/jbc.M411147200. [DOI] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Fan Y, Hida A, Anderson DA, Izumo M, Johnson CH. Cycling of CRYPTOCHROME Proteins Is Not Necessary for Circadian-Clock Function in Mammalian Fibroblasts. Curr Biol. 2007;17:1091–1100. doi: 10.1016/j.cub.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Yagita K, Okamura H. Does mPER2 protein oscillate without its coding mRNA cycling?: post-transcriptional regulation by cell clock. Genes Cells. 2006;11:525–530. doi: 10.1111/j.1365-2443.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Hao H, Glossop NR, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Forster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes & Development. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annual review of genomics and human genetics. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Mesaki M, Ishida N. Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol Cell Biol. 2001;21:6651–6659. doi: 10.1128/MCB.21.19.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numano R, Yamazaki S, Umeda N, Samura T, Sujino M, Takahashi R, Ueda M, Mori A, Yamada K, Sakaki Y, et al. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc Natl Acad Sci U S A. 2006;103:3716–3721. doi: 10.1073/pnas.0600060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, Stark GR, Chumakov PM, Gudkov AV. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci U S A. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Edery I, Hall JC, Rosbash M. The analysis of new short-period circadian rhythm mutants suggests features of D. melanogaster period gene function. J Neurogenet. 1992;8:101–113. doi: 10.3109/01677069209084155. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep. 2005;6:S9–S13. doi: 10.1038/sj.embor.7400424. Spec No. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000a;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000b;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta -TRCP controls clock-dependent transcription via casein kinase 1 dependent degradation of the mammalian period-1 (PER1) protein. J Biol Chem. 2005 doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeth E, Thurber DB, Smith CL. The viral transactivator E1A regulates the mouse mammary tumor virus promoter in an isoform- and chromatin-specific manner. J Biol Chem. 2002;277:19847–19854. doi: 10.1074/jbc.M200629200. [DOI] [PubMed] [Google Scholar]
- Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Finding new clock components: past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. Embo J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Yagita K, Okamura H. Role of cyclic mPer2 expression in the mammalian cellular clock. Mol Cell Biol. 2005;25:1912–1921. doi: 10.1128/MCB.25.5.1912-1921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka I, Koinuma S, Shigeyoshi Y, Uchiyama Y, Yagita K. Presence of robust circadian clock oscillation under constitutive over-expression of mCry1 in rat-1 fibroblasts. FEBS Lett. 2007;581:4098–4102. doi: 10.1016/j.febslet.2007.07.053. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.