Abstract
We have examined the effects of changes in cytosine methylation on DNA repair in UV-irradiated Chinese hamster ovary (CHO) cells. A hypomethylated derivative of the CHO K1B11 line, B11aza, was established by passaging B11 cells over several months in increasing concentrations of 5-azacytidine; greater than 60% demethylation was consistently demonstrated in these conditioned cells. Following a UV dose of 10 J/m2, the amount of repair replication performed within 24 h was approximately twofold higher in B11aza cells than in control B11 cells. Removal of T4 endonuclease V-sensitive sites (ESS) from specific restriction fragments within and around the dihydrofolate reductase (DHFR) gene was then examined in B11aza cells and compared with that in B11 cells. Although demethylation had little or no effect on repair in the 5' half of the DHFR gene, within a nontranscribed sequence immediately downstream from the gene, or within an extragenic region further downstream from the DHFR gene, significant increases in repair were observed at the 3' end of the DHFR gene and within an extragenic region upstream of the DHFR gene. However, the increases in DNA repair were not accompanied by any changes in overall cellular resistance to UV when colony-forming ability was assayed. We suggest that the level of DNA methylation may play an indirect role in the regulation of DNA repair, perhaps through an effect on chromatin structure or transcriptional activity.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball D. J., Gross D. S., Garrard W. T. 5-methylcytosine is localized in nucleosomes that contain histone H1. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5490–5494. doi: 10.1073/pnas.80.18.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V. A., Okumoto D. S., Hanawalt P. C. Survival of UV-irradiated mammalian cells correlates with efficient DNA repair in an essential gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3830–3833. doi: 10.1073/pnas.83.11.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
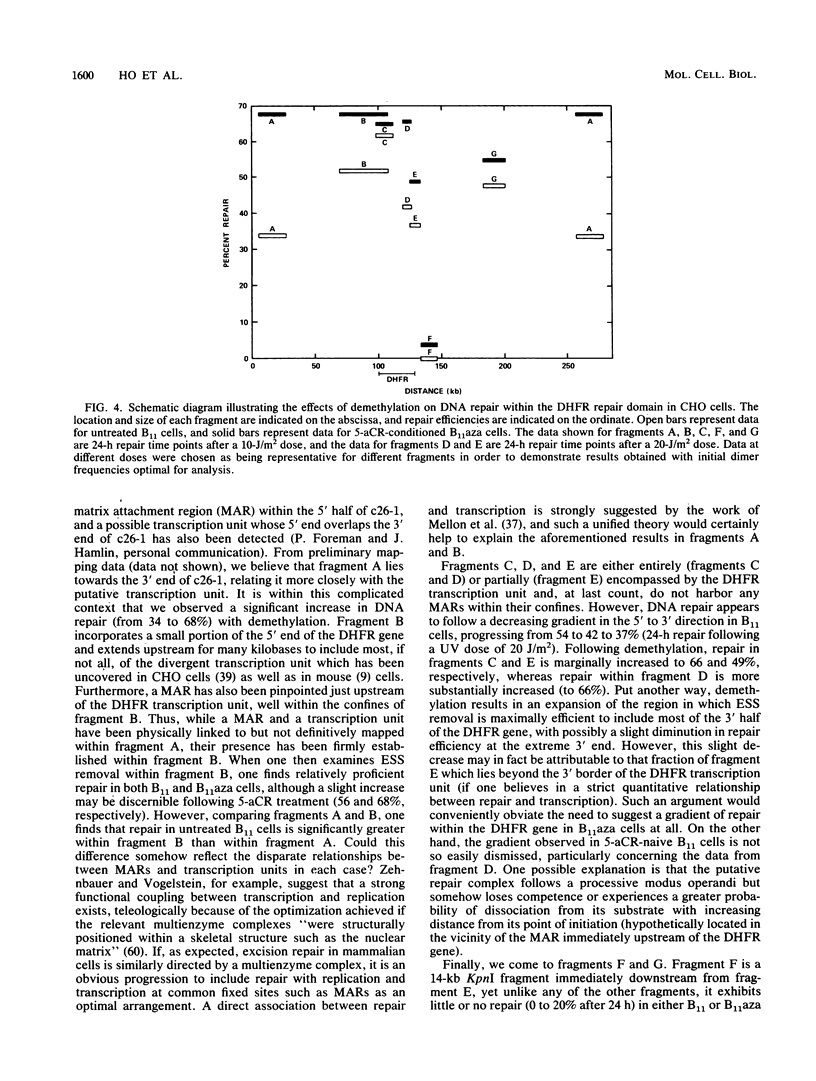
- Bohr V. A., Okumoto D. S., Ho L., Hanawalt P. C. Characterization of a DNA repair domain containing the dihydrofolate reductase gene in Chinese hamster ovary cells. J Biol Chem. 1986 Dec 15;261(35):16666–16672. [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Leys E. J., McEwan R. N., Frayne E. G., Kellems R. E. Analysis of the mouse dhfr promoter region: existence of a divergently transcribed gene. Mol Cell Biol. 1985 Aug;5(8):1847–1858. doi: 10.1128/mcb.5.8.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau E., Gonzales F. A., Michalowsky L. A., Jones P. A. DNA methylation in 5-aza-2'-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol Cell Biol. 1984 Oct;4(10):2098–2102. doi: 10.1128/mcb.4.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C. Studies on the substrate specificity of the T 4 excision repair endonuclease. Mutat Res. 1972 Jun;15(2):113–123. doi: 10.1016/0027-5107(72)90024-3. [DOI] [PubMed] [Google Scholar]
- Glazer P. M., Sarkar S. N., Chisholm G. E., Summers W. C. DNA mismatch repair detected in human cell extracts. Mol Cell Biol. 1987 Jan;7(1):218–224. doi: 10.1128/mcb.7.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata K., Oikawa A. Chromatin structure interferes with excision of abnormal bases from DNA. Biochim Biophys Acta. 1982 Jul 30;698(1):15–21. doi: 10.1016/0167-4781(82)90178-6. [DOI] [PubMed] [Google Scholar]
- Jeggo P. A., Holliday R. Azacytidine-induced reactivation of a DNA repair gene in Chinese hamster ovary cells. Mol Cell Biol. 1986 Aug;6(8):2944–2949. doi: 10.1128/mcb.6.8.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. N., Beverley S. M., Schimke R. T. Rapid spontaneous dihydrofolate reductase gene amplification shown by fluorescence-activated cell sorting. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3711–3715. doi: 10.1073/pnas.80.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M. B., Gowans B. J., Lieberman M. W. Methylation of deoxycytidine incorporated by excision-repair synthesis of DNA. Cell. 1982 Sep;30(2):509–516. doi: 10.1016/0092-8674(82)90248-3. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J., Schimke R. T. Amplification and loss of dihydrofolate reductase genes in a Chinese hamster ovary cell line. Mol Cell Biol. 1981 Dec;1(12):1069–1076. doi: 10.1128/mcb.1.12.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Lee T. T., Karon M. R. Inhibition of protein synthesis in 5-azacytidine-treated HeLa cells. Biochem Pharmacol. 1976 Aug 1;25(15):1737–1742. doi: 10.1016/0006-2952(76)90407-x. [DOI] [PubMed] [Google Scholar]
- Lieberman M. W., Beach L. R., Palmiter R. D. Ultraviolet radiation-induced metallothionein-I gene activation is associated with extensive DNA demethylation. Cell. 1983 Nov;35(1):207–214. doi: 10.1016/0092-8674(83)90223-4. [DOI] [PubMed] [Google Scholar]
- Looney J. E., Ma C., Leu T. H., Flintoff W. F., Troutman W. B., Hamlin J. L. The dihydrofolate reductase amplicons in different methotrexate-resistant Chinese hamster cell lines share at least a 273-kilobase core sequence, but the amplicons in some cell lines are much larger and are remarkably uniform in structure. Mol Cell Biol. 1988 Dec;8(12):5268–5279. doi: 10.1128/mcb.8.12.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. J., Randerath K. Mechanism of 5-azacytidine-induced transfer RNA cytosine-5-methyltransferase deficiency. Cancer Res. 1980 Aug;40(8 Pt 1):2701–2705. [PubMed] [Google Scholar]
- Lu L. W., Chiang G. H., Medina D., Randerath K. Drug effects on nucleic acid modification. I. A specific effect of 5-azacytidine on mammalian transfer RNA methylation in vivo. Biochem Biophys Res Commun. 1976 Feb 23;68(4):1094–1101. doi: 10.1016/0006-291x(76)90308-9. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Bohr V. A., Hanawalt P. C. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell. 1986 May 9;45(3):417–423. doi: 10.1016/0092-8674(86)90327-2. [DOI] [PubMed] [Google Scholar]
- McClelland M., Ivarie R. Asymmetrical distribution of CpG in an 'average' mammalian gene. Nucleic Acids Res. 1982 Dec 11;10(23):7865–7877. doi: 10.1093/nar/10.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Bohr V. A., Smith C. A., Hanawalt P. C. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Minton K., Durphy M., Taylor R., Friedberg E. C. The ultraviolet endonuclease of bacteriophage T4. Further characterization. J Biol Chem. 1975 Apr 25;250(8):2823–2829. [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Zavala M., Hamlin J. L. Similar 150-kilobase DNA sequences are amplified in independently derived methotrexate-resistant Chinese hamster cells. Mol Cell Biol. 1985 Apr;5(4):619–627. doi: 10.1128/mcb.5.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., McClelland M. The effect of site-specific methylation on restriction-modification enzymes. Nucleic Acids Res. 1987;15 (Suppl):r219–r230. doi: 10.1093/nar/15.suppl.r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Okumoto D. S., Bohr V. A. DNA repair in the metallothionein gene increases with transcriptional activation. Nucleic Acids Res. 1987 Dec 10;15(23):10021–10030. doi: 10.1093/nar/15.23.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman M., Penman S. The mechanism of inhibition of protein synthesis by 5-azacytidine in HeLa cells. Biochim Biophys Acta. 1973 Oct 12;324(2):282–289. doi: 10.1016/0005-2787(73)90145-7. [DOI] [PubMed] [Google Scholar]
- Shimada T., Nienhuis A. W. Only the promoter region of the constitutively expressed normal and amplified human dihydrofolate reductase gene is DNase I hypersensitive and undermethylated. J Biol Chem. 1985 Feb 25;260(4):2468–2474. [PubMed] [Google Scholar]
- Solage A., Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978 Jul 11;17(14):2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Kas E., Chasin L. A., Funanage V. L., Myoda T. T., Hamlin J. Effect of gamma rays at the dihydrofolate reductase locus: deletions and inversions. Somat Cell Mol Genet. 1986 Nov;12(6):555–566. doi: 10.1007/BF01671941. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Zhang X. Y., Ehrlich M. A human DNA-binding protein is methylation-specific and sequence-specific. Nucleic Acids Res. 1986 Feb 25;14(4):1599–1614. doi: 10.1093/nar/14.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Y., Zhang X. Y., Khan R., Zhou Y. W., Huang L. H., Ehrlich M. Methylated DNA-binding protein from human placenta recognizes specific methylated sites on several prokaryotic DNAs. Nucleic Acids Res. 1986 Dec 22;14(24):9843–9860. doi: 10.1093/nar/14.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. W., Pitot H. C. Alteration of ribosomal precursor RNA in Novikoff hepatoma cells by 5-azacytidine. Studies on methylation of 45S and 32S RNA. Arch Biochem Biophys. 1974 Dec;165(2):588–596. doi: 10.1016/0003-9861(74)90286-0. [DOI] [PubMed] [Google Scholar]
- Weiss J. W., Pitot H. C. Effects of 5-azacytidine on nucleolar RNA and the preribosomal particles in Novikoff hepatoma cells. Biochemistry. 1975 Jan 28;14(2):316–326. doi: 10.1021/bi00673a018. [DOI] [PubMed] [Google Scholar]
- Williams J. I., Friedberg E. C. Deoxyribonucleic acid excision repair in chromatin after ultraviolet irradiation of human fibroblasts in culture. Biochemistry. 1979 Sep 4;18(18):3965–3972. doi: 10.1021/bi00585a019. [DOI] [PubMed] [Google Scholar]
- Wilson V. G. Methylation of specific cytosine residues enhances simian virus 40 T-antigen binding to origin region DNA. J Virol. 1987 Jul;61(7):2344–2348. doi: 10.1128/jvi.61.7.2344-2348.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelle B., Reynolds R. J., Kottenhagen M. J., Schuite A., Lohman P. H. The influence of the wavelength of ultraviolet radiation on survival, mutation induction and DNA repair in irradiated Chinese hamster cells. Mutat Res. 1980 Aug;72(3):491–509. doi: 10.1016/0027-5107(80)90121-9. [DOI] [PubMed] [Google Scholar]
- van Zeeland A. A., Smith C. A., Hanawalt P. C. Sensitive determination of pyrimidine dimers in DNA of UV-irradiated mammalian cells. Introduction of T4 endonuclease V into frozen and thawed cells. Mutat Res. 1981 Jun;82(1):173–189. doi: 10.1016/0027-5107(81)90148-2. [DOI] [PubMed] [Google Scholar]