Abstract
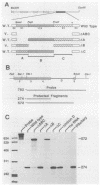
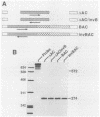
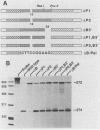
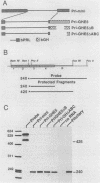
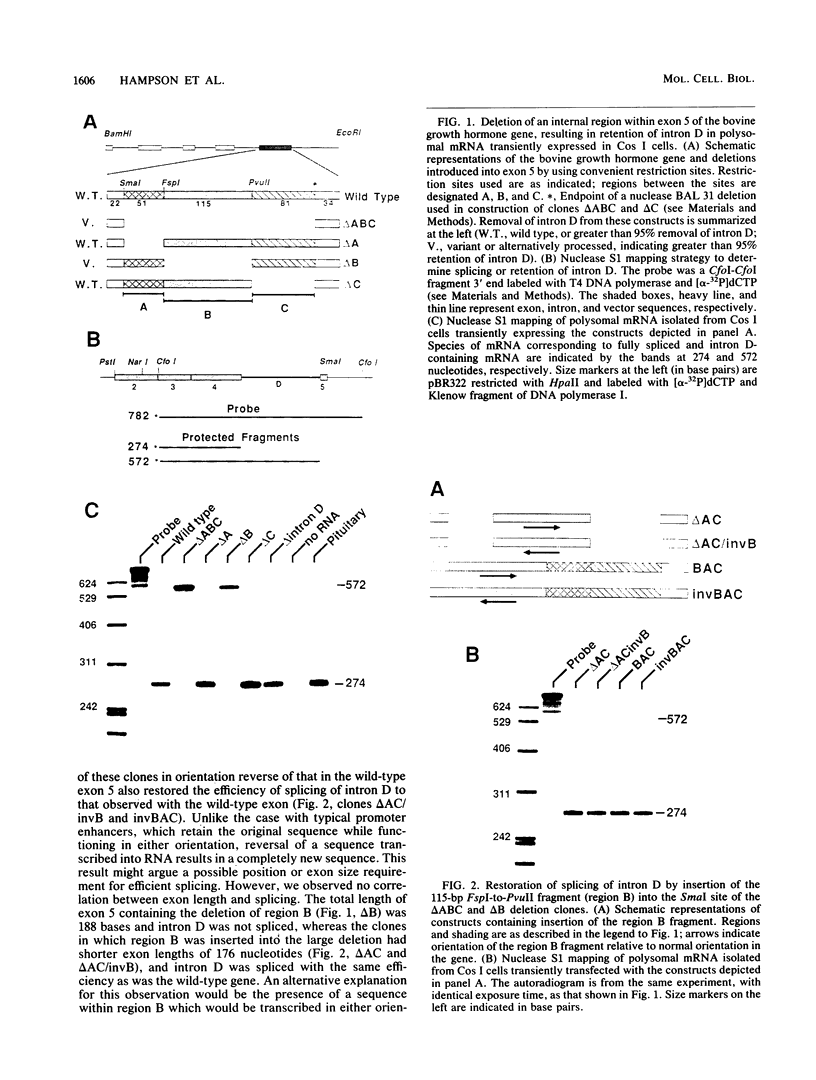
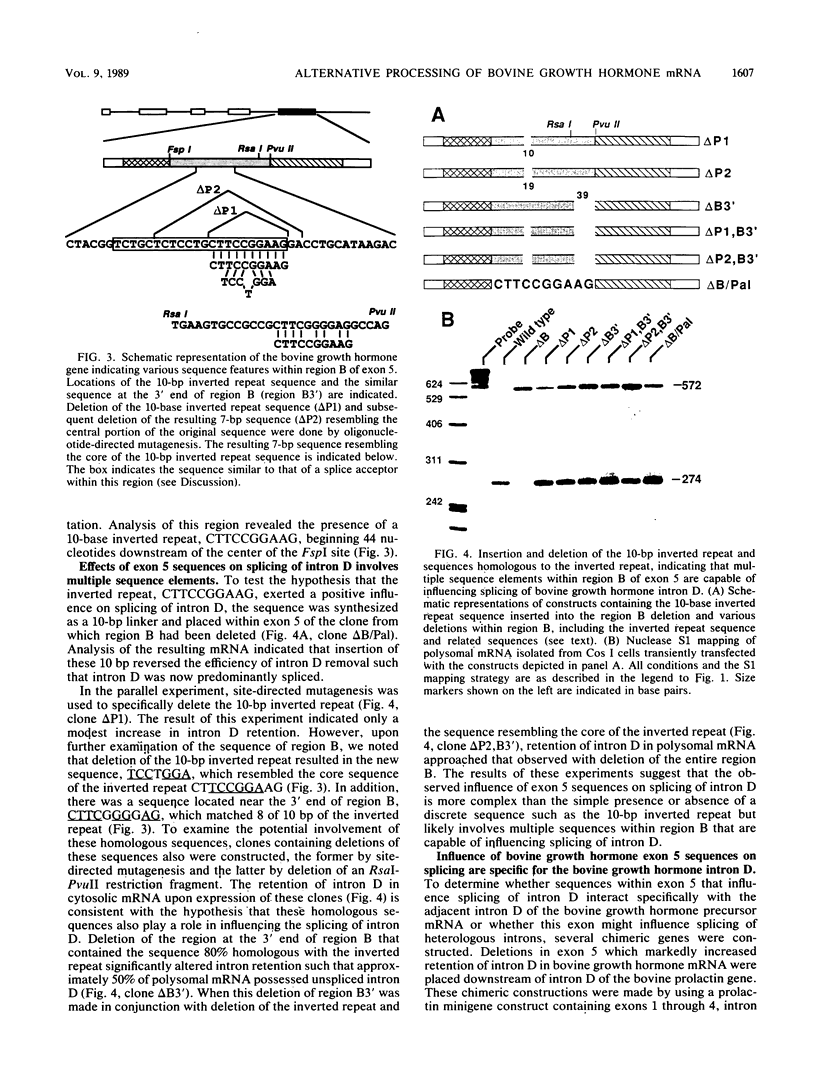
In a previous report, we described the presence, in pituitary tissue, of an alternatively processed species of bovine growth hormone mRNA from which the last intron (intron D) has not been removed by splicing (R. K. Hampson and F. M. Rottman, Proc. Natl. Acad. Sci. USA 84:2673-2677, 1987). Using transient expression of the bovine growth hormone gene in Cos I cells, we observed that splicing of intron D was affected by sequences within the downstream exon (exon 5). Deletion of a 115-base-pair FspI-PvuII restriction fragment in exon 5 beginning 73 base pairs downstream of the intron 4-exon 5 junction resulted in cytoplasmic bovine growth hormone mRNA, more than 95% of which retained intron D. This contrasted with less than 5% of the growth hormone mRNA retaining intron D observed with expression of the unaltered gene. Insertion of a 10-base-pair inverted repeat sequence, CTTCCGGAAG, which was located in the middle of this deleted segment, partially reversed this pattern, resulting in cytosolic mRNA from which intron D was predominantly removed. More detailed deletion analysis of this region indicated that multiple sequence elements within the exon 5, in addition to the 10-base-pair inverted repeat sequence, are capable of influencing splicing of intron D. The effect of these exon sequences on splicing of bovine growth hormone precursor mRNA appeared to be specific for the growth hormone intron D. Deletions in exon 5 which resulted in marked alterations in splicing of growth hormone intron D had no effect on splicing when exon 5 of bovine growth hormone was placed downstream of the heterologous bovine prolactin intron D. Deletions in exon 5 which resulted in marked alterations in splicing of growth hormone intron D had no effect on splicing when exon 5 of bovine growth hormone was placed downstream of the heterologous bovine prolactin intron D. The results of this study suggest a unique interaction between sequences located near the center of exon 5 and splicing of the adjacent intron D.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Birkenmeier C. S. Inhibition of intractable nucleases with ribonucleoside--vanadyl complexes: isolation of messenger ribonucleic acid from resting lymphocytes. Biochemistry. 1979 Nov 13;18(23):5143–5149. doi: 10.1021/bi00590a018. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Camper S. A., Yao Y. A., Rottman F. M. Hormonal regulation of the bovine prolactin promoter in rat pituitary tumor cells. J Biol Chem. 1985 Oct 5;260(22):12246–12251. [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Cooke N. E., Ray J., Emery J. G., Liebhaber S. A. Two distinct species of human growth hormone-variant mRNA in the human placenta predict the expression of novel growth hormone proteins. J Biol Chem. 1988 Jun 25;263(18):9001–9006. [PubMed] [Google Scholar]
- Crenshaw E. B., 3rd, Russo A. F., Swanson L. W., Rosenfeld M. G. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987 May 8;49(3):389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., Arenas J., Perkins K. K., Furneaux H. M., Pick L., Young B., Roberts R. J., Hurwitz J. In vitro formation of a lariat structure containing a G2'-5'G linkage. J Biol Chem. 1987 Mar 25;262(9):4267–4273. [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988 Feb;8(2):860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Gordon D. F., Quick D. P., Erwin C. R., Donelson J. E., Maurer R. A. Nucleotide sequence of the bovine growth hormone chromosomal gene. Mol Cell Endocrinol. 1983 Nov;33(1):81–95. doi: 10.1016/0303-7207(83)90058-8. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., Rottman F. M. Alternative processing of bovine growth hormone mRNA: nonsplicing of the final intron predicts a high molecular weight variant of bovine growth hormone. Proc Natl Acad Sci U S A. 1987 May;84(9):2673–2677. doi: 10.1073/pnas.84.9.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988 May;8(5):2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Lathe R., Kieny M. P., Skory S., Lecocq J. P. Linker tailing: unphosphorylated linker oligonucleotides for joining DNA termini. DNA. 1984;3(2):173–182. doi: 10.1089/dna.1984.3.173. [DOI] [PubMed] [Google Scholar]
- Law R., Kuwabara M. D., Briskin M., Fasel N., Hermanson G., Sigman D. S., Wall R. Protein-binding site at the immunoglobulin mu membrane polyadenylylation signal: possible role in transcription termination. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9160–9164. doi: 10.1073/pnas.84.24.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Legrain P., Seraphin B., Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988 Sep;8(9):3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W., Maroney P. A. Translational efficiency of cMyc mRNA in Burkitt lymphoma cells. Mol Cell Biol. 1984 Oct;4(10):2235–2238. doi: 10.1128/mcb.4.10.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Parent A., Zeitlin S., Efstratiadis A. Minimal exon sequence requirements for efficient in vitro splicing of mono-intronic nuclear pre-mRNA. J Biol Chem. 1987 Aug 15;262(23):11284–11291. [PubMed] [Google Scholar]
- Plotch S. J., Krug R. M. In vitro splicing of influenza viral NS1 mRNA and NS1-beta-globin chimeras: possible mechanisms for the control of viral mRNA splicing. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5444–5448. doi: 10.1073/pnas.83.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Woychik R. P., Camper S. A., Lyons R. H., Horowitz S., Goodwin E. C., Rottman F. M. Cloning and nucleotide sequencing of the bovine growth hormone gene. Nucleic Acids Res. 1982 Nov 25;10(22):7197–7210. doi: 10.1093/nar/10.22.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik R. P., Lyons R. H., Post L., Rottman F. M. Requirement for the 3' flanking region of the bovine growth hormone gene for accurate polyadenylylation. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]