Abstract
A method to determine the rate of protein breakdown in individual proteins was developed and tested in rats and confirmed in humans, using administration of deuterium oxide and incorporation of the deuterium into alanine that was subsequently incorporated into body proteins. Measurement of the fractional breakdown rate of proteins was determined from the rate of disappearance of deuterated alanine from the proteins. The rate of disappearance of deuterated alanine from the proteins was calculated using an exponential decay, giving the fractional breakdown rate (FBR) of the proteins. The applicability of this protein-specific FBR approach is suitable for human in vivo experimentation. The labeling period of deuterium oxide administration is dependent on the turnover rate of the protein of interest.
Keywords: protein kinetics, protein degradation, fractional breakdown rate, deuterated water
body protein pools undergo continual renewal through a constant cycle of synthesis and degradation. Alterations in the balance between synthetic and degradative rates produce changes in protein pool size that are fundamental for the ability of a biological system to adapt to intrinsic and environmental factors (26). Therefore, accurate methods to assess directly the protein synthetic and breakdown rates are necessary to investigate and understand the regulation of protein balance. The rate of protein synthesis can be measured from the rate of isotope-labeled amino acid incorporation into proteins (45, 47). With technical advances in mass spectrometers and the combination of techniques, the determination of the synthesis rate of small quantities of individual proteins is now possible (21, 24). The need for a comparable approach to measure protein-specific breakdown rates is also important for in vivo human research. However, the assessment of protein breakdown is more troublesome (18). Currently, the in vivo approach used routinely relies on isotope dilution and arterial-venous catheterization (2, 6). More pools have been added to the method, improving validity (17), and the latest approach has been reworked and further refined by Zhang and colleagues (51–53). However, the same principle remains, i.e., to measure the dilution of free amino acid tracer enrichment at arterial, venous, and intracellular sites as a consequence of release of tracee amino acids when proteins degrade. Although this technique has substantial utility, the protein origin of the amino acids remains unknown. Hence, it does not permit estimation of the breakdown rate of specific proteins as it does for the direct tracer incorporation approach for protein synthesis measurements. For this reason we developed a fractional breakdown rate (FBR) method in rats, allowing the measurements of FBR of individual proteins, and demonstrated the applicability of this approach in humans.
To meet the requirement of protein specificity, the analytical target in the protein breakdown approach must be the proteins themselves. If protein can be prelabeled with one or more labeled amino acids, then the rate of disappearance of labeled amino acids from protein can be measured to determine protein breakdown (14, 32, 34, 46, 49). Historically, this approach has been used in vitro on bacteria (14, 27, 36, 46), in cell cultures (41), and in chick skeletal muscle cells (49) as well as in human muscle cells (34). In all approaches, an isotope-labeled amino acid was introduced into proteins, and the average rate of protein breakdown was obtained by measuring the rate of release of labeled amino acids from the prelabeled protein structure. Although elegant approaches were developed (e.g., see Ref. 49), it became apparent that the reutilization of the recently released tracer by reuptake via new protein synthesis results in an underestimated breakdown rate, which seems hard to circumvent and avoid (1, 34). To avoid reincorporation of tracer amino acids, the tracer amino acids must be quickly removed via metabolism after release from protein breakdown or not reutilized at all. Molnar et al. (32) took advantage of the latter. They exposed growing rats to 18O2 for 36 h in a closed environment, an approach developed some years earlier by Jackson and Heinenger (23), to label hydroxyproline in collagen proteins undergoing posttranslational hydroxylation of peptide-bound proline residues. Because hydroxyproline cannot be reutilized for protein synthesis once it is released from degraded proteins, the hydroxyproline tracer does not recycle. This approach is limited to proteins releasing specific posttranslationally modified amino acids that cannot recycle. Humphrey and Davies (20) tested the option to remove the label quickly from amino acids released from degraded proteins. They used tritiated water (3H2O) to transfer the 3H isotopes to amino acids, which were subsequently incorporated into proteins. Hydrogens attached to oxygen as well as nitrogen in amino acids are nonenzymatically exchanged, and therefore, they are lost in aqueous solutions (38). However, some carbon-bound hydrogens are also exchanged by enzymatic reactions (38). Therefore, administration of deuterium-labeled water (2H2O) will label a range of metabolites, including amino acids (7, 10, 43). The deuterium from 2H2O exchanges with the α-carbon on amino acids that undergo transamination (5, 7, 38) and, in some cases, with carbons on the amino acid side chain. For example, alanine has been shown to exchange three hydrogens on the β-carbon in vivo (35, 44, 48). Furthermore, the administration of 2H2O is safe and feasible in humans.
The purpose of the present study was to establish a method to measure the FBR of slow turnover proteins by the use of deuterated water as a label donor. To provide proof of principle and work through various methodological issues for use in humans, we utilized a rat model. Our goals with this approach were 1) to define the most suitable deuterated amino acid tracers, 2) to evaluate the degree of labeling in the precursor pool as well as in various proteins during prolonged exposure to deuterium oxide, 3) to follow the decay kinetics of deuterated water and amino acids following cessation of 2H2O administration, and 4) to measure protein breakdown rates from the disappearance rate of deuterium-labeled amino acids from specific proteins. Finally, upon satisfactory establishment of the method in rats, we evaluated its utility in humans.
MATERIALS AND METHODS
Mathematical Derivation and Assumptions
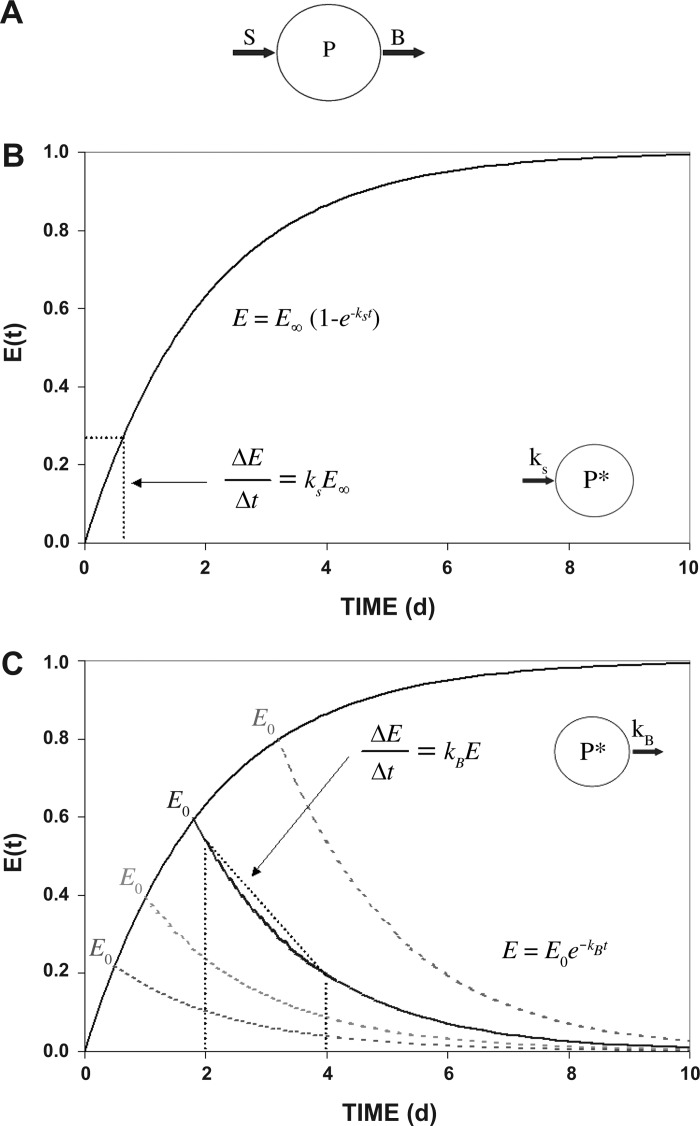
The mathematical derivation and assumptions are outlined in Fig. 1. In a pool of proteins (P), continuous processes of synthesis (S) and breakdown (B) of individual proteins allow remodeling and renewal (Fig. 1A).
Fig. 1.
Model of protein kinetics. A: model of protein synthesis and breakdown. The mass of any protein pool (P) is defined by the rate of protein accretion by synthesis (S) and loss by breakdown processes (B). At tissue protein steady state, P is constant and S and B rates are equal. B: the exponential rise of tracer enrichment over time obtained by incorporation into a protein pool during constant infusion of tracer. The fractional synthesis rate (FSR; kS) can be assessed by determining the initial rate of tracer enrichment increase in protein (ΔE) during a short period of time (Δt), where the precursor tracer enrichment is steady and equal to E∞ (enrichment at infinity). Circle shows tracer incorporated into the protein pool (P*). C: time course of disappearance of tracer from the protein pool after input of tracer is stopped at various time points. The fractional breakdown rate (FBR; kB) of a prelabeled protein pool can be calculated by measuring the rate of tracer disappearance from the protein pool (ΔE) over Δt. It is assumed that no released tracer is reincorporated into the protein.
Assumption 1.
Both the rate of incorporation of label into a protein and the disappearance of label from the protein follow first-order kinetics (as described in Fig. 1, B and C, respectively). For measures of synthesis (Fig. 1B), the appearance of label in a protein pool [given by the enrichment of tracer in the protein (E)] will depend on the synthesis rate (kS):
| (1) |
where E∞ is the enrichment at steady state. To measure the protein fractional synthesis rate (FSR), incorporation of tracer is measured typically by the change in enrichment (ΔE) over a known period of time (Δt) (Fig. 1B).
Correspondingly, a tracer will disappear from a protein pool following the equation
| (2) |
where kB is the fractional protein breakdown rate and E0 is the label enrichment in the protein when the precursor label is removed. Our measurement of protein FBR measures the rate of label removal from the protein that has been labeled previously. As indicated in Fig. 1C, proteins that turn over faster will become labeled to a greater degree than proteins that turn over slower during a period of labeling by a constant delivery of labeled amino acid precursor (greater E0 values). The goal of our method is to then measure the rate of tracer disappearance from the protein (ΔE) over a period of time (Δt) after removal of labeled precursor.
Assumption 2.
The protein pool mass of interest, P, does not change significantly during the period of measurement, which is an underlying assumption also for the precursor-product approach to determine the FSR.
According to assumptions 1 and 2, the first equation can be written as
| (3) |
where rS is the synthesis rate, kB is the rate constant of the protein breakdown, and P is the size of the protein pool.
Equation 4 describes how the protein with labeled amino acids, P*, loses the label, depending on the protein breakdown rate, kB:
| (4) |
Dividing by P we get
Setting E = P*/P, we get
| (5) |
where E is the tracer/tracee (TTR) enrichment ratio.
Thus, Eq. 5 shows that the disappearance of label from a protein follows a mono-exponential decay with the rate constant, kB, as depicted in Fig. 1C.
Assumption 3.
When a labeled amino acid tracer is released from a protein via protein breakdown, reutilization of newly released labeled amino acid can be ignored if the label in free alanine is quickly removed from that amino acid upon release. For the present FBR method to work, sufficient label has to remain in the protein at the time of measurement. On the other hand, the label has to be removed from the free pool of amino acids. Hence, only proteins with a significantly slower turnover rate than the label can be measured using this approach. The fraction of 2H label remaining in the free pool after the administration of 2H2O is stopped can be measured in a preliminary study before any actual measurements of protein breakdown are done. The exponential disappearance of tracer is measured from the free alanine pool over time, and the rate constant of disappearance is (kAla) determined:
| (6) |
where Et(Ala) is the free alanine 2H enrichment at time t and E0(Ala) is the fitted enrichment at time t = 0. Assumption 3 requires that enough time elapse after 2H2O administration is stopped for the alanine 2H enrichment to decline to a minimal level in the free pool. The minimum time (tmin) for that decline to occur can be calculated by rearrangement of Eq. 6:
| (7) |
A maximum threshold is defined [Et(Ala)/E0(Ala)], and from that threshold, the time needed to wait after discontinuing the 2H2O is calculated (tmin). For example, if we defined a maximal residual enrichment in the free alanine pool to be 10% [i.e., Et(Ala)/E0(Ala) = 0.1], then the minimum time to wait after discontinuing 2H2O would be tmin = 2.303/kAla.
Animals and Procedures
Male Sprague-Dawley rats (3 wk old; Charles River Laboratories, St. Constant, QC, Canada) were housed in pairs with a 12:12-h light-dark cycle [controlled temperature: 18–26°C (average: ∼20°C); humidity: 30–70% (average: ∼55%)] and positive pressure. Food (Prolab RMH 3000, Lab Diet; Purina Mills Laboratory, St. Louis, MO), water, and physical activity were accessible ad libitum. Rats were allowed 5 days of acclimatization before any intervention was initiated. At the time of intervention (time zero), all rats were 26 days old and had an average body weight of 76.7 ± 0.7 g (means ± SE). All experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and followed protocols approved by the Institutional Animal Care and Use Committee of the University of Vermont.
The deuterium oxide was administered to the rats via their drinking water, which was mixed with 2H2O as 8% of total volume. Prior to the oral access to the deuterium oxide containing drinking water, we primed all rats at time zero with an intraperitoneal injection of pure sterilized 2H2O (average dose: 2.51 ± 0.02 ml/rat, calculated as 0.040 ml 2H2O/g body wt). All blood samples were obtained in the saphenous vein with the rats restrained. The vein was visualized by prior shaving and gentle occlusion of their thigh. Blood was allowed to clot at room temperature in 500-μl vials containing serum separation gel before centrifugation at 2,500 g for 10 min.
Euthanization of the rats was performed by CO2 inhalation, followed by puncture of the diaphragm and blood draining directly from the heart. Thereafter, the heart was removed and a midpiece of the left ventricle cut out and frozen in liquid nitrogen. A central portion of the midlobe of the liver was then isolated and frozen. Thereafter, the soleus and extensor digitorum longus (EDL) muscles were isolated and cleaned from tendon and visible fascia before freezing. Finally, the patella tendon without bony ends was cut out and frozen. All samples were stored at −80°C until further processing.
Chemicals and Reagents
The following reagents and chemicals were purchased: deuterium oxide (2H2O) (99.8% deuterium; Cambridge Isotopes Laboratories, Andover, MA), acetone and acetonitrile (Fisher Scientific, Fair Lawn, NJ), N-methyl-N-(tert-butyldimethylsilyl) trifluoroacetamide + 1% tert-butyldimethylchlorosilane (MTBSTFA + 1% TBDMCS; Regis Technologies, Morton Grove, IL), and cation exchange resin AG 50W-X8 [100–200 mesh (106–250 μm, hydrogen form), catalog no. 142-1441; Bio-Rad Laboratories, Hercules, CA].
Experimental Design: Rats
Experiment 1.
First, we wanted to determine the time course of the serum water enrichments and the serum-free and serum protein-bound amino acid 2H enrichments by obtaining blood samples from subgroups of rats on days 1, 7, 14, 21, 28, 35, and 42 during continuous administration of deuterated water. These data allowed us to determine the number of hydrogens exposed for exchange in in vivo conditions between water and various amino acids from serum-free amino acids and serum protein-bound amino acids under steady enrichment of serum water. Finally, we wanted to confirm that rats receiving 8% 2H2O in their drinking water would grow normally by comparing them with a group of rats drinking plain tap water.
Based on the measured number of deuterium atoms exposed for exchange (Table 1), we selected those amino acids with the highest number of exchanged deuteriums for the subsequent experiments. We also determined the rate (enrichment) of labeled amino acid tracer incorporated into slow turnover protein in structural tissues as a function of time of deuterium oxide exposure by euthanizing four rats after 42, 49, 56, and 63 days on 8% 2H2O, and tissues of interest were harvested, directly frozen in liquid nitrogen, and stored at −80°C for later analysis. From these samples we determined the duration of the 2H2O administration period that allowed sufficient tracer incorporation into the slow turnover proteins. Finally, a control group of rats (n = 5) were grown for 56 days on plain tap water and then euthanized to determine the natural background isotope ratio for amino acids derived from the various protein fractions. The natural background isotope ratios were used to calculate the actual enrichments (see Amino acid enrichment TTR calculation).
Table 1.
Serum protein amino acid M + 1 enrichments
| Amino Acid | M + 1 Enrichment (E[M + 1]), % | No. of 2H in Amino Acid (nH) |
|---|---|---|
| Alanine | 20.63 ± 0.13 | 3.97 ± 0.13 |
| Glycine | 9.23 ± 0.10 | 1.78 ± 0.06 |
| Valine | 3.57 ± 0.05 | 0.69 ± 0.02 |
| Leucine | 4.80 ± 0.05 | 0.92 ± 0.03 |
| Proline | 6.21 ± 0.08 | 1.19 ± 0.04 |
| Serine | 11.50 ± 0.13 | 2.21 ± 0.08 |
| Phenylalanine | 2.26 ± 0.10 | 0.44 ± 0.02 |
Values are means ± SE. The M + 1 enrichments of selected serum protein amino acids were measured in samples obtained weekly from days 14 through 42. The number of deuteriums in each amino acid was calculated by dividing the average M + 1 enrichment shown in the table by the mean water enrichment at steady state (E2H2O = 5.20 ± 0.17%).
Experiment 2.
Using what was learned in experiment 1, we then administered 2H2O to rats to label body proteins with deuterium-labeled amino acids, and then we measured the disappearance rate of the deuterium from body water, from free amino acids, and from serum protein-derived amino acids. Eight rats were grown for 42 days on 8% vol 2H2O-enriched drinking water. Thereafter, the rats returned to plain, unlabeled tap water for ≤38 days. During the washout period, frequent blood samples (every 4th day in subgroups of 4 rats) were obtained to follow the disappearance of the deuterium from serum water, free amino acids in serum, and serum protein amino acids. In addition, the rats were weighed every week to determine when the weekly weight gain started to level off (to comply with the assumption of a steady protein mass). The rats were euthanized and tissues harvested after 35 (n = 4) and 38 days (n = 4) on tap water.
Experiment 3.
At this point, we had determined the amount of time necessary to label the slow turnover tissue proteins (experiment 1) and the time necessary to allow the tracer to disappear from the precursor pool (experiment 2). The purpose of the third experiment was to measure the disappearance rate of deuterium-labeled amino acid tracer from various tissue proteins and calculate the FBR of slow turnover structural proteins. Thirty-two young rats at the age of 26 days were primed with 2H2O and grown on 8% volume deuterium oxide in their drinking water for 42 days. On day 42, the four subgroups of rats were returned drinking plain tap water for another 21 days (age: 89 days), at which point groups of eight rats were euthanized and tissues harvested on four different days (days 0, 1, 2, and 4). The protein-bound amino acid tracer enrichments in the various tissues at these time points were used to calculate the FBRs (see Calculation of rat FBR).
Rat Sample Preparation and Analysis
Serum water sample preparation.
Serum was obtained from whole blood that was allowed to clot and subsequently spun (6 min at 3,200 g) in Vacuette tubes (Z serum sep clot activator; Greiner Bio-one, Kremsmünster, Germany). After testing several methods to obtain serum water, we ended up simply diluting aliquots of serum (∼5 mg) 1:250 by weight with tap water to obtain a deuterium enrichment within the measurable range for isotope ratio mass spectrometer (IRMS) analysis.
Serum Water Deuterium Abundance Analysis By Gas Chromatograph-P-IRMS
Determination of 2H/H ratios in body water was performed using an Agilent 6890A gas chromatograph (GC) coupled with a PDZ Europa (Cheshire, UK) model 20/20 IRMS via a pyrolysis reactor and a PDZ Europa Orchid GC module. Conditions used were GC inlet temperature of 200°C, injection split 30:1 onto a 5-m deactivated silica column kept isothermal at 200°C, and a helium carrier gas flow of 2 ml/min, which served to isolate water vapor and carry it to the pyrolysis reactor (350-mm length, 0.8-mm ID aluminum oxide tube-packed 0.1-mm nickel wires held at 1,050°C). Prior to each day's deuterium water analyses, the pyrolysis reactor was conditioned by injection of 10 μl of hexane to provide a carbon source for water pyrolysis to hydrogen gas and carbon monoxide. The pyrolysis gas product subsequently entered the Orchid module containing a Nafion water trap (Permapure, Toms River, NJ) and a Poraplot Q column (Varian, Palo Alto, CA) that transferred the hydrogen on to the IRMS. The IRMS was calibrated for 2H enrichment measurement by injection of water standards of known 2H content before each day's analyses. All samples were run seven times, and all enrichments (in ppm 2H) are the mean of the last four analyses.
Serum water enrichment TTR calculation.
The deuterium enrichment as TTR of 2H-enriched water was calculated by first subtracting the abundance of deuterium in unlabeled water from that in the enriched sample and subsequently correcting the enrichment (in ppm) with a factor derived from the daily measured deuterium abundances in the standards and then multiplying by 250 (the dilution value of the serum). This 2H enrichment value (in ppm) was finally converted to TTR in percent.
Serum free amino acid preparation.
A 40-μl aliquot of serum was added to 400 μl of ice cold acetone to precipitate serum proteins and then centrifuged. The supernatant was removed and evaporated at 70°C under a stream of nitrogen. The amino acids were then derivatized using MTBSTFA + 1% TBDMCS, and acetonitrile was mixed 1:1 and heated at 70°C for 30 min to allow maximal tert-butyldimethylsilyl (TBDMS) derivatization.
Serum protein-bound amino acid preparation.
The acetone-precipitated serum proteins were transferred to glass vials that contained a 1-ml slurry of cation exchange resin and 1 ml of 0.1 M HCl. The vials were capped and left overnight (∼15 h) at 110°C to hydrolyze proteins. The liberated free amino acids were purified over columns containing washed and acidified cation exchange resin and eluted with 2 × 1 ml of 4 M NH4OH, which was subsequently evaporated under a stream of nitrogen at 70°C. Amino acids were derivatized as the TBDMS derivative, as described above.
Tissue protein-bound amino acid preparation.
Frozen (−80°C) skeletal muscle, heart muscle, and liver specimens were thawed in 1 ml of ice-cold homogenization buffer (0.15 M NaCl, 0.25 M sucrose, 0.2 mM EGTA, 0.2 mM EDTA, 0.5% Triton X-100, and 0.02 M Tris, pH 7.4) and subsequently homogenized using a Polytron RT-2100 with a dispersing aggregate size 2EC (Kinematica, Luzern, Switzerland). The homogenized tissues were left at 4°C for >1 h. The muscle homogenates were spun (800 g, 4°C, 20 min) to pellet myofibrillar and collagen proteins. Added to the muscle pellets was 1 ml of a myofibrillar protein solubilization buffer (0.7 M KCl, 0.1 M tetrasodium pyrophosphate), which was left for 1 h at 4°C and spun (1,200 g, 4°C, 20 min) to pellet collagen proteins. The supernatant containing the myofibrillar proteins was removed, and 2.3 volumes of ethanol were added to precipitate proteins, and the samples were spun (1,600 g, 4°C, 20 min) to pellet proteins. The pelleted proteins from the liver specimens were precipitated after homogenization by centrifugation (4,000 g, 4°C, 20 min), and the supernatant was discarded. The tendon samples were thawed in 1 ml of collagen precipitation buffer (0.7 M KCl, 0.02 M Tris, pH 7.4), which was left at 4°C for 1 h and spun (3,200 g, 4°C, 20 min) to pellet collagen proteins. All protein isolates were washed once with an ice-cold 80% propanol solution, left for 30 min, and subsequently spun (1,600 g, 4°C, 20 min), and the supernatant was discarded before the samples were hydrolyzed overnight (∼15 h) at 110°C in a 1-ml slurry of cation exchange resin and 1 ml of 0.1 M HCl. The free amino acids were purified and derivatized as the TBDMS derivative, as described above.
Amino acid deuterium abundance analysis by gas chromatography/mass spectrometry.
Rat serum-free and protein amino acids as well as tissue protein amino acid 2H enrichments were determined by gas chromatography/mass spectrometry (GC-MS) (Agilent 5973; Agilent Technologies, Palo Alto, CA). Samples were injected onto a capillary column (29 m × 0.25 mm, film thickness 0.25 μm DB-17, J & W Scientific; Agilent Technologies) that was programmed over a temperature range of 155 to 280°C. The mass spectrometer was operated in electron ionization mode and selected ion monitoring. For most amino acids, the [M-57] ion fragment and one mass higher was monitored, except for leucine, where the [M-159] fragment and one mass higher was used. The specific ions monitored were mass-to-charge ratios 260 and 261 for alanine, 246 and 247 for glycine, 288 and 289 for valine, 200 and 201 for leucine, 286 and 287 for proline, 390 and 391 for serine, and 336 and 337 for phenylalanine. All sample isotope ratios reported are the mean of four runs with an overall 95% confidence interval within the range of 0.065–0.133%. Correspondingly, the pooled inter-rat variation was 0.71–1.92%.
Amino acid enrichment TTR calculation.
The area intensity [M + 1]/M isotope ratio was determined for each amino acid measured by GC-MS where M is the base fragment ion and M + 1 is the fragment one mass heavier. The M + 1 tracer/tracee enrichment ratio (E[M + 1]) of each sample was calculated by subtracting the measured natural background (BKGD) ratio from the labeled sample ratio (LAB):
The number of sites exchanging deuterium atoms in vivo with 2H2O on each individual amino acid (nH) was calculated by dividing the M + 1 enrichment by the water enrichment at equilibrium:
| (8) |
The standard error of the number of exchanged hydrogens for each amino acid (SEnH) was approximated by the equation
| (9) |
where it is assumed that 1) the measurements of the number of exchanged hydrogens of each amino acid are independent and no correlation exists between them, and 2) an equal number (H) of measurements was performed for the enrichment of each amino acid M + 1 and deuterated water (n = 5).
The actual 2H tracer/tracee enrichment ratio of each amino acid (E) was then calculated:
| (10) |
Calculation of rat FBR.
Using rats allowed us to collect tissue at only one time point per animal. Therefore, in experiment 3, we averaged the protein-bound alanine 2H enrichments from the unrelated subgroups (n = 8/group): days 0, 1, 2, and 4 for each protein type. To calculate the disappearance rate of 2H alanine from the tissue proteins, i.e., the FBR (kB), we log transformed the individual protein-bound alanine 2H enrichments from each rat tissue and determined the slope of the line best describing the relation between the log-transformed enrichments and time. The standard error of the FBR value was calculated from the standard error of the slope of the line.
Experimental Design: Humans
Experiment 4.
The purpose of this experiment was to evaluate the utility of the technique in humans and measure the FBR of muscle and skin proteins. We wanted to administer the least amount of deuterated water needed to label the proteins sufficiently to allow determination of loss of labeled amino acids over a reasonable time period (∼5–10 days). The period of deuterium oxide exposure and thus labeling of slow turnover proteins was estimated using an average turnover rate of body water of 9%/day (42) and slow turnover myofibrillar protein of 1%/day (33). First, we modeled the decay in body water 2H enrichment after ingestion of deuterated water at a single occasion. Subsequently, the descending body water enrichment was used as an estimate of the precursor enrichment for the incorporation of deuterated amino acid into slow turnover proteins. The model suggested that by ∼40 days there would be maximal and measurable 2H alanine in slow turnover proteins and that after an additional 40 days (i.e., 80 days after intake of deuterated water) body water and serum-free alanine 2H enrichments would be negligible.
Seven healthy men (age: 31 ± 2 yr; height: 184 ± 2 cm; body mass index: 22.3 ± 0.7 kg/m−2; lean body mass: 66.1 ± 2.3 kg) gave their written consent to participate in the study that was approved by the local ethics committee of Copenhagen municipal (H-1-2010-007). On day 0, all subjects drank a total of 233 ± 8 ml of 2H2O [estimated to be ∼0.5% of total body water (9, 42)] mixed 1:2 with tap water given as six divided hourly oral doses. All subjects had a blood sample taken prior to administration of the first bolus of 2H water. In four subjects, blood was drawn hourly until 1 h after the last bolus. Blood samples were obtained from all subjects on days 1, 2, 3, 7, 14, 17, 24, 41, 50, 59, 70, 80, and 85. Muscle specimens were collected from five subjects and skin biopsies from two subjects to determine the abundance of labeled alanine in different tissue proteins at various time points. All subjects had biopsies taken on day 42 to verify the proximity between the alanine 2H enrichment in proteins and precursor. Tissue biopsies were obtained again on days 80, 85, and 90. Tissue sample sites were prepared with local anesthesia (1% lidocaine). Skin was taken from the posterior hip with a sterile 4-mm disposable Biopsy Punch (Miltex, York, PA), and muscle was sampled from vastus lateralis with a 14-gauge disposable core tissue biopsy needle (MN1410) using a Biopsy Instrument (MG1522, Bard Magnum; Bard, Covington, GA). All tissue samples were immediately frozen in liquid nitrogen and saved at −80°C until later analysis.
Human serum-free and protein-bound alanine preparation.
Aliquots (100 μl) of serum were added to 500 μl of 50% acetic acid left for 30 min and then poured over acidified AG 50W-X8 cation resin columns. The amino acids were eluted by adding 2 × 1 ml of 4 M NH4OH to the columns. The eluent was collected and evaporated under a stream of nitrogen at 70°C. TBDMS derivatization was performed as described previously.
Serum-free deuterated alanine enrichment analysis by GC-MS.
The alanine abundances in human serum samples were determined by GC-MS (Finnigan Mat Automass II MS; Finnigan), as described for the rat samples with small changes that allowed for the use of larger samples.
Protein-bound alanine preparation.
Amino acids from serum proteins were isolated in a similar manner as per the rat samples, with small changes that allowed for use of larger samples. Proteins were isolated from 400-μl aliquots of serum by precipitation by adding 1 ml of ice-cold acetone with subsequent centrifugation. Amino acids were isolated from proteins in frozen muscle and skin specimens after homogenization (FastPrep 120A-230; Thermo Savant, Holbrook, NY) in 1 ml of ice-cold homogenization buffer, as used with the rat samples. Homogenates were treated as per the rat samples to isolate myofibrillar proteins. These protein isolates and the serum protein isolates were hydrolyzed overnight at 110°C in 1 ml of 6 M HCl. The free amino acids were purified over cation resin columns and derivatized as the NAP derivative (11).
Deuterium-labeled protein-bound alanine enrichment analysis by GC-P-IRMS.
The alanine abundances in human serum, myofibrillar, and skin collagen proteins were analyzed on a GC-P-IRMS system (GC Combustion III, Delta Plus XL; Thermo Finnigan). The NAP-amino acid samples were injected in the PTV mode with the GC inlet initially at 45°C for 1 min then elevated to 280°C with 20°C/min. The GC column (30 m × 0.25 mm film thickness, 1 μm DB-1701; J & W Scientific) was ramped from 45 to 280°C. All GC column effluent passed into a high-temperature reduction reactor (1,450°C) where the organic compounds were reduced to hydrogen gas and carbon monoxide before entering the IRMS.
Calculation of human protein FBR.
For the human experiment, we obtained paired samples and were able to determine the FBR of each individual's muscle protein fraction. By log transformation of the measured enrichment values, we determined the slope, kB, of the ln(E) vs. time curve as an average across the time between the latter three biopsies (10 days) as well as the adjacent biopsies (5 days).
Statistics
Changes in body weights in deuterium-labeled and control rats were compared by a two-factor (water and time) ANOVA with repeated measures. The level of deuterium enrichments in serum water and in free and protein-bound alanine turned out not to be normally distributed and thus was tested using a nonparametric one-way ANOVA (Kruskal-Wallis) test followed by Dunn's multiple comparison post hoc test. Finally, the alanine 2H enrichments in the various tissue protein fractions obtained in experiment 3 were compared using a one-way ANOVA. When a significant effect was seen, the Holm-Sidak post hoc test was applied to compare individual enrichments. For the human experiment 4, the differences in tissue protein-bound alanine were compared by a one-way ANOVA with repeated measures, applying a Holm-Sidak post hoc test. A value of P < 0.05 was defined as significant. Unless otherwise stated, all values are means ± SE. All statistical tests were performed in SigmaPlot 11 (Systat Software, San Jose, CA).
RESULTS
Experiment 1
Rat growth while 2H2O is consumed.
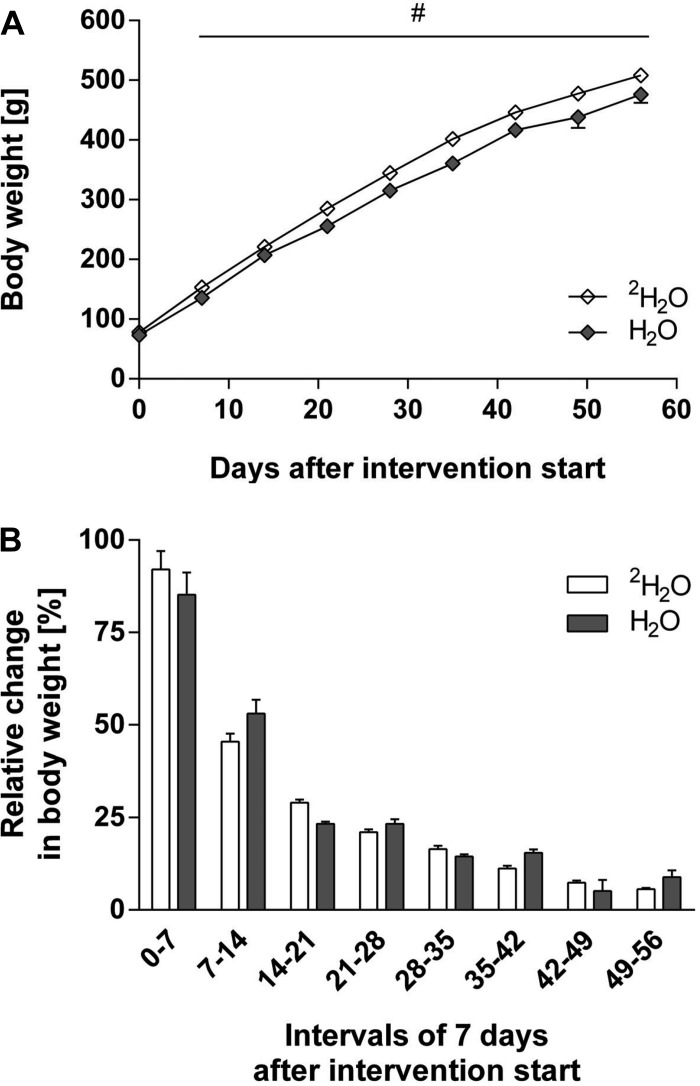
Rats at the age of 26 days were randomized to receive deuterium-labeled water (2H2O; n = 16) or plain tap water (H2O; n = 5). The starting body weights were similar (2H2O: 78.4 ± 1.5 g; H2O: 73.4 ± 2.1 g; P > 0.05). During the following 56 days, rats fed 2H2O were slightly, but statistically, heavier than the control rats after 7 days of being fed deuterium oxide (P < 0.05; Fig. 2A). However, the relative growth rate across 7 days was not statistically different between the groups (Fig. 2B). As expected, the rate of growth diminished as the rats got older.
Fig. 2.
Change in body weight of rats consuming either unlabeled or deuterated water. A: rat body weight in g (values are means ± SE, although the error bars are typically smaller than the symbols). Rats were 26 days old on day 0 on the graph. Rats received either 8% volume of deuterium oxide in drinking water (2H2O; ◇) (n = 16) or plain tap water (H2O; gray diamonds) (n = 5). Rats were weighed every 7th day until euthanization (no. of rats remaining: day 42, n = 16; day 49, n = 12; day 56, n = 8). #Rats receiving deuterium oxide were slightly but significantly (2-way repeated-measures ANOVA) heavier than the control rats. B: weekly relative change in rat body weight (means + SE). There was no significant difference in relative weekly growth (2-way repeated-measures ANOVA).
Deuterium enrichment in the rats fed 2H2O.
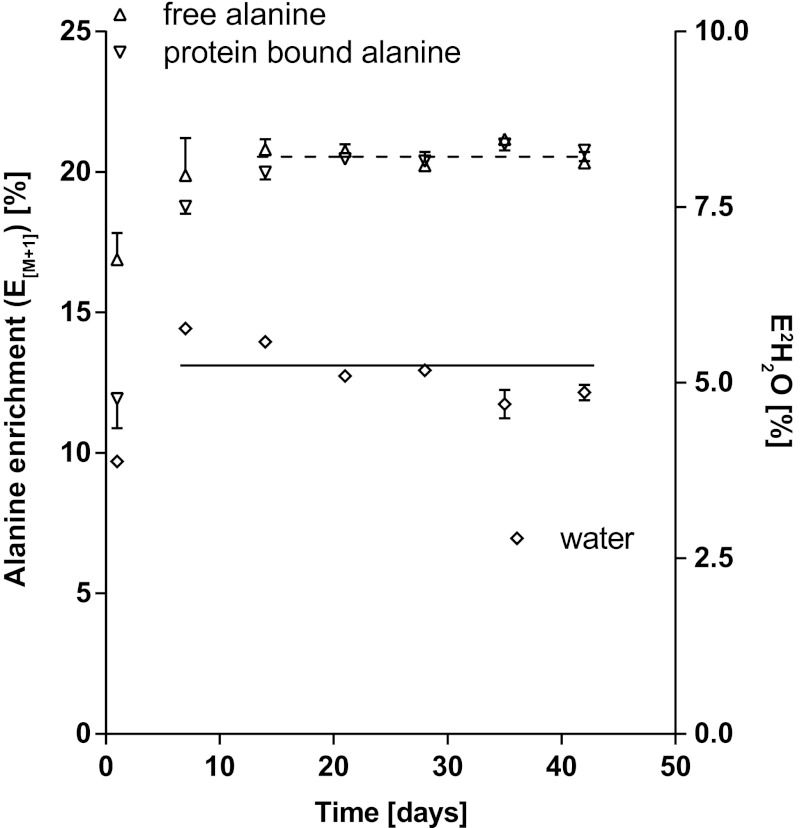
The intraperitoneal priming dose, followed by administration of 2H2O to the rats in their drinking water, elevated the rats' body water deuterium enrichment (as measured from serum samples) to 3.88 ± 0.08% after 1 day and to 5.77 ± 0.05% after 7 days. Any fluctuations in the measured deuterium oxide enrichments after day 7 are ascribed to be a consequence of alterations in water and food intake. The mean body water enrichment during the deuterium labeling period from days 7–42 was 5.20 ± 0.17% (Fig. 3). Figure 3 also shows the M + 1 enrichments measured by GC-MS for free alanine in serum and alanine in serum protein from days 1–42. Both serum-free and serum protein alanine enrichments rose from days 0-14 (1-way ANOVA on ranks, Kruskal-Wallis, P < 0.001) and then reached a plateau.
Fig. 3.
Deuterium enrichments in serum-free and protein-bound alanine (E[M + 1]) and in serum water (E2H2O). The symbols are M + 1 enrichments of free alanine in serum (△; n = 2–4/time point at left y-axis) and M + 1 enrichments of serum protein-bound alanine (▽; n = 4–12/time point at left y-axis) and deuterium enrichment of serum water (◇; n = 4–12/time point at right y-axis) from rats grown for 42 days on 8% volume deuterium-enriched drinking water. Values are means ± SE, although the error bars may be smaller than the symbols. A nonparametric 1-way ANOVA (Kruskal-Wallis) test was performed to define the time point when 2H enrichment was in isotopic steady state in each pool. Alanine deuterium enrichments were in isotopic steady state from days 14 to 42 in serum both free and protein bound, whereas the water enrichment fluctuated slightly more but had equilibrated in the body water pool by day 7. The mean deuterium enrichment for serum-free alanine at steady state is shown as a dashed line (20.66 ± 0.17%) and for body water as a solid line (5.20 ± 0.17%).
The measurement of free alanine enrichment from serum was difficult because of the limited volume of serum available, hence resulting in incomplete data sets. However, a small serum sample contained more than adequate alanine in serum proteins for measurement. Therefore, after establishing that free alanine and serum protein alanine had equivalent deuterium enrichments at isotopic steady state, we used serum protein alanine enrichments as a measurement of free alanine enrichment in subsequent studies. Anticipating this to be true for other amino acids in the fast-turning over serum proteins, we also used serum protein as the proxy source for enrichment measurements for other amino acids.
The M + 1 enrichments measured by GC-MS of serum protein alanine, glycine, valine, leucine, proline, serine, and phenylalanine at isotopic steady state as well as the number of hydrogens that are exposed for exchange between water and each amino acid in vivo are shown in Table 1. Valine, leucine, proline, and phenylalanine had one hydrogen exposed for exchange, which would be the α-hydrogen that would exchange during transamination. Serine and glycine had two exchangeable hydrogens, and alanine exchanged four hydrogens, similar to the data of Previs et al. (38). As per the thinking of Previs et al. (38), we chose alanine as the tracer for determining the FBR of proteins because it has the highest number of exchangeable hydrogens and, therefore, produces the greatest M + 1 enrichment measurement by GCMS, giving the highest analytical sensitivity.
Deuterated alanine enrichment in rat proteins.
The labeling of alanine with deuterium in tissue proteins was assessed by analyzing tissue samples from rats in the 2H2O group euthanized after 42, 49, 56, and 63 days. Table 2 shows the alanine 2H enrichments in various protein fractions obtained from different tissues and serum protein. As expected, deuterium enrichment continued to rise in slow turnover proteins but found that 42 days of 2H2O consumption was sufficient time to allow enough deuterium enrichment in alanine in all proteins of interest in these Sprague-Dawley rats.
Table 2.
Deuterium enrichment in body water and in protein-bound alanine in serum and in various tissues
| Days on 8% Volume 2H2O | Body Water | Serum Protein | Liver | Soleus Mix | LV Myofibrillar | LV Collagen | Tendon |
|---|---|---|---|---|---|---|---|
| 42# | 4.69 ± 0.20 | 5.24 ± 0.11 | 5.22 ± 0.08 | 5.20 ± 0.07 | 4.93 ± 0.11 | 4.77 ± 0.06 | 4.29 ± 0.05 |
| 49## | 4.86 ± 0.11 | 5.63 ± 0.08 | 5.43 ± 0.06 | 5.29 ± 0.09 | 5.06 ± 0.07 | 4.93 ± 0.06 | 4.52 ± 0.01 |
| 56 | 5.47 ± 0.06 | 6.01 ± 0.12 | 5.88 ± 0.14 | 5.80 ± 0.08 | 5.46 ± 0.11 | 5.25 ± 0.07 | 4.84 ± 0.09 |
| 63 | 5.60 ± 0.16 | 5.66 ± 0.04 | 5.56 ± 0.03 | 5.84 ± 0.03 | 5.24 ± 0.05 | 5.05 ± 0.02 | 4.78 ± 0.06 |
Values are means ± SE; n = 4/time point. LV, left ventricle. Alanine 2H enrichments (%) are E = E[M + 1]/nH, where nH = 4 for alanine. Protein fractions are serum protein (mainly albumin), liver mix (midlobe nonsoluble liver proteins), LV myofibrillar (heart left ventricle myofibrillar proteins), LV collagen (heart left ventricle collagen proteins), and tendon (patella tendon collagen protein fraction). #Enrichments on day 42 were different from enrichments on days 56 and 63; ##enrichments on day 49 were different from enrichments on day 56.
Experiment 2
Disappearance of deuterium from body water and serum proteins after removal of 2H2O drinking water.
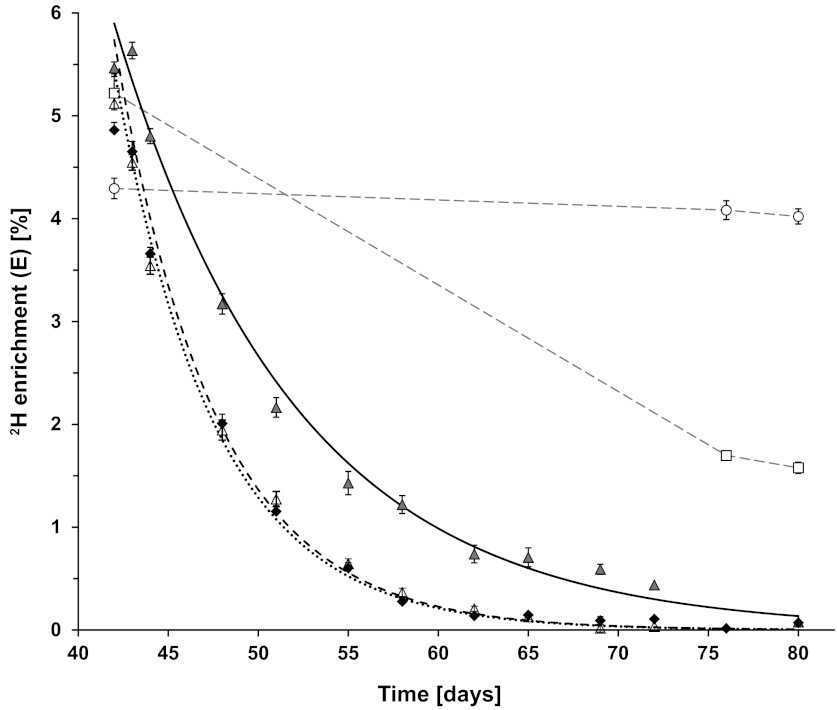
To estimate the minimum number of days that had to elapse after the 2H2O was removed before the FBR could be measured, eight rats were grown first for 42 days on 8% volume deuterium oxide drinking water and then placed on unlabeled drinking water for another 38 days. Figure 4 shows the disappearance of deuterium from serum water, free alanine in serum, and serum protein alanine of the rats (experimental days 42–80). The 2H enrichments decreased monoexponentially as expected in body water, serum free alanine, and serum proteins (Fig. 4). Any remaining deuterium label in water and hence, free alanine after switching the rats from 2H2O to tap water can be reincorporated into the body proteins and thus underestimate the measured FBR. For this purpose, we needed to determine the maximal amount of residual free alanine 2H enrichment allowable based upon the exponential decay constant for serum free alanine 2H enrichment kAla. From this constant, we can calculate the minimum time (tmin) to get the enrichment down below a preset threshold. For example, based upon kAla = 17.9%/day (Fig. 4), we would have to wait 13 days before the residual free alanine 2H enrichment dropped to <10% of its initial value before the FBR could be measured. For convenience, we extended this period to 3 wk, or 21 days, of plain tap water. From Eq. 7 and the measured kAla, we estimate that the residual free alanine 2H enrichment was now only 2% of its initial value. We also note that this time occurs at a rat age where weekly growth was minimal (Fig. 2B).
Fig. 4.
Time course of deuterium enrichments in serum water, free alanine in serum, alanine in serum proteins, and alanine in tendon and liver proteins after removal of 2H water input. All rats consumed 2H2O for the first 42 days and then plain tap water thereafter. The data start on day 42. ◆, Serum water 2H enrichments (n = 4/time point); dotted line is the fitted exponential curve (k = 18.03 ± 0.22%/day). △, Serum-free alanine 2H enrichments (n = 2–3/time point); dashed line is the fitted exponential curve (kAla = 17.94 ± 0.82%/day). Gray triangles, serum protein-bound alanine 2H enrichments (n = 4/time point); solid line is the fitted exponential curve (k = 9.93 ± 0.58%/day). ▫, Tissue protein-bound alanine 2H enrichments for nonsoluble mixed liver proteins. ○, Tissue protein-bound alanine 2H enrichments for tendon collagen proteins. Values are means ± SE, although the error bars may be smaller than the symbols at some time points. The serum water and serum-free alanine enrichments overlap at several time points.
Of the eight rats in experiment 2, four rats were euthanized at 35 days and four rats euthanized at 38 days after return to tap water to harvest tissues (experimental days 77 and 80). The alanine enrichments for nonsoluble mixed liver proteins and tendon collagen protein are shown in Fig. 4 and compared against the alanine enrichments in the same proteins in rats euthanized on the last day of 2H water administration (day 42).
At this point, we had determined that 42 days of deuterium labeling followed by 21 days on unlabeled water was adequate first to label the proteins of interest and subsequently to reduce the deuterium enrichment of free alanine in serum and body water to negligible levels yet have sufficient deuterium retained in various slow turnover proteins. Because the rats were also weight stable on a weekly basis at this age, these conditions were selected for measuring protein breakdown rates in experiment 3.
Experiment 3
Measurement of the rat tissue protein FBRs.
Rats (n = 32) were euthanized on days 0, 1, 2, and 4, corresponding to 21, 22, 23, and 25 days on unlabeled water, respectively, after 42 days of feeding with 2H water (Fig. 5). The mean body weights of the four subgroups of rats were not different at euthanization [1-way ANOVA, P > 0.05, grand mean all groups: 520 ± 5 g (means ± SE)]. The serum water 2H enrichments were low and equal on the four days (1-way ANOVA, P > 0.05, grand mean all groups: 0.104 ± 0.014%).
Fig. 5.
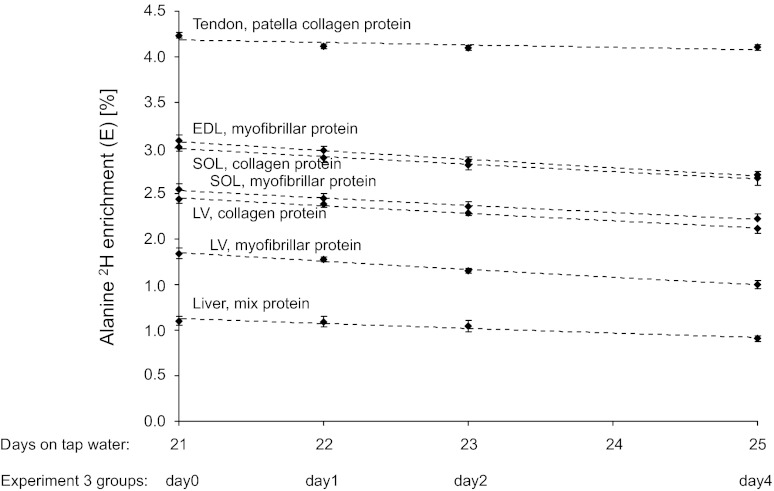
Enrichment of 2H alanine in slow turnover tissue proteins. Points are the means ± SE of alanine 2H enrichments for 8 rats euthanized on each day. Dashed lines represent the exponential decay fitted to the 4 mean enrichments for each protein. The x-axis is the no. of days on plain tap water after 42 days on deuterated water. The experimental group abbreviations represent days of euthanization for FBR determination (days 0, 1, 2, and 4).
The 2H enrichments of alanine in various tissue proteins are illustrated in Fig. 5. The monoexponential disappearance rates of 2H alanine, depicted by the dotted line in Fig. 5, were fitted to the measured enrichments at the four time points for each protein to calculate the FBR (Table 3). The practical use of this approach allows FBR to be determined only from two enrichment measurements. Although more time points would improve accuracy, it would also require more protein collections and more animals or biopsies. To determine the minimum time needed to detect a significant loss of tracer between two time points, we applied one-way ANOVA on the alanine 2H enrichments in the individual tissue proteins. A significant loss of protein-bound 2H alanine was found after 2 days in heart left ventricle collagen protein, after 3 days in heart left ventricle myofibrillar protein, EDL myofibrillar protein, soleus muscle myofibrillar and collagen proteins, and liver nonsoluble mixed proteins, and after 4 days in patella tendon collagen proteins. Thus, 2–4 days are needed between tissue collections to determine the FBR of slow turnover proteins in rats.
Table 3.
Protein FBRs in rats
| Tissue/Protein | FBR, %/day |
|---|---|
| Skeletal muscle | |
| Myofibrillar, soleus | 3.35 ± 0.79 |
| Myofibrillar, EDL | 3.27 ± 0.66 |
| Collagen, soleus | 2.99 ± 0.66 |
| Heart muscle, left ventricle | |
| Myofibrillar | 5.30 ± 0.88 |
| Collagen | 3.64 ± 0.61 |
| Tendon, collagen, patella | 0.66 ± 0.28 |
| Liver, nonsoluble mix | 5.00 ± 1.50 |
Values are means ± SE. FBR, fractional breakdown rate; EDL, extensor digitorum longus. The FBR values are the slopes of the linear relation between the log-transformed 2H alanine enrichments determined on days 0, 1, 2, and 4. The soleus muscle in rats consists of 80% myosin heavy chain (MHC) I, and the remaining 20% is MHC IIa, whereas EDL consists of 10% MHC IIa and 90% MHC IIb/x fiber type II. The mixed liver proteins were the nonsoluble proteins pelleted by centrifugation after homogenization.
Experiment 4: Human Experiment
Human free and protein-bound alanine enrichments.
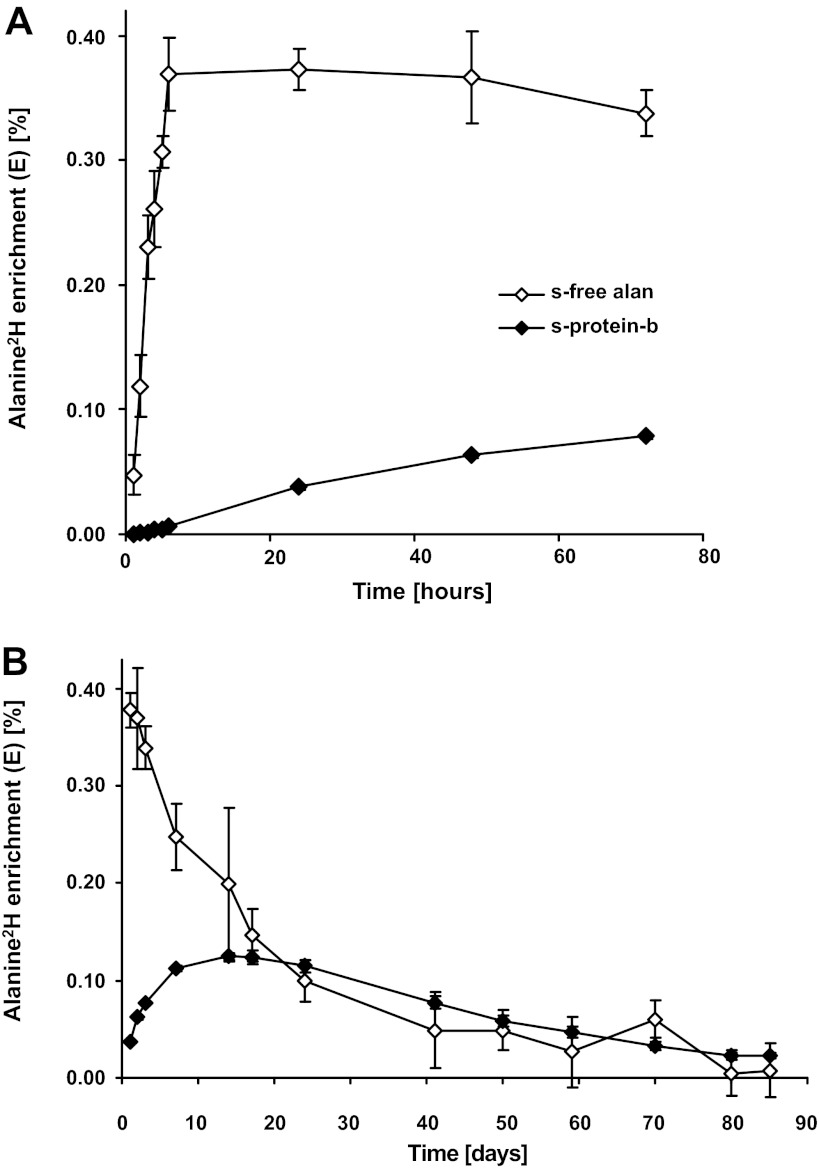
All subjects experienced a mild vertigo starting ∼30 min after intake of the first bolus of deuterated water and continuing until 2–3 h after the last bolus. Otherwise, no other side effects were experienced. One hour after drinking the first bolus of deuterated water, a marked elevation in deuterated alanine was present in the serum, and it continued to rise (n = 4; Fig. 6A). One hour after intake of the last 2H2O bolus, the alanine 2H enrichment reached a plateau and remained unchanged through day 2 (Fig. 6A). The alanine 2H enrichment in serum began to decline by day 3. The alanine 2H enrichment rose in serum protein exponentially at a slower rate over the 3-day measurement period (Fig. 6A). During the subsequent washout period, the serum-free alanine 2H enrichment declined exponentially (Fig. 6B) and returned to background levels after ∼80 days. As expected, alanine 2H enrichments in serum protein rose slower and to a lesser peak enrichment than in the serum-free pool (Fig. 6B).
Fig. 6.
Deuterium enrichments in alanine in human serum. ◇, Free alanine 2H enrichments in serum; ◆, alanine 2H enrichments in serum proteins. Data points are means ± SE, although the SE values may be smaller than the symbols. A: initial labeling period, days 0–3 after administration of deuterated water (n = 4). B: time course out to 85 days after the initial intake of deuterated water (n = 7).
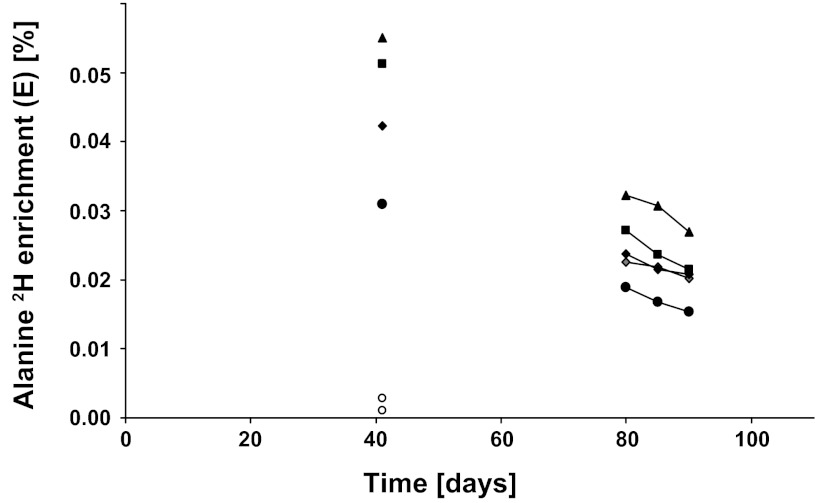
At 42 days after the initial administration of the deuterated water, we obtained vastus lateralis muscle biopsies in four subjects and skin biopsies in two subjects. We found that the alanine 2H enrichment in myofibrillar protein was 0.0449 ± 0.0054% (Fig. 7), which was similar to the enrichment in serum-free alanine (0.0486 ± 0.0389%; Fig. 6B). At the same time, the skin-derived collagen proteins had an enrichment of 0.0019 ± 0.0009% (Fig. 7).
Fig. 7.
Deuterium enrichments in alanine in human myofibrillar and skin collagen proteins. Values are alanine 2H enrichments for individual subjects in myofibrillar proteins (▲, ■, ◆, ●, and gray diamonds; n = 4 on day 42 and n = 5 on days 80, 85, and 90) and in skin collagen protein (○; n = 2). By day 80, alanine 2H enrichment was not detectable in skin collagen, and no collagen points are shown.
On days 80, 85, and 90, we obtained muscle vastus lateralis biopsies from five subjects and skin biopsies from two subjects. The alanine 2H enrichment declined over time in the myofibrillar protein (Fig. 7). However, the alanine 2H enrichment in skin collagen protein had diminished to background levels (data not shown) by day 80. Thus, there was significant 2H enrichment remaining in the myofibrillar proteins to determine FBR but insignificant 2H enrichment in either serum protein or skin collagen protein alanine at this point in time. We fit the alanine 2H enrichment in the myofibrillar proteins to a monoexponential decay and calculated the FBRs of the myofibrillar proteins over 5-day periods: 80–85 days, 1.68 ± 0.348%/day; 85–90 days: 1.76 ± 0.261%/day; and over the 10-day period (80–90 days): 1.73 ± 0.185%/day.
DISCUSSION
This study establishes an approach allowing the in vivo determination of protein FBR of slow turnover proteins. We used a sequential experimental setup to design the final approach: 1) incorporation of stable isotopically labeled amino acids into proteins using deuterium oxide as label donor, 2) withdrawal of the labeled amino acids from the precursor pool, and 3) tissue/protein collection at two time points to determine the disappearance rate of protein-bound labeled amino acids. Compared with the classic tracer dilution principle approaches, which are the most often used alternatives to determine protein breakdown, this method is distinct because the FBR determination is based on the determination of tracer abundance in individual proteins of interest. Hence, the breakdown rate can be measured on specific proteins, protein fractions, or mixtures and is analogous to the direct incorporation model by which the fractional synthesis rate is measured.
Deuterium Labeling of Amino Acids
In our initial studies, we determined the number of labeling sites on various amino acids with the purpose to select amino acids exchanging the highest number of deuteriums, giving the highest analytical sensitivity. Previously, Previs et al. (38) and Rachdaoui et al. (39) determined in both humans and rodents, respectively, that alanine exchanges essentially four carbon-bound hydrogens in vivo. We also note almost complete exchange (3.97 ± 0.13) of these four hydrogens in alanine. Furthermore, the exchange of deuterium between water and alanine via the transamination reaction is very fast both during administration of 2H water (Fig. 3) and when rats were returned to unlabeled drinking water (Fig. 4), which is in accord with previous reports (5, 13).
An important point for measuring the rate of fractional protein breakdown by our method compared with measuring fractional synthetic rates of proteins is that the FBR measurement does not depend upon knowing the deuterium labeling of a precursor species. The FBR measurement requires only measurement of the rate of loss of deuterated amino acid enrichment from a protein. Alanine was chosen for use here because 1) alanine is relatively abundant in proteins, 2) alanine is readily measured by both GC-MS and GC-P-IRMS, 3) alanine hydrogens exchange rapidly with water hydrogens, resulting in quick deuterium equilibration with water, and 4) alanine has the highest number of exchangeable hydrogens of the amino acids measured and, therefore, provides the greatest analytical sensitivity.
We expected that one hydrogen would exchange for the essential amino acids that undergo transamination in vivo, i.e., leucine, isoleucine (not measured), valine, and phenylalanine. The exchange for leucine and valine was ∼0.9 and ∼0.7 hydrogens, respectively (Table 1) but only ∼0.4 hydrogens for phenylalanine. This pattern of exchange in our rats is similar but higher than the pattern of exchange reported by Rachdaoui et al. (39) in mice.
As would be expected, glycine and serine each exchanged about two hydrogens. These results are consistent with what Rachdaoui et al. (39) found in mouse liver for glycine, but they found less than one hydrogen exchanged for serine. We did not measure glutamate or glutamine in protein because hydrolysis combines the two. But Rachdaoui et al. (39) measured free glutamate and glutamine in mouse liver and found the exchange of hydrogen to be ∼3.5 and 3.0, respectively. Although proline is synthesized from glutamate, we found that the number of hydrogens exchanged in proline to be only 1.2. Rachdaoui et al. (39) found 2.2 protons exchanged in mouse liver-free proline, still lower than expected if it was synthesized solely from glutamate, suggesting that de novo proline synthesis from glutamate is a minor component of proline turnover via protein synthesis and breakdown. All in all, our conclusion from the results reported here and those reported previously (39) is that alanine is the only amino acid to exchange consistently the majority of its exposed hydrogens. The reason that this result is possible is that reversible transamination is a critical part of alanine metabolism, providing high throughput in both directions, which is in agreement with previous studies in humans using different labeled alanines (48).
Water Enrichment Determines the Tracer Enrichment
A crucial assumption in the model is that the amount of tracer amino acid in the free pool is negligible during the period of the breakdown measurement and, therefore, cannot return tracer to the protein. For this reason, we completed experiment 2 to determine when the 2H enrichment in body water and intracellular free alanine [measured from serum protein (albumin)-bound alanine] had diminished to inconsequential levels. At the same time, we wanted to keep the maximum amount of 2H labeling in alanine in the proteins for which we were trying to determine FBR to obtain the best analytical sensitivity. The optimal point in time to measure the protein FBR was thus a compromise. In the present rat experiments, we defined that time to be about 21 days after removal of the 2H water intake. At this point, both body water and free alanine 2H enrichments had decreased to negligible levels (≈0.1%) compared with the 2H enrichments of the slow turnover proteins of 1–4% (Fig. 5). This scheme works only for slow turnover proteins. Proteins that turnover quickly will lose 2H labeling, similar to what is seen for serum proteins (Fig. 4), and cannot have their FBR measured by this method.
An important advantage of using 2H water to label alanine is that labeling occurs quickly and that when 2H water is removed, the alanine labeling quickly disappears through exchange with unlabeled water. Thus, recycling and reincorporation of alanine 2H labeling during measurement of FBR will be negligible once the water labeling is relatively absent.
Comparing the FBRs With Existing FSRs
To validate the measured FBR values in the rats, we compared them with existing FSR values in comparable animals. For example, the FSR of fast fiber type muscles in adult Lister rats of similar size as our rats was 3.5 ± 0.4%/day, using a continuous infusion of radiolabeled tyrosine (3), which was comparable with our FBR on EDL muscle (Table 3). Furthermore, for mixed muscle protein in meal-fed rats, the FSR is reported to be in the range of 4.6–8.9%/day (3, 15, 22). Using deuterated water as label donor to amino acids but still measuring mixed muscle protein synthesis over short (4- to 6-h) intervals, Gasier et al. (16) and Yuan et al. (50) reported conflicting values ranging from 8 to 19%/day. Thus, even in the literature, the muscle FSR values of comparable animals seem to span a range where our FBR values fit in the lower end. This may be related to inherent differences between methods and the fact that prolonged measurements of FSR seem to reduce the values (16). Muscle-derived collagen turnover rates in smaller rodents are more sparsely investigated than for contractile proteins. Laurent (28) as well as McAnulty and Laurent (29) have reported synthesis rates for mature collagen from rabbits and rats, respectively, in heart and skeletal muscle (5.2 ± 0.7 and 2.2 ± 0.3%/day, respectively) that are similar to our breakdown rates (Table 3).
By the use of the flooding dose tracer method, heart myofibrillar proteins have been shown to have a slightly higher protein synthesis rate (17.5 ± 0.6%/day) compared with skeletal muscle myofibrillar proteins (plantaris myofibrillar of 14.3 ± 1.1%/day) (37). We verified this difference between heart and skeletal muscle protein turnover rates, albeit with markedly lower values (Table 3). However, note that the reported skeletal muscle myofibrillar FSR is markedly higher in that reference than in some of the previous studies discussed above, emphasizing that it may be hard to compare the numerical values from study to study. Similarly, from liver we isolated a nonsoluble structural protein fraction containing slow turnover proteins and measured a much lower breakdown rate than reported for synthesis of mixed hepatic proteins, ranging from 23%/day in young, growing Holtzman albino rats (22) to 20–50%/day in male Wistar rats (15) and ≤90%/day in other rat strains (8, 30). Such differences between the turnover rates between studies are probably due to the abundance of different proteins in the tissue homogenates used. The FSR measurement relies on measuring incorporation of tracer into proteins and will include the weighted average of all the different proteins in that pool. If the pool of proteins includes some fast turnover proteins, then the FSR measured will be higher than if specific proteins of slower rate were measured alone, which the results from Gasier et al. (16) support. However, in the FBR measurement, faster turnover proteins that lose their alanine 2H label by the beginning of the sample collection window will not contribute to disappearance of [2H]alanine during the sample collection window and will not contribute to the FBR rate. Thus, the FBR measurement using 2H2O will measure slow turnover protein rates even with some contamination of faster turnover proteins in the sample. This divergence between FSR and FBR measurements can be avoided only by isolating single proteins and measuring their turnover rate, similar to work by Jaleel and colleagues (24, 25).
Human Data
We administered an average of 233 ml of deuterated water per adult subject to add about 0.5% deuterium to the total body water pool. The maximal measured free alanine 2H enrichment was 0.37 ± 0.02% 1 day after the administration (Fig. 6), corresponding to ∼70–75% of the administered 2H dose remaining in the free pool. A similar dilution of the administered water enrichment was seen in the rat experiments, where 8% deuterium oxide in the drinking water elevated body water enrichment to 5.2%, i.e., ∼70–75% labeling. This dilution is due to metabolic processes that constantly remove hydrogen from water and release hydrogen into body water from compounds not yet labeled (4).
In humans, free alanine became labeled with 2H, quickly approaching plateau enrichment shortly after intake of deuterated water began (Fig. 6A). During the subsequent 80-day washout period, no restrictions were given to the subjects, who varied in daily physical activity level. Three performed endurance-like training three to five times a week, one did resistance training, and the remaining subjects were not involved in organized exercise. These differences presumably resulted in some interindividual temporal variation in the turnover rate of body water (42) and hence, in free alanine 2H enrichment. However, there were measurable amounts of 2H remaining in the alanine in muscle contractile proteins at 80 days. We cannot translate these results to all populations, especially those that may be critically ill, very old, or especially sedentary. For these groups, pilot measurements should be performed to determine the adequate amount of deuterated water necessary to label the proteins of interest as well as the time adequate to allow the free label in alanine and/or water to disappear.
After ∼40 days, we found the alanine 2H enrichment in plasma and in myofibrillar protein to be about 0.045%. Whereas the 2H enrichment in the free alanine pool was descending, the appearance of labeled alanine in the slow turnover myofibrillar proteins had accumulated up to this point in time. Hereafter, a net loss of label from these proteins starts, although some reincorporation will take place as long as the precursor contains label. However, only a very low amount of 2H had accumulated in skin-derived collagen protein-bound alanine (0.002% at 42 days). This low labeling limited our ability to detect 2H and, therefore, measure FBR in these very slow turnover proteins. For these proteins, a longer labeling period with 2H water with larger, repeated doses of deuterated water is required.
The FBR of vastus lateralis myofibrillar protein determined as an average over 10 days was 0.072 ± 0.008%/h. This value corresponds to the rate of myofibrillar protein synthesis that our group reported earlier using the direct incorporation precursor-product approach (12, 19, 31) and is similar in magnitude to the 8-wk synthesis rates reported by Robinson et al. (40) in humans.
Practicalities
Our approach described here is suitable only for proteins that have a markedly slower turnover rate than body water, because the 2H label has to be removed from and equilibrate with the free pool of tracer amino acid to prevent reincorporation into protein at the time when FBR is measured. Furthermore, the loading period of 2H alanine incorporation into proteins should be tailored to suit both the species and the turnover rates of the proteins to be measured. Because we are trying to measure the FBR while the protein pool is constant in mass, we want to minimize the sample collection window, that is, the time between the two biopsies obtained to determine protein alanine 2H enrichment needed to calculate FBR. On the other hand, a longer sample collection window is required to allow the detection of the loss of tracer enrichment in the very slow turnover proteins. Thus, the length of the sample window can be optimized to fit the turnover rate of the protein of interest. If done appropriately, the protein mass will undergo minimal change because the change in the mass of a protein is dependent on the protein's turnover rate. Therefore, planning experiments require first estimating the turnover rates of the proteins measured. From this information and the sensitivity of the mass spectrometry method that is used for sample analysis, the loading period to enrich the protein to be measured, the time for the intrinsic enrichment of the free (precursor) 2H alanine to diminish after stopping administration of the 2H2O, and the sample collection window can all be calculated.
GRANTS
This work was supported by H:S (Region Hovedstaden); a foreign exchange stipend from the Faculty of Health Sciences, University of Copenhagen; Anti Doping Denmark; the Danish Council for Independent Research (09-073587); and National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK-038429.
DISCLOSURES
The authors declare no conflicts of interest, and there are no financial interests to disclose.
AUTHOR CONTRIBUTIONS
L.H., N.-H.H.-R., and D.E.M. contributed to the conception and design of the research; L.H., M.J.T., and R.B. performed the experiments; L.H., B.O., and D.E. analyzed the data; L.H., N.-H.H.-R., and D.E.M. interpreted the results of the experiments; L.H. and D.E.M. prepared the figures; L.H. and M.K. drafted the manuscript; L.H., B.O., D.E., M.J.T., R.B., N.-H.H.-R., M.K., and D.E.M. approved the final version of the manuscript; M.J.T., R.B., N.-H.H.-R., and D.E.M. edited and revised the manuscript.
ACKNOWLEDGMENTS
We give a special thank to Dr. Ruth Blauwiekel and the staff at the animal facility at the University of Vermont for help and assistance with the rat housing and treatment procedures. We thank Jennifer Dykhuizen for assisting with the rat care and for serum sample preparation. Additionally, we thank Dr. Stephen Bell at the Department of Medicine and Cardiology Unit, University of Vermont, for teaching animal dissection procedures and engineer Flemming Jessen, Clinical Metabolomics Core Facility, Rigshospitalet, Copenhagen, Denmark, for help with measurement of deuterium enrichment by mass spectrometry.
REFERENCES
- 1. Alemany M. Effect of amino acid reutilization in the determination of protein turnover in mice. Horm Metab Res 8: 70–73, 1976 [DOI] [PubMed] [Google Scholar]
- 2. Barrett EJ, Revkin JH, Young LH, Zaret BL, Jacob R, Gelfand RA. An isotopic method for measurement of muscle protein synthesis and degradation in vivo. Biochem J 245: 223–228, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bates PC, Millward DJ. Myofibrillar protein turnover. Synthesis rates of myofibrillar and sarcoplasmic protein fractions in different muscles and the changes observed during postnatal development and in response to feeding and starvation. Biochem J 214: 587–592, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bederman IR, Dufner DA, Alexander JC, Previs SF. Novel application of the “doubly labeled” water method: measuring CO2 production and the tissue-specific dynamics of lipid and protein in vivo. Am J Physiol Endocrinol Metab 290: E1048–E1056, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Belloto E, Diraison F, Basset A, Allain G, Abdallah P, Beylot M. Determination of protein replacement rates by deuterated water: validation of underlying assumptions. Am J Physiol Endocrinol Metab 292: E1340–E1347, 2007 [DOI] [PubMed] [Google Scholar]
- 6. Biolo G, Chinkes D, Zhang XJ, Wolfe RR. Harry M. Vars Research Award. A new model to determine in vivo the relationship between amino acid transmembrane transport and protein kinetics in muscle. JPEN J Parenter Enteral Nutr 16: 305–315, 1992 [DOI] [PubMed] [Google Scholar]
- 7. Busch R, Kim YK, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim Biophys Acta 1760: 730–744, 2006 [DOI] [PubMed] [Google Scholar]
- 8. Chiku K, Mochida H, Yamamoto M, Natori Y. Amino acids suppress intracellular protein degradation in rat liver during parenteral nutrition. J Nutr 123: 1771–1776, 1993 [DOI] [PubMed] [Google Scholar]
- 9. Chumlea WC, Guo SS, Zeller CM, Reo NV, Baumgartner RN, Garry PJ, Wang J, Pierson RN, Jr, Heymsfield SB, Siervogel RM. Total body water reference values and prediction equations for adults. Kidney Int 59: 2250–2258, 2001 [DOI] [PubMed] [Google Scholar]
- 10. Collins ML, Eng S, Hoh R, Hellerstein MK. Measurement of mitochondrial DNA synthesis in vivo using a stable isotope-mass spectrometric technique. J Appl Physiol 94: 2203–2211, 2003 [DOI] [PubMed] [Google Scholar]
- 11. Corr LT, Berstan R, Evershed RP. Optimisation of derivatisation procedures for the determination of delta13C values of amino acids by gas chromatography/combustion/isotope ratio mass spectrometry. Rapid Commun Mass Spectrom 21: 3759–3771, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Doessing S, Heinemeier KM, Holm L, Mackey AL, Schjerling P, Rennie M, Smith K, Reitelseder S, Kappelgaard AM, Rasmussen MH, Flyvbjerg A, Kjaer M. Growth hormone stimulates the collagen synthesis in human tendon and skeletal muscle without affecting myofibrillar protein synthesis. J Physiol 588: 341–351, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dufner DA, Bederman IR, Brunengraber DZ, Rachdaoui N, Ismail-Beigi F, Siegfried BA, Kimball SR, Previs SF. Using 2H2O to study the influence of feeding on protein synthesis: effect of isotope equilibration in vivo vs. in cell culture. Am J Physiol Endocrinol Metab 288: E1277–E1283, 2005 [DOI] [PubMed] [Google Scholar]
- 14. Epstein I, Grossowicz N. Intracellular protein breakdown in a thermophile. J Bacteriol 99: 418–421, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garlick PJ, Millward DJ, James WP. The diurnal response of muscle and liver protein synthesis in vivo in meal-fed rats. Biochem J 136: 935–945, 1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gasier HG, Riechman SE, Wiggs MP, Previs SF, Fluckey JD. A comparison of 2H2O and phenylalanine flooding dose to investigate muscle protein synthesis with acute exercise in rats. Am J Physiol Endocrinol Metab 297: E252–E259, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gore DC, Wolfe RR, Chinkes DL. Quantification of amino acid transport through interstitial fluid: assessment of four-compartment modeling for muscle protein kinetics. Am J Physiol Endocrinol Metab 292: E319–E323, 2007 [DOI] [PubMed] [Google Scholar]
- 18. Holm L, Kjaer M. Measuring protein breakdown rate in individual proteins in vivo. Curr Opin Clin Nutr Metab Care 13: 526–531, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm L, van Hall G, Rose AJ, Miller BF, Doessing S, Richter EA, Kjaer M. Contraction intensity and feeding affect collagen and myofibrillar protein synthesis rates differently in human skeletal muscle. Am J Physiol Endocrinol Metab 298: E257–E269, 2010 [DOI] [PubMed] [Google Scholar]
- 20. Humphrey TJ, Davies DD. A new method for the measurement of protein turnover. Biochem J 148: 119–127, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ilchenko SA, Rachdaoui N, Li L, Wiilard B, Previs SF, Kasumov T. Application of a high-resolution mass spectrometry for estimation of proteome dynamics (Abstract). FASEB J. 24 Experimental Biology Meeting Abstract, Anaheim, CA, 2010, p. 670.4 [Google Scholar]
- 22. Ip C, Harper AE. Protein synthesis in liver, muscle, and brain of rats fed a high tyrosine-low protein diet. J Nutr 105: 885–893, 1975 [DOI] [PubMed] [Google Scholar]
- 23. Jackson SH, Heininger JA. A study of collagen reutilization using an 18O2 labeling technique. Clin Chim Acta 51: 163–171, 1974 [DOI] [PubMed] [Google Scholar]
- 24. Jaleel A, Nehra V, Persson XM, Boirie Y, Bigelow M, Nair KS. In vivo measurement of synthesis rate of multiple plasma proteins in humans. Am J Physiol Endocrinol Metab 291: E190–E197, 2006 [DOI] [PubMed] [Google Scholar]
- 25. Jaleel A, Short KR, Asmann YW, Klaus KA, Morse DM, Ford GC, Nair KS. In vivo measurement of synthesis rate of individual skeletal muscle mitochondrial proteins. Am J Physiol Endocrinol Metab 295: E1255–E1268, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev 84: 649–698, 2004 [DOI] [PubMed] [Google Scholar]
- 27. Koch AL, Levy HR. Protein turnover in growing cultures of Escherichia coli. J Biol Chem 217: 947–957, 1955 [PubMed] [Google Scholar]
- 28. Laurent GJ. Rates of collagen synthesis in lung, skin and muscle obtained in vivo by a simplified method using [3H]proline. Biochem J 206: 535–544, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McAnulty RJ, Laurent GJ. Collagen synthesis and degradation in vivo. Evidence for rapid rates of collagen turnover with extensive degradation of newly synthesized collagen in tissues of the adult rat. Coll Relat Res 7: 93–104, 1987 [DOI] [PubMed] [Google Scholar]
- 30. McNurlan MA, Tomkins AM, Garlick PJ. The effect of starvation on the rate of protein synthesis in rat liver and small intestine. Biochem J 178: 373–379, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mikkelsen UR, Schjerling P, Helmark IC, Reitelseder S, Holm L, Skovgaard D, Langberg H, Kjaer M, Heinemeier KM. Local NSAID infusion does not affect protein synthesis and gene expression in human muscle after eccentric exercise. Scand J Med Sci Sports 21: 630–644, 2011 [DOI] [PubMed] [Google Scholar]
- 32. Molnar JA, Alpert N, Burke JF, Young VR. Synthesis and degradation rates of collagens in vivo in whole skin of rats, studied with 1802 labelling. Biochem J 240: 431–435, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore DR, Tang JE, Burd NA, Rerecich T, Tarnopolsky MA, Phillips SM. Differential stimulation of myofibrillar and sarcoplasmic protein synthesis with protein ingestion at rest and after resistance exercise. J Physiol 587: 897–904, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neville HE, Neville MC, Harrold S, Farrell R. Measurement of steady state protein degradation in cultured human muscle cells. Anal Biochem 134: 424–438, 1983 [DOI] [PubMed] [Google Scholar]
- 35. Oshima T, Tamiya N. Mechanism of transaminase action. Biochem J 78: 116–119, 1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pine MJ. Metabolic control of intracellular proteolysis in growing and resting cells of Escherichia coli. J Bacteriol 92: 847–850, 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Preedy VR, Garlick PJ. Inhibition of protein synthesis by glucagon in different rat muscles and protein fractions in vivo and in the perfused rat hemicorpus. Biochem J 251: 727–732, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Previs SF, Fatica R, Chandramouli V, Alexander JC, Brunengraber H, Landau BR. Quantifying rates of protein synthesis in humans by use of 2H2O: application to patients with end-stage renal disease. Am J Physiol Endocrinol Metab 286: E665–E672, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Rachdaoui N, Austin L, Kramer E, Previs MJ, Anderson VE, Kasumov T, Previs SF. Measuring proteome dynamics in vivo: as easy as adding water? Mol Cell Proteomics 8: 2653–2663, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson MM, Turner SM, Hellerstein MK, Hamilton KL, Miller BF. Long-term synthesis rates of skeletal muscle DNA and protein are higher during aerobic training in older humans than in sedentary young subjects but are not altered by protein supplementation. FASEB J 25: 3240–3249, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rubinstein N, Chi J, Holtzer H. Coordinated synthesis and degradation of actin and myosin in a variety of myogenic and non-myogenic cells. Exp Cell Res 97: 387–393, 1976 [DOI] [PubMed] [Google Scholar]
- 42. Shimamoto H, Komiya S. Comparison of body water turnover in endurance runners and age-matched sedentary men. J Physiol Anthropol Appl Human Sci 22: 311–315, 2003 [DOI] [PubMed] [Google Scholar]
- 43. Strawford A, Antelo F, Christiansen M, Hellerstein MK. Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am J Physiol Endocrinol Metab 286: E577–E588, 2004 [DOI] [PubMed] [Google Scholar]
- 44. Walter U, Luthe H, Soling HD. Hydrogen exchange at the b-carbon of amino acids during transamination. Eur J Biochem 59: 395–403, 1975 [DOI] [PubMed] [Google Scholar]
- 45. Waterlow JC, Stephen JM. The effect of low protein diets on the turn-over rates of serums, liver and muscle proteins in the rat, measured by continuous infusion of l-[14C]lysine. Clin Sci 35: 287–305, 1968 [PubMed] [Google Scholar]
- 46. Willetts NS. Intracellular protein breakdown in non-growing cells of Escherichia coli. Biochem J 103: 453–461, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolfe RR, Chinkes DL. Measurement of the synthesis of specific proteins. In: Isotope Tracers in Metabolic Research: Principles and Practice of Kinetic Analysis, edited by Wolfe RR, Chinkes DL. Hoboken, NJ: Wiley-Liss, 2005, p. 325–360 [Google Scholar]
- 48. Yang RD, Matthews DE, Bier DM, Lo C, Young VR. Alanine kinetics in humans: influence of different isotopic tracers. Am J Physiol Endocrinol Metab 247: E634–E638, 1984 [DOI] [PubMed] [Google Scholar]
- 49. Young RB, Dombroske OC. Metabolism of myosin heavy chain in steady-state chick skeletal muscle cultures. Biochem J 194: 241–247, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan CL, Sharma N, Gilge DA, Stanley WC, Li Y, Hatzoglou M, Previs SF. Preserved protein synthesis in the heart in response to acute fasting and chronic food restriction despite reductions in liver and skeletal muscle. Am J Physiol Endocrinol Metab 295: E216–E222, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang XJ, Chinkes DL, Herndon DN, Wolfe RR. Measurement of protein fractional synthesis and breakdown rates in the skin of rabbits using a subflooding dose method. Metabolism 58: 1239–1247, 2009 [DOI] [PubMed] [Google Scholar]
- 52. Zhang XJ, Chinkes DL, Sakurai Y, Wolfe RR. An isotopic method for measurement of muscle protein fractional breakdown rate in vivo. Am J Physiol Endocrinol Metab 270: E759–E767, 1996 [DOI] [PubMed] [Google Scholar]
- 53. Zhang XJ, Chinkes DL, Wolfe RR. Measurement of muscle protein fractional synthesis and breakdown rates from a pulse tracer injection. Am J Physiol Endocrinol Metab 283: E753–E764, 2002 [DOI] [PubMed] [Google Scholar]