Abstract
To determine placental microRNA (miRNA) expression at different gestational age, total RNA from six first and six third trimester placentas was isolated. miRNA expression was analyzed by Affymetrix miRNA microarray, and miRNA clusters were identified by web-based programs MirClust and miRGen Cluster. qRT-PCR was carried out to validate miRNA expression, and in situ hybridization (ISH) was performed to determine compartmental localization of miRNAs within villous tissue. A total of 208 miRNA transcripts, which represent 191 mature miRNAs, were found differently expressed between first and third trimester placentas. miRNAs within the miR-17-92 cluster, C14MC, miR-371 cluster, and C19MC were significantly upregulated in the first trimester placentas. In contrast, miRNAs of the let-7 family, miR-34 family, miR-29a cluster, miR-195 cluster, and miR-181c cluster were significantly upregulated in the third trimester placentas. Increased miR-371–5p, miR-17-3p, and miR-708–5p expression and decreased miR-125b-5p and miR-139–5p expression in the first trimester placentas were confirmed by qRT-PCR. Different expression pattern for miR-371-5p and miR-125b-5p within villous tissue was demonstrated by ISH. Distinct miRNA cluster expression profiles between the first and third trimester placentas were identified. miRNAs that regulate innate/adaptive immune responses are strongly expressed in both first and third trimester placentas. miRNAs that exert oncogenic, angiogenic, and antiapoptotic properties are dominantly expressed in the first trimester placentas, whereas miRNAs that promote cell differentiation and function as tumor suppressors are strongly expressed in the third trimester placentas. These results indicate that miRNAs play critical roles in placental development.
Keywords: miRNA, placenta, pregnancy, development
micrornas (mirnas) have emerged as important posttranscriptional regulators of gene expression. miRNAs induce translational repression, target degradation, and gene silencing by binding to complementary sequences on target messenger RNA (mRNA) transcripts. It has been estimated that ∼30–60% of mRNAs are regulated by miRNAs, which virtually control almost every aspect of cellular events from stem cell differentiation (4, 14) and organ development and formation (2, 6), and aging (15) to physiological/pathophysiological processes in cancer cell growth and metastasis (19, 39), genetic and cardiovascular diseases (13, 20, 38), and metabolic disorders (34, 36).
The placenta is the first organ formed during pregnancy. It connects the developing fetus to the mother's uterine wall. Normal placental development is critical for a healthy pregnancy outcome to both the mother and the fetus. Although the cellular and molecular mechanisms in the regulation of placental development are largely unknown, recent studies suggest that deregulation of miRNA expression in placental trophoblasts may contribute to pregnancy complications that are associated with placental insufficiency including preeclampsia and intrauterine growth restriction (24, 33). The placental-specific miRNAs, chromosome 14 miRNA cluster (C14MC) and chromosome 19 miRNA cluster (C19MC), have also been described (29). Particularly, the C14MC miRNAs are imprinted from maternal chromosome, and C19MC miRNAs are imprinted from paternal chromosome. Both C14MC and C19MC miRNAs are believed to play important roles in the regulation of cellular differentiation and immunomodulation during pregnancy. The detection of trophoblast-derived miRNAs in the maternal circulation (11, 22) further suggests their potential role as valuable biomarkers of placental function and the likelihood of molecular regulators acting on maternal systemic vasculature during pregnancy.
Recently, Morales-Prieto et al. (27) reported different miRNA expression profiles between primary isolated placental trophoblasts in culture and immortalized trophoblast HTR-8/SVneo cells and JEG-3 choriocarcinoma cells, suggesting the involvement of miRNAs in trophoblast behavior and characteristics. In the present study, miRNA expression profile was determined in villous tissue from first and third trimester placentas. Whole villous tissue contains cytotrophoblasts, syncytiotrophoblasts, mesenchymal/stromal cells, and villous core fetal vessel endothelium, etc. A miRNA expression profile in whole villous tissue would closely represent miRNA expression without disruption of tissue integrity in an in vivo situation. We also determined expression and localization of miR-371-5p and miR-125b-5p by in situ hybridization in the first and third trimester placental villous tissue sections. Distinct miRNA expression profiles, miRNA clusters, and specific miRNA localization were found between the first and third trimester placentas.
MATERIALS AND METHODS
Placenta tissue collection.
Twelve placentas were used in this study, six from third trimester and six from first trimester pregnancies. Third trimester/term placentas from normotensive pregnant women (30–38 wk) were collected from the main hospital of Louisiana State University Health Sciences Center, Shreveport (LSUHSC-Sh), LA. First trimester placental tissues (6–8 wk) were collected from selective pregnancy termination at the Department of Obstetrics and Gynecology, the First Hospital of Harbin Medical University, China. Tissue collection was approved by both institutions. Study subjects had normal maternal blood pressure < 140/90 mmHg without obstetrical and medical complications. None of the patients had signs of infection, nor were they smokers. Freshly obtained placental villous tissues excluding chorion, amnion, and decidua were immediately processed and either preserved in RNA later/snap-frozen for RNA isolation or fixed in 10% formalin and then embedded in paraffin.
Total RNA isolation.
Total RNA was isolated from placental villous tissues with TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction. RNA quality and quantity were assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
miRNA microarray analysis.
Affymetrix GeneChip 2.0 miRNA Microarray (Santa Clara, CA) was used for miRNA expression profile analysis. Briefly, 1 μg of total RNA of each sample was subjected to a tailing reaction labeled with the Flashtag RNA labeling kit (Genisphere, Hatfield, PA) followed by ligation of the biotinylated signal molecule to the RNA sample according to the manufacturer's instructions. Each sample was then hybridized to a GeneChip 2.0 miRNA Array at 48°C for 16 h and then washed and stained on a Fluidics Station 450. After staining, the chip was scanned on a GeneChip Scanner 3000 7G. Expression levels of miRNA transcripts were captured through the probe set by Command Console 3.2 (Affymetrix). The expression values were summarized and normalized with the robust multichip analysis program Expression Console v. 1.2.0.20 (Affymetrix).
miRNA expression by quantitative real-time PCR.
Quantitative real-time PCR (qRT-PCR) was performed to confirm the upregulation and downregulation of miRNAs in the first and third trimester placental tissues. Five miRNAs were examined, including miR-17, miR-371, miR-708, miR-125b, and miR-139. miRNA primers hsa-miR-17-3p (PM12246), hsa-miR-371a-5p (PM12791), hsa-miR-708-5p (PM11161), hsa-miR-125b-5p (PM10148), and hsa-miR-139-5p (PM12466) were purchased from Invitrogen (Carlsbad, CA). Reverse transcription was performed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). TaqMan Universal Master Mix II (AB Applied Biosystems) was used for miRNA amplification. qRT-PCR was performed using an Applied Biosystems 7900HT (PerkinElmer, Foster City, CA). Expression of small nuclear RNA (snRNA) U6 was also determined for each samples and used as an endogenous control. The threshold cycle (5) value (CT) was defined as the fractional cycle number at which signal passes a fixed threshold. The relative amount of each target miRNA to U6 was calculated using the equation 2−ΔCT, where ΔCT = CT miRNA − CT U6.
miRNA expression by in situ hybridization.
miRNA expression for miR-371 and miR-125b was further determined by in situ hybridization (ISH) on formalin-fixed, paraffin-embedded placental tissue sections from first and third trimester placentas. miRCURY LNA 5′-DIG- and 3′-DIG-labeled detection probe specific to hsa-miR-371-5p (38554-15) and hsa-miR-125b-5p (18022-15) was used. 5′-DIG-labeled LNA U6 snRNA control probe (5′-DIG/cacgaatttgcgtgtcatcctt/-3′) was used as positive control, and 5′-DIG- and 3′-DIG-labeled LNA scrambled miRNA control probe (5′-DIG/gtgtaacacgtctatacgccca/DIG-3′) was used as negative control. All the probes and the miRCURY LNA Optimization Kit were purchased from Exiqon (Vedbaek, Denmark) and used according to the manufacturer's instructions. The sections were counterstained with Nuclear Fast Red (Vector Laboratories, Burlingame, CA). Stained slides were then reviewed under microscope, and images were captured by PictureFrame computer software (Uptronics, Sunnyvale, CA) and recorded to a microscope-linked PC computer.
Data analysis.
Statistical analysis of miRNA data was performed using the web-based computer software GeneSifter v. 4.0 (www.genesifter.com, VizX Labs, Seattle, WA). A two-tail t-test with Benjamini and Hochberg correction was used. All data were log transformed. From the set of 1,105 total miRNA transcripts on the GeneChip 2.0 miRNA Array, a set of miRNAs with more than twofold upregulation and more than twofold downregulation with a 95% significance (P < 0.05) was identified between the third trimester and first trimester placentas. miRNA clusters were identified by web-based programs MirClust (http://fgfr.ibms.sinica.edu.tw/MetaMirClust/MetaMirClustSearch.php) and miRGen Cluster (www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Cluster.cgi). For qRT-PCR data, the value of fold change was presented by 2−ΔCT ± SE. Statistical analysis was carried out by Mann-Whitney test. A probability level of < 0.05 was considered statistically significant.
RESULTS
miRNA expression profiles and clusters in first and third trimester placentas.
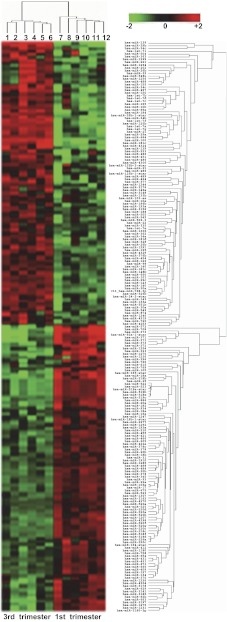
RNA samples from 12 placentas were analyzed by Affymetrix miRNA microarray, six from third trimester and six from first trimester pregnancies. Based on a twofold change cutoff with 95% significance (P < 0.05), a total of 208 miRNA transcripts were found differently expressed between the third and first trimester placentas, with 104 transcripts significantly upregulated and 104 transcripts significantly downregulated in the third trimester vs. first trimester placentas or vice versa. These transcripts represent a total of 191 mature miRNAs, since several miRNAs have more than one transcript in the miRNA array 2.0 platform. Table 1 is the list of 97 miRNAs from 104 transcripts that are upregulated in the first trimester compared with the third trimester placentas (Table 1). Table 2 is the list of 94 miRNAs from 104 transcripts that are significantly upregulated in the third trimester compared with the first trimester placentas (Table 2). Hierarchical clustering analysis of miRNAs expression between the first and third trimester placentas is shown in Fig. 1. Complete array data are accessible through GEO Series accession number GSE42915, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42915).
Table 1.
miRNAs upregulated in first trimester placentas compared with third trimester placentas (n = 97)
| Name | Fold Change | Name | Fold Change |
|---|---|---|---|
| Hsa-mir-708 | 22.84 | Hsa-mir-1292 | 2.83 |
| Hsa-mir-92a-1* | 16.38 | Hsa-mir-17 | 2.83 |
| Hsa-mir-371 | 14.95 | Hsa-mir-106a | 2.82 |
| Hsa-mir-372 | 9.35 | Hsa-mir-370 | 2.82 |
| Hsa-mir-373 | 7.50 | Hsa-mir-378 | 2.76 |
| Hsa-mir-25 | 7.20 | Hsa-mir-654 | 2.73 |
| Hsa-mir-296 | 7.10 | Hsa-mir-365* | 2.72 |
| Hsa-mir-518b | 6.77 | Hsa-mir-758 | 2.71 |
| Hsa-mir-1254 | 6.52 | Hsa-mir-193a | 2.67 |
| Hsa-mir-1226 | 6.47 | Hsa-mir-422a | 2.66 |
| Hsa-mir-18a | 6.46 | Hsa-mir-205 | 2.55 |
| Hsa-mir-519c | 5.10 | Hsa-mir-105 | 2.53 |
| Hsa-mir-886 | 5.10 | Hsa-mir-421 | 2.53 |
| Hsa-mir-519b | 5.08 | Hsa-mir-409 | 2.51 |
| Hsa-mir-519a* | 5.07 | Hsa-mir-378c | 2.49 |
| Hsa-mir-518e | 4.97 | Hsa-mir-665 | 2.48 |
| Hsa-mir-523 | 4.96 | Hsa-mir-760 | 2.48 |
| Hsa-mir-522 | 4.89 | Hsa-mir-4270 | 2.45 |
| Hsa-mir-934 | 4.70 | Hsa-mir-134 | 2.44 |
| Hsa-mir-1275 | 4.30 | Hsa-mir-19b-1* | 2.43 |
| Hsa-mir-1270 | 4.07 | Hsa-mir-2277 | 2.43 |
| Hsa-mir-182 | 3.98 | Hsa-mir-1180 | 2.42 |
| Hsa-mir-3180-3p | 3.93 | Hsa-mir-520a | 2.40 |
| Hsa-mir-20a | 3.87 | Hsa-mir-671 | 2.40 |
| Hsa-mir-520f | 3.68 | Hsa-mir-1285 | 2.38 |
| Hsa-mir-520c | 3.53 | Hsa-mir-455 | 2.36 |
| Hsa-mir-378 | 3.52 | Hsa-mir-520 h | 2.34 |
| Hsa-mir-675 | 3.49 | Hsa-mir-127 | 2.32 |
| Hsa-mir-323 | 3.47 | Hsa-mir-3162 | 2.32 |
| Hsa-mir-520 g | 3.42 | Hsa-mir-30b | 2.30 |
| Hsa-mir-4298 | 3.37 | Hsa-mir-663 | 2.28 |
| Hsa-mir-1231 | 3.36 | Hsa-mir-19a | 2.27 |
| Hsa-mir-466 | 3.36 | Hsa-mir-4304 | 2.27 |
| Hsa-mir-518f | 3.32 | Hsa-mir-498 | 2.27 |
| Hsa-mir-518c | 3.18 | Hsa-mir-412 | 2.24 |
| Hsa-mir-18b | 3.06 | Hsa-mir-431 | 2.23 |
| Hsa-mir-3197 | 3.03 | Hsa-mir-484 | 2.23 |
| Hsa-mir-93 | 2.99 | Hsa-mir-20b | 2.20 |
| Hsa-mir-3178 | 2.98 | Hsa-mir-31 | 2.18 |
| Hsa-mir-518d | 2.96 | Hsa-mir-520d | 2.16 |
| Hsa-mir-1307 | 2.94 | Hsa-mir-92b | 2.14 |
| Hsa-mir-1207 | 2.93 | Hsa-mir-525 | 2.13 |
| Hsa-mir-526a | 2.93 | Hsa-mir-1247 | 2.09 |
| Hsa-mir-615 | 2.92 | Hsa-mir-1910 | 2.08 |
| Hsa-mir-3141 | 2.89 | Hsa-mir-194* | 2.08 |
| Hsa-mir-345 | 2.89 | Hsa-mir-520a | 2.08 |
| Hsa-mir-629 | 2.88 | Hsa-mir-210 | 2.07 |
| Hsa-mir-92a | 2.88 | Hsa-mir-769 | 2.06 |
| Hsa-mir-3185 | 2.85 |
miRNA underlined has more than one transcript in the data sets.
Table 2.
miRNAs upregulated in third trimester placentas compared with first trimester placentas (n = 94)
| Name | Fold Change | Name | Fold Change |
|---|---|---|---|
| Hsa-mir-139 | 42.05 | Hsa-mir-140 | 3.13 |
| Hsa-mir-29b | 22.08 | Hsa-mir-1299 | 3.11 |
| Hsa-mir-100 | 12.43 | Hsa-mir-26b | 3.10 |
| Hsa-mir-34c | 11.62 | Hsa-mir-542 | 3.09 |
| Hsa-mir-181c | 9.70 | Hsa-mir-486 | 3.07 |
| Hsa-let-7f | 9.34 | Hsa-mir-874 | 3.06 |
| Hsa-mir-29b-1* | 8.43 | Hsa-mir-4315 | 3.05 |
| Hsa-mir-125b | 8.33 | Hsa-mir-1294 | 2.95 |
| Hsa-mir-184 | 8.04 | Hsa-mir-146a | 2.94 |
| Hsa-mir-4324 | 7.71 | Hsa-mir-376b | 2.91 |
| Hsa-let-7 g | 7.05 | Hsa-mir-148b | 2.86 |
| Hsa-mir-21 | 6.86 | Hsa-mir-32 | 2.84 |
| Hsa-mir-497 | 6.71 | Hsa-mir-3156 | 2.82 |
| Hsa-mir-34b | 6.41 | Hsa-mir-4306 | 2.82 |
| Hsa-let-7a | 6.37 | Hsa-mir-3148 | 2.73 |
| Hsa-mir-29a | 5.92 | Hsa-mir-10a | 2.66 |
| Hsa-mir-221 | 5.88 | Hsa-mir-125b-1* | 2.62 |
| Hsa-mir-150 | 5.80 | Hsa-mir-1291 | 2.62 |
| Hsa-let-7i | 5.67 | Hsa-mir-98 | 2.58 |
| Hsa-mir-1244 | 5.43 | Hsa-mir-489 | 2.57 |
| Hsa-mir-664 | 5.28 | Hsa-mir-4253 | 2.54 |
| Hsa-mir-10b | 5.13 | Hsa-mir-660 | 2.47 |
| Hsa-mir-504 | 4.84 | Hsa-mir-133b | 2.45 |
| Hsa-mir-29c | 4.72 | Hsa-mir-505 | 2.45 |
| Hsa-mir-451 | 4.70 | Hsa-mir-140 | 2.44 |
| Hsa-mir-526b | 4.52 | Hsa-mir-22 | 2.43 |
| Hsa-mir-195 | 4.51 | Hsa-mir-720 | 2.42 |
| Hsa-mir-363 | 4.32 | Hsa-mir-103-as | 2.40 |
| Hsa-mir-3176 | 4.28 | Hsa-mir-335 | 2.32 |
| Hsa-mir-24-2* | 4.05 | Hsa-mir-454 | 2.29 |
| Hsa-mir-424 | 4.04 | Hsa-mir-548u | 2.29 |
| Hsa-let-7d | 4.03 | Hsa-mir-34a | 2.28 |
| Hsa-mir-30a | 4.00 | Hsa-mir-1306 | 2.26 |
| Hsa-mir-551b | 4.00 | Hsa-mir-155 | 2.24 |
| Hsa-mir-143 | 3.97 | Hsa-let-7b | 2.23 |
| Hsa-mir-4329 | 3.96 | Hsa-mir-181d | 2.23 |
| Hsa-mir-99a | 3.94 | Hsa-mir-509-3 | 2.21 |
| Hsa-let-7c | 3.84 | Hsa-mir-126 | 2.20 |
| Hsa-mir-768-3p | 3.75 | Hsa-mir-572 | 2.20 |
| Hsa-mir-328 | 3.64 | Hsa-mir-1256 | 2.18 |
| Hsa-mir-377 | 3.56 | Hsa-mir-362 | 2.13 |
| Hsa-mir-101 | 3.49 | Hsa-mir-1246 | 2.07 |
| Hsa-mir-133a | 3.34 | Hsa-let-7e | 2.06 |
| Hsa-mir-202 | 3.20 | Hsa-mir-21 | 2.06 |
| Hsa-mir-488 | 3.18 | Hsa-mir-188 | 2.05 |
| Hsa-mir-125b-2* | 3.15 | Hsa-mir-196b | 2.04 |
| Hsa-mir-223 | 3.15 | Hsa-mir-193a | 2.03 |
miRNA underlined has more than one transcript in the data sets.
Fig. 1.
Hierarchical clustering analysis of miRNAs that are significantly upregulated or significantly downregulated in 3rd trimester placentas compared with 1st trimester placentas. Comparison was made as at least >2-fold changes, P < 0.05. Lanes 1–6: 3rd trimester placentas; lanes 7–12: 1st trimester placentas. Since a number of miRNAs have more than one transcript in the microarray platform, several miRNAs are shown more than once in the hierarchical clustering analytic result. The level of miRNA expression is color coded: red, upregulation; green, downregulation.
Among the 191 miRNAs, several miRNA clusters were identified and differentially expressed between the first and third trimester placentas. Clusters of miRNAs that were found significantly upregulated in the first trimester compared with the third trimester placentas are presented in Table 3. Interestingly, 33 of 97 miRNAs (34%) that are upregulated in the first trimester placentas are localized on chromosomes 14 and 19 (Table 3). In addition, eight miRNAs within the miR17-92 cluster on chromosome 13q31.3 were also found strongly expressed in the first trimester placentas (Table 3). miRNA families and clusters that are upregulated in the third trimester placentas are listed in Table 4, which include eight members of let-7 family miRNAs, and miRNAs of miR-29 cluster on chromosome 7q32.3, miR-34 cluster on chromosome 11q23.1, miR-195 cluster on chromosome 17q13.3, and miR-181c cluster on chromosome 19q13.13.
Table 3.
miRNA clusters upregulated in first trimester placentas compared with third trimester placentas
| Cluster | Chromosome Location | miRNAs | Functions | Refs. |
|---|---|---|---|---|
| 1. miR-25 cluster | 7q22.1 | Hsa-miR-25, Hsa-miR-93 | modulate TGFβ signaling, oncogenic, antiapoptotic | 20, 21 |
| 2. miR-17-92 cluster | 13q31.3 | Hsa-miR-17, Hsa-miR-18a, Hsa-miR-19a, Hsa-miR-19b-1*, Hsa-miR-20a, Hsa-miR-20b, Hsa-miR-92a, miR-92a-1* | immune tolerance, oncogenic, angiogenic, proliferation, antiapoptotic | 20, 22, 24 |
| 3. C14MC cluster | 14q32.2 | Hsa-miR-127, Hsa-miR-345, Hsa-miR-370, Hsa-miR-431, Hsa-miR-665 | immune suppressive, anti-inflammatory response, protect ischemic/hypoxia injury | 25, 27 |
| 14q32.31 | Hsa-miR-134, Hsa-miR-323, Hsa-miR-409, Hsa-miR-412, Hsa-miR-654, Hsa-miR-758 | immune suppressive, protect ischemic/hypoxia injury, lipid metabolism, angiogenic associated with neuron function | 28, 29 | |
| 4. miR-371 cluster | 19q13.42 | Hsa-miR-371, Hsa-miR-372, Hsa-miR-373 | stem cell signature, oncogenic, immune suppressive | 30, 31 |
| 5. C19MC cluster | 19q13.42 | Hsa-miR-498, Hsa-miR-518b, Hsa-miR-518c, Hsa-miR-518d, Hsa-miR-518e, Hsa-miR-518f, Hsa-miR-519a*, Hsa-miR-519b, Hsa-miR-519c, Hsa-miR-520a, Hsa-miR-520c, Hsa-miR-520d, Hsa-miR-520f, Hsa-miR-520 g, Hsa- miR-520 h, Hsa-miR-522, Hsa-miR-523, Hsa-miR-525, Hsa-miR-526a | immune suppressive, innate/adaptive immune responses | 31, 32 |
| 6. miR-106a cluster | Xq26.2 | Hsa-miR-106a, Hsa-miR-18b, Hsa-miR-20b | angiogenic, anti-inflammatory, immunoregulator | 33, 34 |
Table 4.
miRNA clusters/families upregulated in third trimester placentas compared with first trimester placentas
| Cluster | Chromosome Location | miRNAs | Functions | Refs. |
|---|---|---|---|---|
| 1. miR-29 cluster | 7q32.3 | Hsa-miR-29a, Hsa-miR-29b, Hsa-miR-29b-1* | tumor suppressor, innate/adaptive immune responses, differentiation | 35 |
| 2. miR-34 family | 1p36.22 | Hsa-miR-34a | tumor suppressor, apoptosis, onco-immunology | 36, 37 |
| 11q23.1 | Hsa-miR-34b, Hsa-miR-34c | |||
| 3. miR-195 cluster | 17q13.1 | Hsa-miR-195, Hsa-miR-497 | tumor suppressor, differentiation, lymph genic | 38 |
| 4. miR-181c cluster | 19p13.13 | Hsa-miR-181c, Hsa-miR-181d | tumor suppressor, autoimmunity, differentiation, hematopoietic | 39, 40 |
| 5. let-7 family | 11q24.1 | Hsa-let-7a | promote differentiation, tumor suppressor, glucose homeostasis | 41 |
| 22q13.31 | Hsa-let-7b | |||
| 21q21.1 | Hsa-let-7c | |||
| 9q22.32 | Hsa-let-7d | |||
| 19q13.41 | Hsa-let-7e | |||
| 9q22.32 (Xp11.22) | Hsa-let-7f-1 (7f-2) | |||
| 3p21.1 | Hsa-let-7 g | |||
| 12q14.1 | Hsa-let-7i | |||
| let-7a-cluster | 11q24.1 | Hsa-let-7a, Hsa-miR-100, Hsa-miR-125b | tumor suppressor, differentiation | 41 |
| let-7c-cluster | 21q21.1 | Hsa-let-7c, Hsa-miR-99a | tumor suppressor, differentiation | 41 |
Based on miRNA microarray data, five miRNAs were chosen to confirm their expression differences between the first and third trimester placentas by qRT-PCR, including miR-708, miR-371, miR-17, miR-139, and miR-125. miR-708 has the highest significant increase in the first trimester compared with the third trimester (Table 1), whereas miR-139 has the highest significant increase in the third trimester compared with the first trimester (Table 2). miR-371 and miR-17 were randomly chosen from the miR-371 cluster and the miR-17-92 cluster (Table 3), respectively. These two clusters were both significantly increased in the first trimester placentas. miR-125 was chosen because it targets vitamin D receptor (VDR) (26), and VDR expression was significantly reduced in placental trophoblasts in preeclampsia (23). Consistent with the microarray data, expressions of miR-17-3p, miR-371-5p, and miR-708-5p were significantly higher in the first trimester than in the third trimester placentas. In contrast, expressions of miR-125b-5p and miR-139-5p were significantly higher in the third trimester than the first trimester placentas. The fold change for expression of miR-17-3p, miR-371-5p, miR-708-5p, miR-125b-5p, and miR-139-5p is shown in Fig. 2.
Fig. 2.
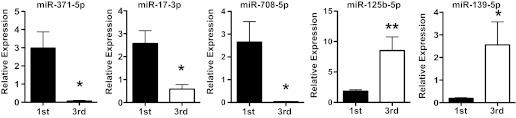
Quantitative RT-PCR analysis of miRNA expression for miR-17-3p, miR-371-5p, miR-708-5p, miR-125b-5p, and miR-139-5p between placental villous tissues from 1st and 3rd trimester placentas (n = 6 in each group). Data are expressed as fold change of mean 2−ΔCT ± SE of each target miRNA after being normalized with U6 expression. Expressions of miR-371-5p, miR-17-3p and miR-708-5p were significantly downregulated in 3rd vs. 1st trimester placentas, *P < 0.05. In contrast, expressions of miR-125b-5p and miR-139-5p were significantly upregulated in 3rd vs. 1st trimester placentas, *P < 0.05, **P < 0.01, respectively.
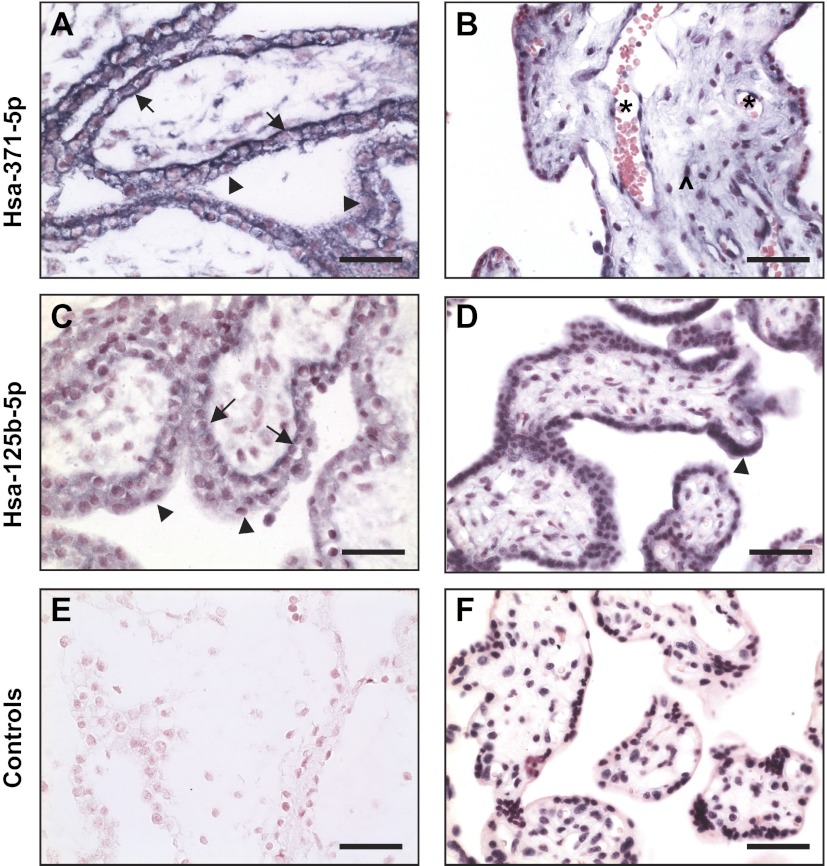
Villous tissue expression of miR-371-5p and miR-125b-5p by ISH.
Previous studies revealed different miRNA expression profiles in trophoblasts isolated from first and third trimester placentas (27). Placental villous tissue is composed of different cell types, including cytotrophoblasts, syncytiotrophoblasts, mesenchymal stromal cells, fibroblasts, and fetal vessel cells of smooth muscle cells and endothelial cells. First trimester placental villous tissues are enriched with cytotrophoblasts and mesenchymal/stromal cells but contain very few villous core fetal vessels compared with third trimester villous tissues. In contrast, abundant syncytiotrophoblasts and a plethora of villous core vessels containing endothelial cells are major components of third trimester placental villous tissue. To determine the possibility of cell-specific miRNA expression within placental villous tissue, miR-371-5p and miR-125b-5p expression was examined by ISH. Results from microarray analysis and qRT-PCR studies showed that miR-371 expression was significantly upregulated in the first trimester placentas and miR-125b expression was significantly upregulated in the third trimester placentas. Different expression patterns between miR-371-5p and miR-125b-5p were observed by ISH. Representative images for miR-371-5p and miR-125b-5p expression are shown in Fig. 3. miR-371-5p is strongly expressed in trophoblasts, especially in the basal membrane of cytotrophoblasts in the first trimester placentas (Fig. 3A). In the third trimester placentas, miR-371-5p expression was also detected in villous stromal cells, fetal vessel smooth muscle cells, and endothelial cells (Fig. 3B). In contrast, miR-125b-5p was mainly expressed in trophoblasts in the third trimester placentas (Fig. 3D) compared with the first trimester placentas (Fig. 3C). The different expression patterns of miR-137-5p and miR-125b-5p between the first and third trimester placentas suggest that miRNA expression in the placental tissue is developmentally dependent during pregnancy.
Fig. 3.
Representative images of miR-371-5p and miR-125b-5p expression in 1st and 3rd trimester placental villous tissues detected by in situ hybridization. A and B: miR-371-5p. C and D: miR-125b-5p. A and C: 1st trimester. B and D: 3rd trimester. miR-371-5p is intensively expressed in cyto- and syncytiotrophoblasts in 1st trimester placentas (A) but also expressed in stromal cells, and fetal vessel smooth muscle cells and endothelial cells in 3rd trimester placentas (B). miR-125b-5p is mainly expressed in trophoblasts with intensive expression in 3rd trimester placentas (D) compared with 1st trimester placentas (C). E: negative control. F: U6, positive control. Arrowhead, syncytiotrophoblasts; arrow, cytotrophoblasts. *villous core fetal vessels; ^stromal cells. Bar, 50 μm.
DISCUSSION
To date, numerous miRNAs have been identified to play important roles in the regulation of cell proliferation and differentiation and in organ development. Recent findings support the idea that miRNAs are key regulators of placental angiogenic and immune tolerant activities (18). In the present study, through microarray analysis we found that a total of 191 miRNAs were differently expressed between the first and third trimester placentas. We further noticed that 39 of 97 miRNAs that are upregulated in the first trimester placentas are localized on chromosomes 13, 14, and 19 and belong to four miRNA clusters. A total of 94 miRNAs were found upregulated in the third trimester, and a number of them have been identified as having immunological and angiogenic functions and characterized as tumor suppressors.
Four clusters of miRNAs that are significantly more highly expressed in the first trimester than in the third trimester placentas are the miR17-92 cluster on chromosome13q31.3, miRNAs within C14MC on chromosome 14q32, and miRNAs within C19MC and the miR-371 cluster on chromosome 19q13.42 (Table 3). The miR17-92 cluster comprises six miRNA genes (pre-/pri-miRNAs), and potentially 12 mature miRNAs would be produced. Our results showed that eight members of miR17-92 cluster miRNAs were significantly upregulated in the first trimester placentas, including miR-17, miR-18a, miR-19a, miR-19b-1-star, miR-20a, miR-20b, miR-92a, and miR-92a-1-star. A previous study showed that miR-17-92 cluster miRNAs play critical roles in promoting T cell proliferation and antiapoptosis to resist cell death (43). Therefore, the miR-17-92 cluster miRNAs are very likely involved in immune tolerance and cell survival in both trophoblasts and non-trophoblasts during placental development, especially during the first trimester.
Our results showed that 11 of 34 miRNAs within C14MC and 19 of 54 miRNAs within C19MC are significantly upregulated in the first trimester placentas, which suggests that a larger portion of miRNAs in the C14MC and C19MC clusters decreases with advancement of gestation age. C14MC is the largest imprinted, maternally expressed human miRNA cluster in the placenta (40). C19MC is the largest imprinted, paternally expressed human miRNA cluster in the placenta (29). Imprinted genes are expressed in a parent-of-origin-specific manner; e.g., for a given locus, the paternal allele is transcriptionally active, whereas the maternal allele remains silent, or vice versa (29). Thus, our results support the concept that both C14MC and C19MC miRNAs probably engage in immune suppressive and innate/adaptive immune responses to protect a fetus from maternal immune insults. A study by Morales-Prieto et al. (27) showed that expression of miRNAs within C19MC increases significantly from the first to the third trimester trophoblast, whereas that of C14MC members decreases. The discrepancy of C19MC miRNA expression in the first and third trimester placentas between our study and that of Morales-Prieto et al. is not known but could be due to the sample difference. Whole villous tissue without disruption of tissue integrity was used in our study, whereas isolated trophoblasts after culture were used in the Morales-Prieto et al. study, suggesting that in vitro cell culture conditions may affect miRNA expression. Nonetheless, placenta/trophoblast-derived C19MC miRNAs may contribute to maternal C19MC miRNA levels during pregnancy, since expression of C19MC miRNAs was found in trophoblast exosomes (7) and maternal plasma concentrations of cell-free C19MC miRNAs was significantly increased in pregnant women toward term (25).
Although we did not specifically examine the localization of C14MC and C19MC miRNAs within villous tissue, our miRNA-371-5p ISH data suggest that the expression level of an individual miRNA may depend on the developmental stage. The miRNA-371 cluster is located on chromosome 19, in close proximity to C19MC miRNAs. We found that miRNA-371-5p is predominantly expressed in both cyto- and syncytiotrophoblasts in the first trimester placentas and is markedly expressed in syncytiotrophoblasts, villous stromal cells, and fetal vessel endothelium in the third trimester placentas. Abundant expression of C19MC in placenta-derived stromal cells (10) supports our findings. In contrast, the pattern for trophoblast expression of miR-125b-5p is different, in which miRNA-125b-5p is strongly expressed in trophoblasts of the third trimester compared with the first trimester placentas. The different expression pattern of individual miRNA in different cell types within villous tissue between first trimester and third trimester placentas further suggests that on and off of an individual miRNA expression/production within villous tissue and its roles during pregnancy is dynamic and developmental dependent.
Many miRNAs that are strongly expressed in the first trimester placentas are associated with oncogenic/angiogenic and tissue remodeling. For example, miR-708 acts as an oncogene to promote cell proliferation, migration, and invasion (16). miR-675 regulates type II collagen expression (8). miR-210 is considered a pleiotropic hypoxamir (3). Increased miR-210 expression in the first trimester placenta reflects a physiological hypoxic condition in the early placental development. On the other hand, several miRNAs that are increased in the term placentas exert immunological and hematopoietic activity. For instance, miR-181 impacts the development of NK cells from CD34(+) hematopoietic progenitor cells and influences IFNγ production in primary CD56(+) NK cells (5). miR-424 regulates monocyte and macrophage differentiation (35). miR-125b regulates proliferation of hematopoietic stem cells and also affects the balance of cell fates during lymphoid development, in part, probably, by acting as a lineage-specific antiapoptotic factor (30).
We also noticed that eight members of the let-7 family miRNAs that were reported in humans (37) were upregulated in the third trimester placentas compared with the first trimester placentas, including let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, and let-7i. Let-7 family miRNAs have been intensively studied in cancer cells and tumor tissues. They are characterized as key miRNA regulators in development and cancer biology (17). Two major function of let-7 family miRNAs have been identified: 1) promotion of cell differentiation and 2) tumor suppression. It was revealed that in C. elegans, let-7 upregulation mediates terminal differentiation “larval-to-adult switch” (1). Let-7 expression is upregulated during mouse brain development (42). In HeLa cells, overexpression of let-7 inhibits cell proliferation and promotes the G1 to S transition via regulation of key cell cycle protooncogenes, including RAS, CDC25a, CDK6, and cyclin D (18). In prostate cancer cells, overexpression of let-7 induces G2-M phase cell-cycle arrest (21). In contrast, downregulation or lack of expression of let-7 family miRNAs has been demonstrated in several types of cancers, including prostate cancer (28), breast cancer (44), lung cancer (31), and ovarian cancer (12), etc. The finding of upregulation of let-7 family miRNA expression in the third trimester/term placentas is important, suggesting that the homeostasis of let-7 family miRNAs likely plays a critical role in regulating placental cell differentiation or placenta maturation.
In addition to the let-7 family microRNAs, several miRNAs and miRNA clusters that are strongly expressed in the term placentas have also been reported to have tumor-suppressive effects. These miRNAs include miR-125b, miR-181c, miR-195, and miRNAs within the miR-34 family (miR-34a, miR-34b, miR-34c) (41) and the miR-29 cluster (miR-29a, miR-29b, miR-29b-1-star) (9, 32). Expressions of miR-21 and miR-221 are also increased in the third trimester/term placentas; both of them are considered inhibitory miRNAs (17). Expression of tumor suppressor miRNAs in the term placenta provides a logical rationale that tumor suppressor miRNAs participate in placental maturation and play a role in limiting placental (vascular) tissue growth when pregnancy toward term. It is speculated that specific miRNA expression/production may also be involved in initiating the parturition process.
miRNAs exhibit spatial, temporal, and tissue/cell specificities that result in their involvement in tissue morphogenesis, developmental timing, and cell differentiation. They regulate temporal transitions in gene expression associated with cell fate progression and differentiation throughout development. The placenta is a unique organ during pregnancy with invasive characteristics of trophoblasts and their ability to modulate the mother's immune system. In the present study, we identified different patterns of miRNA expression profiles between the first and third trimester placentas. As summarized in Tables 3 and 4, a number of miRNAs that have oncogenic and immunologic suppressive characteristics such as miRNAs within the miR-17-92 cluster, the miR-371 cluster, C14MC, and C19MC are strongly expressed in first trimester placentas. In contrast, tumor suppressor miRNAs and miRNAs that promote differentiation and exert hematopoietic activities including let-7 family miRNAs and miRNAs within the miR-29 and miR-34 clusters are strongly expressed in third trimester/term placentas. Although how miRNAs regulate placental and innate/adaptive immune function is largely unknown, it is believed that the endogenous autoregulatory circuit that controls miRNA production is dynamic and regulated on many levels during placental development. Thus, aberrant placental miRNA expression would no doubt have an impact on pregnancy disorders associated with placental deficiency such as intrauterine growth restriction and preeclampsia.
GRANTS
This study was supported in part by grants from National Institutes of Health, National Institute of Child Health and Human Development HD-36822 and National Heart, Lung, and Blood Institute HL-65997 to Y. Wang.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: Y.G. and J.S. performed experiments; Y.G. and Y.W. analyzed data; Y.G. and Y.W. drafted manuscript; Y.G. and Y.W. edited and revised manuscript; Y.G., J.S., L.J.G., and Y.W. approved final version of manuscript; J.S., L.J.G., and Y.W. conception and design of research; Y.W. interpreted results of experiments; Y.W. prepared figures.
REFERENCES
- 1. Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev 21: 511–517, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boettger T, Braun T. A new level of complexity: the role of microRNAs in cardiovascular development. Circ Res 110: 1000–1013, 2012 [DOI] [PubMed] [Google Scholar]
- 3. Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle 9: 1072–1083, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol 187: 6171–6175, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cochella L, Hobert O. Diverse functions of microRNAs in nervous system development. Curr Top Dev Biol 99: 115–143, 2012 [DOI] [PubMed] [Google Scholar]
- 7. Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, Sadovsky Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod 18: 417–424, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem 285: 24381–24387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 104: 15805–15810, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flor I, Neumann A, Freter C, Helmke BM, Langenbuch M, Rippe V, Bullerdiek J. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem Biophys Res Commun 422: 411–416, 2012 [DOI] [PubMed] [Google Scholar]
- 11. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS One 3: e3148, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helland Å, Anglesio MS, George J, Cowin PA, Johnstone CN, House CM, Sheppard KE, Etemadmoghadam D, Melnyk N, Rustgi AK, Phillips WA, Johnsen H, Holm R, Kristensen GB, MJ; B, Australian Ovarian Cancer Study Group, Pearson R.B, Børresen-Dale AL, Huntsman DG, deFazio A, Creighton CJ, Smyth GK, Bowtell DD. Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian. PLoS One 6: e18064, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henrion-Caude A, Girard M, Amiel J. MicroRNAs in genetic disease: rethinking the dosage. Curr Gene Ther 12: 292–300, 2012 [DOI] [PubMed] [Google Scholar]
- 14. Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell 5: 351–358, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Inukai S, de Lencastre A, Turner M, Slack F. Novel MicroRNAs differentially expressed during aging in the mouse brain. PloS One 7: e40028, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H, Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, Park JY, Harris CC, Yang P, Jen J. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers. Clin Cancer Res 18: 3658–3667, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jérôme T, Laurie P, Louis B, Pierre C. Enjoy the silence: the story of let-7 microRNA and cancer. Curr Genomics 8: 229–233, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67: 7713–77122, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. MicroRNAs: molecular features and role in cancer. Front Biosci 17: 2508–2540, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol 6: 419–429, 2009 [DOI] [PubMed] [Google Scholar]
- 21. Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res 72: 3393–3404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, Ohkuchi A, Matsubara S, Takeshita T, Takizawa T. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod 81: 717–729, 2009 [DOI] [PubMed] [Google Scholar]
- 23. Ma R, Gu Y, Zhao S, Sun J, Groome LJ, Wang Y. Expressions of vitamin D metabolic components VDBP, CYP2R1, CYP27B1, CYP24A1, and VDR in placentas from normal and preeclamptic pregnancies. Am J Physiol Endocrinol Metab 303: E928–E935, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci 18: 46–56, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, Masuzaki H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem 56: 1767–1771, 2010 [DOI] [PubMed] [Google Scholar]
- 26. Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer 125: 1328–1333, 2009 [DOI] [PubMed] [Google Scholar]
- 27. Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S, Schneider U, Herrmann J, Gruhn B, Markert UR. MicroRNA expression profiles of trophoblastic cells. Placenta 33: 725–734, 2012 [DOI] [PubMed] [Google Scholar]
- 28. Nadiminty N, Tummala R, Lou W, Zhu Y, Shi XB, Zou JX, Chen H, Zhang J, Chen X, Luo J, deVere White RW, Kung HJ, Evans CP, Gao AC. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS One 7: e32832, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefèvre A, Coullin P, Moore GE, Cavaillé J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 19: 3566–3582, 2010 [DOI] [PubMed] [Google Scholar]
- 30. Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA 107: 21505–21510, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osada H, Takahashi T. Let-7 and miR-17-92: small-sized major players in lung cancer. Cancer Sci 102: 9–17, 2011 [DOI] [PubMed] [Google Scholar]
- 32. Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget 1 224–227, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P, Hassan SS, Kim CJ. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol 196: 261: e261–e266, 2007 [DOI] [PubMed] [Google Scholar]
- 34. Rayner KJ, Fernandez-Hernando C, Moore KJ. MicroRNAs regulating lipid metabolism in atherogenesis. Thromb Haemost 107: 642–647, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA 104: 19849–19854, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13: 239–250, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol 18: 505–506, 2008 [DOI] [PubMed] [Google Scholar]
- 38. Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutr Metab Cardiovasc Dis 22: 937–43, 2012 [DOI] [PubMed] [Google Scholar]
- 39. Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27: 5959–5974, 2008 [DOI] [PubMed] [Google Scholar]
- 40. Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 14: 1741–1748, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong KY, Yu L, Chim CS. DNA methylation of tumor suppressor miRNA genes: a lesson from the miR-34 family. Epigenomics 3: 83–92, 2011 [DOI] [PubMed] [Google Scholar]
- 42. Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 21: 415–426, 2007 [DOI] [PubMed] [Google Scholar]
- 43. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9: 405–414, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y, Deng C, Wang J, Xiao J, Gatalica Z, Recker RR, Xiao GG. Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer. Breast Cancer Res Treat 127: 69–80, 2011 [DOI] [PubMed] [Google Scholar]