Abstract
Once mRNAs are transcribed, spliced and transported to the cytoplasm, their fate is determined by the complex interplay of RNA binding proteins (RBPs) and microRNAs (miRNAs) that act on regulatory elements within the transcripts. The importance of post-transcriptional regulatory mechanisms in angiogenesis is underscored by the observation that perturbations in miRNAs and/or RBPs lead to profound phenotypic alterations in vascular development, homeostasis and disease, with current data suggesting that mRNAs for key angiogenic regulators (secreted factors and intracellular signaling intermediates) are subject to stringent post-transcriptional regulation by both RBPs and miRNAs. In addition, an intricate network of miRNAs and RBPs allow robust gene regulation in vascular cells. This review focuses on the miRNAs and RBPs which often cooperate to achieve precise spatial and temporal control of angiogenic regulatory genes.
Vascular development and angiogenesis
The vascular system in vertebrates is multifunctional – it provides all organ systems with nutrients and oxygen, a nonthrombogenic surface (see Glossary), controls vascular tone, blood flow, blood pressure and selective transport of substances [1,2]. In addition, vascular cells communicate with surrounding tissues via inductive signals called ‘angiocrine factors’ which are essential for organ formation during development, hematopoiesis and liver regeneration [3,4]. Thus, the proper functioning of the vascular system is essential not only for embryonic development but also for physiological homeostasis in the adult. Indeed, abnormal vascular function and growth contributes to many pathological conditions including tumor growth and metastasis, rheumatoid arthritis, psoriasis, diabetic retinopathy and peripheral vascular disease [1,2,5,6].
The development of the vascular system is generally classified into two mechanistically distinct processes [2,5,7,8]. Vasculogenesis, the de novo generation of blood vessels from mesenchymal cells, leads to the formation of a primary vascular network. By contrast, angiogenesis, or formation of new vessels from pre-existing vasculature, occurs by a highly orchestrated series of cellular sprouting events by so-called ‘tip’ cells, differentiation into a nascent vascular sprout and fusion of these sprouts to form a new vascular network. Subsequently, neovessels become ensheathed with pericytes and/or vascular smooth muscle cells, which allows further vascular stabilization and maturation. Once formed, blood vessels remain quiescent and are essential for physiological homeostasis and tissue integrity. However, the failure of normal vascular development results in the abnormal vasculature often observed in pathological states such as chronic inflammation and cancer [5,6]. Indeed, it is thought that inhibition of pathological angiogenesis or phenotypic modulation of neovessels (so-called ‘vascular normalization’) might result in therapeutic benefit for diseases such as cancer [9]. For example, normalized vessels have better blood flow, and thus the effectiveness of anticancer therapeutics is enhanced. Therefore, better understanding of the angiogenic process is of great importance in both normal physiology and abnormal pathology.
Angiogenesis is initiated by endothelial cells which are activated by numerous soluble (growth factors, cytokines, bioactive lipids), gaseous (hypoxia, NO) as well as solid-state (i.e. extracellular matrix-derived) signals. The prevailing paradigm suggests that extracellular stimuli activate intracellular signaling pathways which transmit the signal into the nucleus to regulate endothelial cell gene expression. Given the importance of transcriptional mechanisms in tissue-specific and inducible gene expression scenarios, it is generally assumed that the primary result of extracellular signals is to influence changes in gene transcription [8]. Indeed, analysis of transcriptional elements of differentiated vascular phenotypes support this model and have identified several critical transcriptional regulators including Ets, TFII-1, GATA2, Prox1, KLF factors and the Id family of bHLH regulators [8,10,11]. However, independent lines of investigations have suggested an equally important step in the regulation of vertebrate gene expression is post-transcriptional regulation of mRNA fate by microRNAs (miRNAs) and RNA binding proteins (RBPs).
The discovery of small RNAs and noncoding RNAs that profoundly influence biological events as well as specific RBPs that are required for major physiological and pathological phenomena have all supported the concept that RNA regulatory mechanisms are critically important in the fidelity and robustness of gene expression patterns in defined biological states [12–18]. However, post-transcriptional gene regulatory mechanisms are not fully understood in angiogenesis. We will discuss recent findings which suggest a profound regulatory function for miRNAs and RBPs in angiogenesis. We will also review the emerging mechanistic paradigm of gene regulation by the cooperative actions of miRNAs and RBPs.
miRNAs and RBPs as major regulators of mRNA fate
Eukaryotic mRNAs are derived from splicing of the primary transcript in the nucleus. Following transport of the mature mRNA into the cytoplasm as mRNPs (messenger ribonu-cleoprotein particles), the mRNA can undergo several fates. It can associate with polyribosomes to produce proteins, be stored as a translationally inactive mRNA in P bodies or degraded in the exosome [12]. In many cases, specific mRNAs must be transported to defined locations in the cell, for example targeting of β-actin mRNA to the lamellipodia in migrating cells [19]. Such processes are regulated by specific RBPs and regulatory mRNAs (such as miRNAs) that respond to cell intrinsic and extracellular cues.
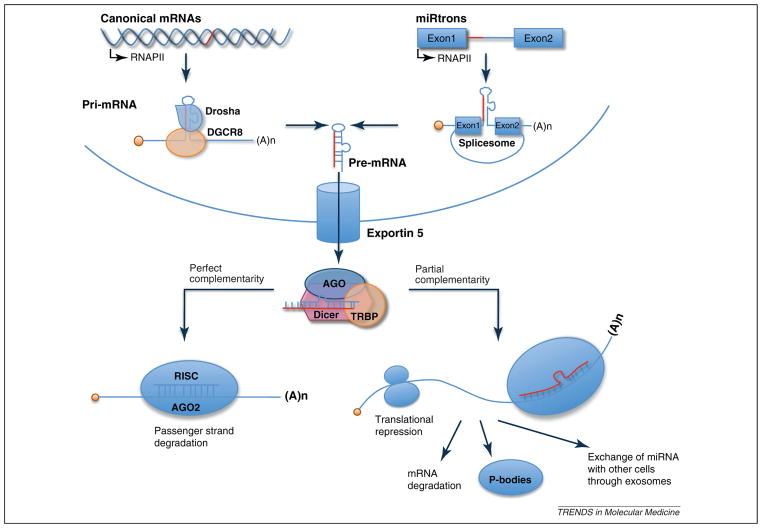
miRNAs are a class of small (~20 nt), noncoding RNAs that target 3′-untranslated regions (3′UTR) in a sequence-specific manner, and in most cases repress translation or induce degradation of the target mRNA [20–23]. miRNAs originate from primary miRNAs (pri-miRNAs) that are transcribed by RNA polymerase II and cleaved by Drosha, a ribonuclease III, to produce an ~70 nt pre-miRNA in the nucleus. This pre-miRNA is exported to the cytoplasm by exportin 5 and cleaved by a second ribonuclease III called Dicer to produce ~20 nt double-stranded (ds) miRNAs. These short dsRNAs bind to argo-naute (AGO) and other regulatory factors, thus forming the RNA-induced silencing complex (RISC) (Figure 1). After removal of the passenger RNA strand (the strand that does not pair with the mRNA), single-stranded miRNA associate with the 3′UTR elements of mRNAs through partial complementarity with a seed sequence, leading to the repression of protein synthesis. A single miRNA can repress multiple targets to regulate protein synthesis and mRNA levels [22–25], and some regulatory genes possess binding sites for multiple miRNAs. Although in the majority of cases miRNA repress gene expression, in some cases translational activation by the AGO2–miRISC complex was observed [26]. Furthermore, it is becoming increasingly clear that miRNA activity can be regulated by interacting with RBPs, another key factor that interacts with regulatory elements of the transcript [27,28]. Although miRNA-mediated gene regulatory mechanisms are intensively studied, the current data suggest that miRNAs fine-tune gene expression and are required to provide ‘robustness’ to regulatory systems.
Figure 1.
microRNA (miRNA) biogenesis. miRNAs are processed from primary miRNAs (pri-miRNAs), which are transcribed by RNA polymerase II from independent miRNA genes or from portions of introns of protein coding genes (miRtrons) in the nucleus. The maturation of pri-miRNAs is mediated by two RNase III-like endonucleases, namely Drosha and Dicer. Drosha functions with DGCR8, containing a double-stranded RNA (dsRNA)-binding domain, to process pri-miRNAs to ~70 nt pre-miRNAs. Some pre-miRNAs are produced from very short introns (miRtrons) as a result of splicing and debranching, bypassing the Drosha–DGCR8 step. Pre-miRNAs are exported to the cytoplasm via exportin 5 and cleaved by Dicer within complexes with transactivation responsive RNA binding protein (TRBP), producing a ~20 bp miRNA duplex. One strand of the miRNA duplex that has partial complementarity with sequences in the 3′-untranslated region (3′UTR) of target mRNA is preferentially incorporated into a miRNA-induced silencing complex (miRISC), whereas the passenger strand is released and degraded. The activity of miRISC leads to translational repression or mRNA degradation. Repressed mRNA could be moved to P bodies for either degradation or storage. Alternatively, miRNA could be transferred to other cells through exosome-dependent mechanisms [73].
Numerous RBPs interact with regulatory elements on mRNAs and regulate gene expression by modulating mRNA stability and/or translation [29,30]. The best known regulatory element is the AU-rich element (ARE), an RNA specific sequence motif located in the 3′UTR of the transcript that controls mRNA stability and translation by interacting with ARE-BPs (ARE-binding proteins) [29,30]. Around 20 ARE-BPs have been identified; well-known members include AUF1 (hnRNP D), the ELAV family (HuR, HuB, HuC, HuD), the tristetraprolin (TTP) family [which will be referred to as the zinc finger protein-36 (Zfp36) family], TIA, TIAR and the KH splicing regulatory protein (KSRP) [31]. Zfp36, AUF1 and KSRP ARE-BPs are thought to promote the rapid decay of target transcripts, whereas HuR generally stabilize the ARE-containing mRNA. Although sequence specificity of different ARE-BPs has been identified, distinct ARE-BPs appear to have competitive, additive or redundant functions on the same mRNA target. Thus, several ARE-BPs appear to interact with numerous mRNAs in a cell-type or stimulus-dependent manner. For example, AUF1 and Zpf36 appear to antagonize the p38 stress-activated protein kinase-dependent stabilization of cytokine mRNAs [32]. By contrast, HuR is known to stabilize the cell stress inducible mRNAs such as the cell cycle inhibitor p21cip1/waf1 [33].
A recent systems-level bioinformatics study showed that the ARE motif is over-represented in miRNA target sites of transcripts and might antagonize or cooperate with miRNA-dependent gene regulation [34]. This in silico analysis provides support for other studies in which close interactions between ARE-BPs and miRNAs were found [26,34–37]. For example, the binding of a miRNA-loaded RISC at or near the ARE motif of the tumor necrosis factor α (TNFα) 3′UTR induces rapid decay of TNFα mRNA [36]. These emerging studies strongly suggest that the miRNAs and ARE-BPs are tightly coupled.
In this review, we summarize the functional roles of miRNAs and RBPs, in particular ARE-BPs, in angiogene-sis and provide a general framework for the interconnections between miRNA and ARE-BPs.
Close-knit interactions between miRNAs and ARE-BPs
Although the importance of miRNAs in the regulation of gene expression has been demonstrated in several systems, recent data suggest that miRNA action might occur in conjunction with RNA binding protein/mRNA interactions [26,35–37] (Figure 2). In fact, cooperative interactions between miRNA and ARE-BPs in the modulation of gene expression have been demonstrated; Zfp36, an ARE-BP and miR-16, a miRNA containing a UAAAUAUU sequence that is complementary to the ARE sequence, were shown to depend on each other to efficiently suppress the TNFα gene [36]. The binding of miR-16 to the ARE sequence as well as the Zfp36 interaction with Ago/eiF2C is needed for the rapid decay of TNFα mRNA. This was the first example of a miRNA/RBP interaction in the regulation of TNFα, a major inflammatory cytokine.
Figure 2.
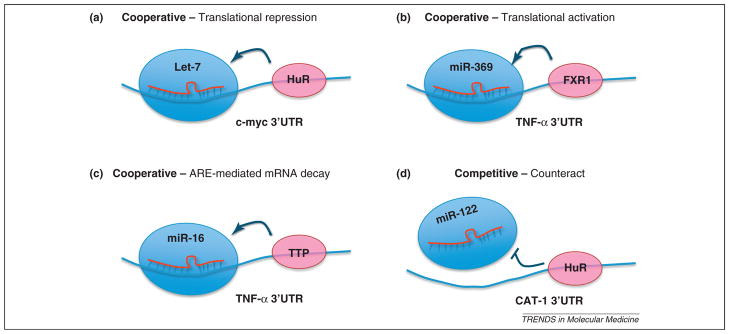
Interplay between microRNA (miRNA) and AU-rich element binding proteins (ARE-BPs). Examples of cooperative interactions between miRNAs and ARE-BPs that facilitate translational repression are as follows: (a) let-7 and HuR in the 3′-untranslated region (3′UTR) of c-Myc, translational activation (b) by miR-369-3 and FXR1 (Fragile X mental retardation syndrome-related 1) in the 3′UTR of tumor necrosis factor α (TNFα) under cell cycle arrest, and ARE-induced mRNA decay by (c) miR-16 and ZFP36 in the 3′UTR of TNFα. Competitive interactions between miRNAs and ARE-BPs counteract miRNA translational repressive activity, as shown in (d), where miR-122 and HuR interact in the 3′UTR of CAT (cationic amino acid transporter).
Another example is miR-369-3 and FXR1 (Fragile X mental retardation syndrome-related 1), an ARE-BP [26]. miR-369-3 interacts with the ARE of TNFα mRNA, which recruits the AGO2–FXR1 complex to the ARE element itself and activates translation under serum starvation conditions. In the third example, HuR cooperates with the let-7 miRNA to repress c-Myc expression [37]. The binding of HuR to the 3′UTR of c-Myc recruits let-7 loaded RISC and inhibits the expression of c-Myc. These studies clearly indicate that ARE-BPs interact with the miRNA machinery to modulate gene expression at the post-transcriptional level.
In some cases, ARE-BPs and miRNA compete for a binding site on the mRNA, thereby counteracting the functions of each other. The binding of HuR to the 3′UTR UTR of cationic amino acid transporter (CAT)-1 mRNA prevents the miR-122 mediated repression of CAT-1 expression, thereby resulting in enhanced expression of the CAT-1 gene [35].
Recent studies have unraveled novel interaction modes between miRNAs and RBPs [17,38]. The interaction between miRNAs and RBPs on the CA-rich element (CARE) of vascular endothelial growth factor (VEGF) mRNA influences translation [38]. The translation of VEGFA mRNA is negatively regulated by CARE-binding to endogenous miR-NAs under basal conditions; however, under hypoxia, heterogeneous nuclear ribonucleoprotein L (hnRNP L) is translocated to the cytoplasm where it binds the 3′UTR CARE of VEGFA, prevents miRNA silencing activity and allows VEGFA translation. In a separate system, a miRNA functions as an RNA decoy that interferes with RBP activity in leukemic cells [17]. The direct binding of miR-328 to hnRNP E2 impedes the interaction of this RBP with a target mRNA, resulting in the release of translational repression.
Such examples illustrate that miRNAs and RBPs cooperate to achieve additional level of control in the regulation of gene expression. This type of mechanism might be widespread to obtain the specificity, plasticity and robustness of post-transcriptional gene regulation.
In addition, miRNA biogenesis is regulated by RBPs [39,40]. Lin-28, a RBP, selectively blocks the processing of pre-let-7 miRNA [40]. This inhibition of let-7 maturation by Lin-28 is essential for maintaining the self-renewal of embryonic stem cells in development and oncogenesis [41]. Overexpression of Lin-28 in human tumors is linked to repression of let-7 family miRNAs and derepression of let-7 target genes, such as K-Ras and c-Myc, which are important for tumorigenesis [42]. In yet another example, KSRP, a key mediator of mRNA decay that interacts with the ARE motif, is found to serve as a component of both Drosha and Dicer complexes and promotes the maturation of a subset of miRNAs [39]. Thus, RBPs can regulate the biogenesis of miRNAs, thereby influencing gene expression, although in an indirect manner.
In addition to miRNAs, other examples of noncoding RNAs, such as pseudogene-encoded RNAs and antisense transcripts, have been shown to regulate gene expression [16,18]. Pseudogene mRNAs compete with mRNAs for miRNA binding, thereby modulating the derepression of miRNA targets [16]. Noncoding antisense RNAs inhibit the expression of the transcription factor PU.1 by modulating mRNA translation [18].
Thus, miRNAs, as well as other classes of noncoding RNAs, together with RBPs regulate gene expression in a widespread manner.
Endothelial miRNA regulation of angiogenesis
The first evidence implicating miRNAs in the regulation of angiogenesis comes from observations that Dicer is essential for normal development [43,44]. Gene deletion of Dicer in mice resulted in very early embryonic lethality due to a defect in gastrulation. However, a hypomorphic Dicer allele caused defective angiogenesis in the embryo proper and yolk sac, concomitant with altered expression of angiogenic regulators such as VEGF, VEGFR1, VEGFR2 and Tie1 [43,44]. Because most miRNAs need to be synthesized by the Dicer pathway, this data implied the essential role played by miRNA-dependent gene regulation in embryonic vascular development. Endothelial specific deletion of Dicer in mice showed reduced VEGF-induced angiogenesis in the mouse ear and in Lewis lung carcinoma xenografts [45]. Total reduction of miRNAs via Dicer silencing in endothelial cells in vitro induced the expression of thrombospondin-1 (TSP-1), a potent inhibitor of angiogenesis, and impaired in cellular processes important for angiogenesis such as proliferation, migration, tube formation and sprouting [45,46].
miRNA expression profiling in human umbilical vein endothelial cells (HUVECs) has been performed by several groups which revealed regulation of abundant miRNAs (Table 1) [45–49]. miRNA expression is regulated by various factors such as hypoxia, VEGF, serum and extracellular matrix-derived signals. For example, many miRNAs are induced by VEGF (miR-191, miR-296, miR-126, miR-155, miR-31 and the miR-17-92 cluster, for example) [45]. Similarly, exposure of endothelial cells to hypoxia triggers the expression of miR-210 [50], and serum induces the expression of miR-130a [47]. As described in detail below, these regulated miRNAs target key angiogenic signaling pathways in a temporal- and spatial-specific manner, leading to the alteration of the angiogenic response (Figure 3). Angiomirs, which are miRNAs that promote angiogenesis, include miR-126, miR-132, miR-130a, miR-210 and miR-296 [47,51–53]. Conversely, miR-221 and miR-222 inhibit angiogenesis [49]. In components of the miR-17-92 cluster, miR-92a serves as an endogenous repressor of the angiogenic program in endothelial cells (ECs) [54], in contrast to the proangiogenic effects of the miR-17-92 cluster in tumor angiogenesis [55]. These studies point to the potent modulatory actions of miRNAs on the angiogenic response. In particular, endothelial-specific miR-126 knockout in mice showed reduced survival and defective angiogenesis following myocardial infarction [51,52]. The induction of miR-132 [56] in the endothelial cells of human breast tumors suggests that miR-132 might be a marker of hyperproliferative or activated endothelium. Specific examples in which miRNAs regulate angiogenesis and endothelial cell functions are described below.
Table 1.
Summary of microRNA involvement in angiogenesis
| Stimulator | miRNA | Function | Targetse | Refs. |
|---|---|---|---|---|
| VEGFa | miR-132b | Inhibition of miR-132 suppresses angiogenesis and decreases tumor burden in an orthotopic xenograft mouse model. | p120RasGAP | [56] |
| miR-296b | Increases the levels of proangiogenic growth factor receptors. Inhibition of miR296 reduces angiogenesis in tumor xenograft. | HGS | [53] | |
| miR-126b | Regulates vascular integrity and angiogenesis. | PI3KR2, SPRED1 | [48,51,52] | |
| miR-92ac | Overexpression of miR-92a in EC blocks angiogenesis. Inhibition of miR-92a enhances angiogenesis and perfusion in limb ischemia and myocardial infarction. | Integrin a5 | [54] | |
| miR-221/222c | Reduce SCFd-induced EC survival/migration/tube formation. | c-Kit | [49] | |
| miR-191, miR-155, miR-31, miR-17-5p, miR-18a, miR-20a | Expressed in VEGF time-dependent manner. Functions are not known. | Not known | [45] | |
| Hypoxia | miR-210b | Enhances EC survival/migration/tube formation. | Ephrin A3 | [50] |
| Serum | miR-130ab | Enhances EC migration/tube formation. | GAX, HOXA5 | [47] |
VEGF, vascular endothelial growth factor.
Indicates miRNAs that promote angiogenesis.
Indicates miRNAs that inhibit angiogenesis.
SCF, stem cell factor.
PI3KR2, phosphatidylinositol 3 kinase regulatory subunit 2; SPRED1, sprouty-related, EVH1 domain containing 1.
Figure 3.
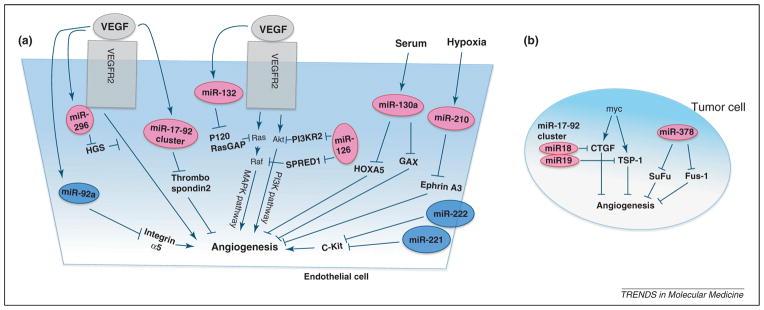
Regulation of angiogenesis by microRNAs (miRNAs). Multiple miRNAs regulate the angiogenic responses to growth factors or hypoxia. (a) Endothelial miRNAs that promote angiogenesis include miR-296, miR-132, miR-126, the miR-17-92 cluster, miR-130a and miR-210. By contrast, endothelial miRNAs that inhibit angiogenesis are miR-92a, miR-221 and miR-222. (b) The miR-17-92 cluster and miR-378 in tumor cells promote angiogenesis by repressing antiangiogenic factors. Suspected targets and regulators for each miRNAs are indicated. The vascular endothelial growth factor (VEGF) pathway can be regulated at multiple levels by miRNAs (a). miR-126 promotes VEGF signaling by repressing SPRED1, a negative regulator of the MAPK pathway, and by repressing PI3KR2, a negative regulator of the PI3K pathway. miR-132 represses the expression of p120RasGAP, which is a negative regulator of Ras and thereby induces neovascularization. miR-296 represses the expression of HGS, which regulates the intracellular sorting of activated VEGFR2 to lysosomes.
miR-296, which was induced in endothelial cells in response to VEGF or glioma-derived factors [53], repressed the expression of hepatocyte growth factor-regulated tyrosine kinase substrate, a molecule that targets ligand/receptor complexes to lysosomes for degradation. This results in the accumulation of VEGFR2 and an increased response to VEGF. An antagomir of miR-296 reduced tumor neovascularization and tumor volume in glioblastoma xenografts.
miR-126, which is expressed in endothelial and hematopoietic progenitor cells, was shown to stimulate angiogenesis and vascular integrity in vivo in response to VEGF [51,52,57]. Knockdown of miR-126 in zebrafish induced hemorrhage and collapse of lumen-containing vascular structures [51]. Endothelial specific deletion of miR-126 in mice causes leaky vessels, hemorrhage and partial embryonic lethality due to a loss of vascular integrity and defects in endothelial cell proliferation, migration and angiogenesis [52]. This proangiogenic function of miR-126 was proposed to be mediated by the repression of SPRED1 (sprouty-related, EVH1 domain containing 1) and PI3KR2 (phosphatidylinositol 3 kinase regulatory subunit 2), negative regulators of the MAPK and PI3K pathways, respectively.
miR-132 acts as an angiogenic switch by targeting p120RasGAP, a negative regulator of Ras, in tumor endothelium [56]. This miRNA is upregulated in tumor endothelium but not detectable in normal endothelium. An antagomir of miR-132 restored p120RasGAP expression in the tumor endothelium, reduced basic fibroblast growth factor induced angiogenesis, suppressed tumor angiogenesis and decreased tumor burden, suggesting a critical role for miR-132 in pathological angiogenesis.
The miR-17-92 cluster was shown to be induced by VEGF in HUVECs [45]. Inhibition of the miR-17-92 cluster in vitro reduced EC sprouting and endothelial cord formation on Matrigel [46]. Transfection of ECs with miR-18a, a component of the miR-17-92 cluster, rescued the phenotype induced by silencing of Dicer; that is, induction of TSP-1, defective EC proliferation and morphogenesis [45]. Thus, the miR-17-92 cluster might be an endogenous repressor of angiogenesis. Indeed, systemic administration of an antagomir of miR-92a led to enhanced blood vessel growth and functional recovery of damaged tissue in mouse models of limb ischemia and myocardial infarction [54].
Hypoxia, a key driver of angiogenesis, works primarily by the induction of angiogenic factors (such as VEGFA) via the prolyl hydroxylase-dependent hypoxia-inducible factor 1α pathway [1,2]. miR-210 was identified as a specific hypoxia-inducible miRNA [50], and miR-210 overexpression under normoxic conditions increased tube formation and VEGF driven cell migration, whereas inhibition of mir-210 with an antagomir under hypoxia inhibited the proangiogenic phenotype. Such proangiogenic effects of miR-210 in hypoxia appear to be mediated via suppression of ephrin A3, a repressor of VEGF signaling.
Nonendothelial miRNAs and angiogenesis
Tumor cells actively induce angiogenesis, in part, through miRNA-dependent mechanisms. One prominent example is a cluster of miRNA, miR17-92, which is termed an ‘oncomir’ and transcribed as a polycistron whose transcription is stimulated by c-myc and upregulated in tumors [58]. Overexpression of the miR17-92 cluster in oncogenic Ras expressing cells promotes tumor angiogenesis via suppression of the antiangiogenic factors TSP-1 and connective tissue growth factor [55]. In another example, miR-378 promotes tumor angiogenesis by targeting the tumor suppressors SuFu and Fus-1 [59]. Cancer cells transfected with miR-378 formed significantly larger tumors that are highly vascularized. These studies highlight the potential importance of miRNA-dependent mechanisms in tumor cells that control angiogenesis by regulating the expression of angiogenic regulators.
Critical angiogenic factors such as VEGF appear to be regulated by miRNAs in nasopharyngeal carcinoma cells in a complex, multifaceted manner. First, miRNAs with independent binding sites can cooperate to regulate VEGF. Second, multiple miRNAs compete with each other for a common binding site. Third, miRNAs coregulate a group of angiogenic regulatory genes, thereby coordinating the angiogenic response. And finally, via differential regulation, where a gene with multiple binding sites for several miR-NAs can be regulated by discrete miRNAs in differing cells under different conditions [60].
RBPs and angiogenesis
Many cytokines and growth factor mRNAs contain various regulatory elements, including AREs in their 3′UTRs, and are regulated at the post-transcriptional level [29,30]. Their production is controlled by the ARE-BPs, with opposing stabilizing and destabilizing activities. In addition, RBP interactions with mRNAs can also regulate translation of mRNAs. For example, VEGF, a critical regulator of angiogenesis, contains a functional ARE motif in its 3′UTR [31]. The ARE-BP Zfp36 (also known as Tis11 or TTP) induces VEGF mRNA degradation by interacting with the ARE. However, upon TNFα stimulation, phosphorylated Zfp36 loses its affinity for the VEGF mRNA and allows the interaction of other RBPs, such as HuR with the VEGF mRNA, thereby enhancing VEGF expression [32,61–63]. An analogous phenomenon was found for a related RBP called Zfp36l1 (also known as Tis11b) [64]. In addition to the regulation of RNA stability, VEGF mRNA is also regulated by hypoxic and inflammatory conditions at the level of translation [31,61]. RBPs such as hnRNP L and a translational regulatory complex called the GAIT complex influence the conformational switch of the VEGF mRNA into translationally competent and translationally repressive states, respectively [65]. This phenomenon is called riboswitch-dependent translational regulation.
Some of these ARE-BPs are shown to be required for normal vascular development and hematopoiesis in vivo (Table 2). Zfp36l1-deficient mice experience lethality at E10.5 with vascular abnormalities and heart defects [66]. Areas of hemorrhage, lack of vascular organization and the paucity of Ter119+ erythroid cells in the yolk sac of Zfp36l1−/− embryos indicate its requirement for extraembryonic vasculogenesis as well as the expansion and differentiation of erythroid cells. An enlarged dorsal aorta and disorganized endocardium in Zfp36l1−/− embryos suggest the role of the RBP Zfp36l1 in the normal development and differentiation of the intraembryonic vasculature and the heart. This abnormal vascular defect is correlated with elevated VEGF expression due to increased VEGF mRNA loading onto polysomes. This suggested a role for Zfp36l1 in the translational repression of VEGF mRNA.
Table 2.
Summary of ARE-BP involvement in angiogenesis
| ARE-BP | mRNA targets | miRNAs | Knockout mice | KO phenotypes | Refs. |
|---|---|---|---|---|---|
| Zfp36 (TTP) | VEGFa, COX2a, c-fos, GM-CSF, TNF-α, IL-2, IL-3 | miR-16c, miR-182, miR-27, miR-29 | Normal at birth but develop autoimmune diseases within 1–8 weeks | Defective hematopoiesis, cachexia, arthritis and autoimmunity | [31,69,79] |
| Zfp36l1 | VEGFa, Stat5b | Die at E10.5 days | Vascular abnormalities, defective erythroid differentiation | [66] | |
| Zfp36l2 | Cxcl1b, Plekha3b, Mllt11b | Die within 2 weeks after birth | Defective hematopoiesis | [67] | |
| Zfp36l1 36l2 | Notch1 | Specific double knockout (dKO) (CD2Cre); 90% of dKO die by 6 months | Perturbed thymopoiesis T-ALLd | [80] | |
| HuR | HIF-1αa, VEGFa, COX2a, TSP1a, c-fos, p21, cyclin A, cyclin D, NOSII, SIRT1, GMCSF, TNFα, Bcl2, c-myc, CAT-1, MKP-1, p53, uPA, uPAR | let-7c, miR-122c, miR-9, miR-133, miR-124, miR-19, miR-182, miR-146, miR-324-5p | Global KO
|
|
[31,70,71, 72,79] |
| AUF1 | COX2a, TNFα, IL-1β | miR-200, miR-141, miR-146 | Normal development | Vascular hemorrhage, coagulation and high mortality under endotoxic challenge | [31,68,79] |
Indicates angiogenic responsive genes.
Indicates the target candidates for Zfp36l2 screened by microarray. All other mRNA targets except b are validated to bind with each ARE-BP.
miRNAs that are validated to interact with each ARE-BP. All other miRNAs are the predicted target by Targetscans, MiRNanda, PicTar [79].
T-ALL: T cell acute lymphoblastic leukemia.
Zfp36l2 (also known as Tis11D) has an essential role in definitive hematopoiesis during mouse development [67]. Zfp36l2 knockout mice die within 2 weeks of birth and exhibit pancytopenia, decreased hematopoietic progenitor cells in the fetal liver and yolk sac, suggesting a key function for post-transcriptional mechanisms in hematopoiesis.
Zfp36 and AUF1 mutant mice have been characterized and revealed the importance of these RBPs in the regulation of the inflammatory response, which might indirectly affect endothelial cell functions and angiogenesis [68,69]. For example, Zfp36 knockout mice appear normal at birth but soon develop several abnormalities such as cachexia and inflammatory arthritis which is primarily due to the stabilization of the TNFα mRNA, leading to TNFα overproduction [69]. By contrast, AUF1 knockout mice did not show major developmental defects other than a smaller size and reduced body weight [68]. However, upon endo-toxin challenge, they exhibited increased intravascular coagulation, capillary leakage and high mortality. The mechanism was attributed to the deregulation of ARE-mRNA stability of inflammatory cytokines such as TNFα and interleukin 1β (IL-1β), which are normally destabilized by AUF1.
In addition to the regulation of the inflammatory response, overexpression of Zpf36 in Ras-dependent tumor cells reduced vascularization with delayed tumor growth in tumor xenografts concomitant with destabilization of VEGF mRNA [32]. Thus, Zpf36 and AUF1-dependent suppression of cytokines and growth factors have a major impact on the inflammatory and angiogenic responses.
HuR (also known as Elavl1) knockout mice are lethal due to a defect in placentation [70]. Inducible deletion of Elavl1 postnatally induced a rapid apoptosis of hematopoietic and intestinal progenitor cells which led to lethality within 10 days [71]. HuR binds and stabilizes Mdm-2, a negative regulator of p53, to keep p53 levels in check in proliferating progenitor cells. By contrast, tissue-specific deletion of Elavl1 in thymocytes resulted in the defective egress of mature thymocytes from the thymus [72]. The function of HuR in vascular cells has not been analyzed.
These studies suggest that RBPs have profound effects on vascular development and function. In many cases, specific mechanisms involved as well as interactions with other signaling pathways are not well understood.
Concluding remarks and perspectives
miRNAs and RBPs are post-transcriptional gene regulators that bind mRNA, regulate mRNA stability and translation in vascular cells, and thus influence angiogenesis. Through the analysis of individual miRNAs and RBPs as well as the machinery involved in miRNA generation, it is clear that post-transcriptional mechanisms play a major role in the angiogenic response by modulating key regulators including cytokines, growth factors and signaling intermediates. Both vascular cell intrinsic as well as paracrine mechanisms via neighboring cells are involved. This new mechanistic knowledge is expanding our approaches in the therapeutic control of angiogenesis in oncology as well as in various other diseases, including rheumatoid arthritis and psoriasis.
Recent studies also reveal that exosome-mediated transfer of mRNAs and miRNAs occurs in vivo, allowing the exchange of genetic material between cells [73]. Indeed, purified exosomes from glioblastoma deliver functional RNA to endothelial cells and stimulate angiogenesis in vitro [74]. Exosomes might be useful as both serum biomarkers and as delivery vehicles for therapeutic RNAs.
Although not covered in this review, differential alternative splicing events in cancer and angiogenesis are increasingly being identified [75,76]. hnRNP proteins including pyrimidine tract binding protein, hnRNP A1 and hnRNP A2 regulate alternative splicing events required for tumor cell proliferation [75]. In addition, the alternative splicing of VEGF into pro- and antiangiogenic isoforms is often deregulated in diseases such as cancer [76]. It is probable that RBPs and miRNAs are involved in these alternative splicing events. In fact, the targeted deletion of Dicer in adult mouse myocardium revealed the role of miRNAs in maintaining the adult splicing program [77].
Future challenges in this emerging field of vascular biology include the characterization of interactions between miRNAs and RBPs on key, regulatory mRNAs. This is necessary for a more complete understanding of the regulation of the angiogenic process. In addition, it is not clear how extracellular cues influence miRNA and RBP-dependent gene expression regulation. For example, even though it is known that some RBPs are phosphorylated by signal-activated MAPK signaling to ultimately affect cytokine expression [32,63,69], such mechanistic information is not available for angiogenic regulators. Furthermore, if critical factors of angiogenesis are indeed regulated by cooperative interactions between RBPs and miRNAs, targeting both pathways might achieve a more effective control of angiogenesis. Indeed, RNA-based therapeutics to control VEGF levels have been successfully implemented in ophthalmology [78]. Further study in this area will likely yield novel therapeutic agents to control angiogenesis in many diseases.
Acknowledgments
This work is supported by National Institutes of Health grants HL49094 and HL89934 to T.H.
Glossary
- Angiocrine factors
factors that are secreted from endothelium to regulate neighboring tissue responses
- Angiogenesis
the process of new blood vessel formation from pre-existing vessels
- Angiomirs, specific miRNAs that regulate angiogenesis
Antagomir, a small synthetic RNA used to silence endogenous miRNA. It is complementary to the specific miRNA target, with modifications to make it resistant to degradation
- Cachexia
a condition of general ill-health, wasting and malnutrition usually associated with cancer or chronic disease
- Dicer
a class 3 ribonuclease III that cleaves a dsRNA and a pre-miRNA stem loop into short dsRNA fragments to produce small interfering RNAs and miRNAs
- Drosha
a class 2 ribonuclease III that bind to dsRNA and processes a pri-miRNA to pre-miRNA
- Endocardium
endothelial cells and connective tissue that line the inner layer of heart chambers
- Endothelial cord
a group of aggregated endothelial cells that form string-like structures in a network, often forming a honeycomb pattern. This pattern is often observed when endothelial cells are cultured on Matrigel
- Endotoxin
lipopolysaccharide from the cell walls of Gram-negative bacteria which induces the inflammatory response
- Hematopoiesis
formation and development of blood cells from hematopoietic stem cells
- Limb ischemia
lack of blood flow to a limb leading to reduced oxygenation of tissue
- Myocardial infarction
occlusion of the coronary vessels leading to ischemic injury of the myocardium
- Nonthrombogenic
prevents the formation of blood clots
- Normoxic
homeostatic oxygen tension of the vascularized tissue
- Oncomir
miRNA that is expressed in tumors and functions to promote tumorigenesis
- Pancytopenia
a deficiency of all types of blood cell lineages
- Paracrine
a form of cell-to-cell signaling in which a signal released from one cell, or cells, acts on neighboring cells
- Pericytes
cells that are closely associated with endothelial cells at the extraluminal side of capillaries, arterioles or venules
- Polysomes
a cluster of ribosomes along a single strand of messenger RNA during the process of protein synthesis
- RISC (RNA-induced silencing complex)
the ribonucleoprotein complex, consisting of one strand of miRNA, the argonaute protein (AGO) and other accessory factors. It facilitates RNA silencing of mRNAs by suppressing translation and/or degradation
- Vascular tone
state of contractile tension in the vessel wall, which is determined by the balance of vasoconstriction and vasodilatation
- Vasculogenesis
the process of blood vessel formation by de novo differentiation of progenitor cells into endothelium
References
- 1.Fraisl P, et al. Regulation of angiogenesis by oxygen and metabolism. Dev Cell. 2009;16:167–179. doi: 10.1016/j.devcel.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 3.Ding BS, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rafii S, Lyden D. Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration. Nat Med. 2003;9:702–712. doi: 10.1038/nm0603-702. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 6.Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flamme I, et al. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J Cell Physiol. 1997;173:206–210. doi: 10.1002/(SICI)1097-4652(199711)173:2<206::AID-JCP22>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 8.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain RK. Molecular regulation of vessel maturation. Nat Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 10.Mammoto A, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyden D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 12.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 13.Lukong KE, et al. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Nicoli S, et al. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Olson EN. AngiomiRs – key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eiring AM, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebralidze AK, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008;22:2085–2092. doi: 10.1101/gad.1654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farina KL, et al. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol J, et al. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 21.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipowicz W, et al. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 24.Guo H, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 26.Vasudevan S, et al. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 27.Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest. 2010;40:370–374. doi: 10.1111/j.1365-2362.2010.02279.x. [DOI] [PubMed] [Google Scholar]
- 28.Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- 29.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 30.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda K, et al. RNA-binding proteins implicated in the hypoxic response. J Cell Mol Med. 2009;13:2759–2769. doi: 10.1111/j.1582-4934.2009.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essafi-Benkhadir K, et al. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol Biol Cell. 2007;18:4648–4658. doi: 10.1091/mbc.E07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lafarga V, et al. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobsen A, et al. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–1019. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya SN, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 36.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 37.Kim HH, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jafarifar F, et al. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30:1324–1334. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan SR, et al. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viswanathan SR, Daley GQ. Lin28: a microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 44.Yang WJ, et al. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 45.Suarez Y, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuehbacher A, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris TA, et al. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poliseno L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 50.Fasanaro P, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wurdinger T, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonauer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 55.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anand S, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivey KN, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee DY, et al. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hua Z, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levy AP. Hypoxic regulation of VEGF mRNA stability by RNA-binding proteins. Trends Cardiovasc Med. 1998;8:246–250. doi: 10.1016/s1050-1738(98)00020-6. [DOI] [PubMed] [Google Scholar]
- 62.Levy NS, et al. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 63.Suswam E, et al. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res. 2008;68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- 64.Ciais D, et al. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 65.Ray PS, et al. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bell SE, et al. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn. 2006;235:3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 67.Stumpo DJ, et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu JY, et al. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 70.Katsanou V, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghosh M, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest. 2009;119:3530–3543. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papadaki O, et al. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 73.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 74.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.David CJ, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ladomery MR, et al. Alternative splicing in angiogenesis: the vascular endothelial growth factor paradigm. Cancer Lett. 2007;249:133–142. doi: 10.1016/j.canlet.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 77.Kalsotra A, et al. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campochiaro PA. Potential applications for RNAi to probe pathogenesis and develop new treatments for ocular disorders. Gene Ther. 2006;13:559–562. doi: 10.1038/sj.gt.3302653. [DOI] [PubMed] [Google Scholar]
- 79.Asirvatham AJ, et al. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodson DJ, et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]