Summary
Transition to flowering in plants is tightly controlled by environmental cues, which regulate the photoperiod and vernalization pathways, and endogenous signals, which mediate the autonomous and gibberellin pathways. In this work, we investigated the role of two Zn2+-finger transcription factors, the paralogues AtVOZ1 and AtVOZ2, in Arabidopsis thaliana flowering. Single atvoz1-1 and atvoz2-1 mutants showed no significant phenotypes as compared to wild type. However, atvoz1-1 atvoz2-1 double mutant plants exhibited several phenotypes characteristic of flowering-time mutants. The double mutant displayed a severe delay in flowering, together with additional pleiotropic phenotypes. Late flowering correlated with elevated expression of FLOWERING LOCUS C (FLC), which encodes a potent floral repressor, and decreased expression of its target, the floral promoter FD. Vernalization rescued delayed flowering of atvoz1-1 atvoz2-1 and reversed elevated FLC levels. Accumulation of FLC transcripts in atvoz1-1 atvoz2-1 correlated with increased expression of several FLC activators, including components of the PAF1 and SWR1 chromatin-modifying complexes. Additionally, AtVOZs were shown to bind the promoter of MOS3/SAR3 and directly regulate expression of this nuclear pore protein, which is known to participate in the regulation of flowering time, suggesting that AtVOZs exert at least some of their flowering regulation by influencing the nuclear pore function. Complementation of atvoz1-1 atvoz2-1 with AtVOZ2 reversed all double mutant phenotypes, confirming that the observed morphological and molecular changes arise from the absence of functional AtVOZ proteins, and validating the functional redundancy between AtVOZ1 and AtVOZ2.
Key words: VOZ, Flowering, Arabidopsis, FLC, MOS
Introduction
Flowering plants possess an intricate regulatory network to properly time the transition to flowering. Floral initiation has evolved over time to optimize the reproductive success of plants in a variety of environments. Flowering is induced in response to environmental and endogenous cues. Genetic studies have defined four major pathways that sense and respond to flowering cues: the photoperiod, autonomous, vernalization, and gibberellin (GA) pathways (Simpson and Dean, 2002; Srikanth and Schmid, 2011). These pathways are tightly connected and converge on a small number of flowering integrators.
Many genes that specifically regulate flowering time have been identified in Arabidopsis thaliana. Mutations in genes that promote flowering result in a late-flowering phenotype. Two major groups of delayed mutants have been categorized in flowering-time studies based on responsiveness to day length (Rédei, 1962; Koornneef et al., 1991). In the first group, late-flowering mutants that are delayed only in long days (LDs) but not in short days (SDs) were proposed to affect the day length sensing pathway, which functions to initiate flowering following exposure to inductive photoperiods. These photoperiod-pathway mutants involve a set of genes that promotes flowering through inductive long-day photoperiods. On the other hand, late-flowering mutants that are delayed in both LDs and SDs involve genes that enable flowering independently of day length. These mutants impact a flowering pathway designated as the autonomous pathway (Rédei, 1962; Koornneef et al., 1991; Simpson and Dean, 2002). In many plant species flowering cannot happen until a progression has been made from juvenile to adult phase through the autonomous pathway, resulting in competence to flower (Poethig, 2003).
Prolonged exposure to cold is known to render plants competent to flower. This process, known as vernalization, is required by winter-annual types of Arabidopsis to prevent flowering in the fall season and promote it in the spring. The vernalization response is conferred by dominant alleles of FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) (Koornneef et al., 1991; Lee and Amasino, 1995; Michaels and Amasino, 1999; Schmitz and Amasino, 2007). The flowering inhibition is largely a result of FRI-mediated upregulation of FLC, which encodes a potent suppressor of flowering. Prolonged cold turns off FLC expression and renders the plant capable of undergoing floral transition. Plants that contain FLC/FRI exhibit delayed flowering that is reversed completely by vernalization (Lee and Amasino, 1995). The late flowering of FRI-carrying lines is phenotypically similar to that of mutants in the autonomous pathway, as their late flowering is abolished by loss of FLC function or vernalization (Koornneef et al., 1991; Michaels and Amasino, 1999; Sheldon et al., 1999; Michaels and Amasino, 2001). While FRI serves to upregulate FLC and repress flowering, the genes in the autonomous pathway, such as FCA, FPA, FVE, FLOWERING LOCUS D (FLD), FLOWERING LOCUS K HOMOLOGY DOMAIN (FLK), LUMINIDEPENDENS (LD) and FY function to promote flowering by suppressing FLC (Lim et al., 2004). The FRI complex is known to supersede the repressive effects of the autonomous pathway (Schmitz and Amasino, 2007).
In this study, we analyzed the involvement of two transcription factors, AtVOZ1 and AtVOZ2, in Arabidopsis flowering and development. Our results show that AtVOZs regulate flowering time and other aspects of plant growth and development. Double atvoz1-1 atvoz2-1 mutant plants exhibit severely delayed flowering time, which correlates with increased mRNA levels of the floral repressor FLC and several FLC activators, as well as MOS3/SAR3, a nuclear pore protein known to play a role in transition to flowering. Both late flowering and upregulation of FLC are completely reversed by vernalization. Double atvoz1-1 atvoz2-1 mutants display additional phenotypes: they are smaller than wild-type (WT) plants, show signs of senescence, exhibit delayed transition from juvenile to adult phase, and show defects in seed production.
Materials and Methods
Plant materials
Experiments were carried out with Arabidopsis ecotype Col-0 (WT) and mutant lines in the same ecotype. Seeds of putative T-DNA insertion mutants of AtVOZ1 (atvoz1-1, WiscDsLox489-492010) and AtVOZ2 (atvoz2-1, salk_115813c) were obtained from ABRC, Ohio State University (http://www.arabidopsis.org) and homozygous lines were identified using genomic PCR and RT-PCR with gene-specific and T-DNA-specific primers listed in supplementary material Table S3. The double atvoz1-1 atvoz2-1 mutant was generated by crossing and identified in the segregating population by PCR with gene-specific and T-DNA-specific primers. To create AtVOZ2; atvoz1-1 atvoz2-1, the atvoz1 atvoz2 double mutant was complemented with a pC-TAP-VOZ2 construct driven by the 35S promoter using the Agrobacterium-mediated transformation (Clough and Bent, 1998; Rubio et al., 2005) and the expression of pC-TAP-VOZ2 was detected with the TAP-specific primer listed in supplementary material Table S3.
Growth conditions, vernalization treatment, phenotypic analyses
Arabidopsis plants were grown in Pro-mix BX Mycorise soil (Premier) in growth chambers with cold fluorescent light under controlled conditions (temperature 22°C, 80% relative humidity and 90–110 µmol photons m−2 s−1 light intensity). Long-day photoperiods consisted of 16 hours of light and 8 hours of dark and short-day conditions were 8 hours of light and 16 hours of dark. To study the effect of vernalization on flowering, seeds were vernalized for 12 weeks and then transferred to normal growth conditions in the growth chamber. Flowering time was measured by recording the number of days from sowing to opening of the first flower and by counting the total number of rosette leaves at bolting. Three experimental replications were performed and statistically analyzed (20 to 60 plants per replication). Trichome analysis was performed on plants as they opened the first flower, with the help of a dissecting microscope. Three replications were performed (17 to 30 plants each) and statistically analyzed. DAPI (4′,6-diamino-phenylindole) staining of pollen and microscopic analysis was performed as described previously (Golovkin and Reddy, 2003). Flowers, anthers, siliques and seeds were visualized and abortive seed count was performed under a dissecting microscope.
Expression analysis of flowering genes
Transcript levels of flowering-time genes were measured in 28-day-old WT and mutant plants by RT-PCR. Aerial parts were collected from plants grown in LD conditions 10 hours after dawn. Total RNA was extracted by using the Omega RNA extraction kit (Omegabiotek). After treatment with RNase-free DNase I (Fermentas) to remove any contaminating genomic DNA, cDNA was synthesized with Superscript II RNase H− reverse transcriptase (Invitrogen) and subjected to PCR. Specific primers for FLC, VIP3, ELF7, PIE1, SEF, FD, FT, CO and MOS3 are described in supplementary material Table S3. Bands were visualized by agarose-gel electrophoresis.
Protein purification
AtVOZ2 in pET-32a (N-terminally His-tagged) and AtVOZ2m in pET-28a (N-terminally His-tagged) were expressed in Escherichia coli BL21 at 30°C by adding IPTG (1 mM final concentration) when the culture density reached OD600∼0.6. Following 4 hours of incubation, bacteria were harvested by centrifugation for 10 minutes at 4°C (5000 g). Pellets were resuspended in Bind/Wash buffer (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.1% Triton X-100) containing 1× Complete protease inhibitor cocktail without EDTA (Roche) and sonicated. The lysate was centrifuged (4°C, 10 minutes, 15000 g) and the supernatant was filtered through a 0.45 µm filter (Life Science Products, Inc.). AtVOZ2 and AtVOZ2-m proteins were isolated from the filtrate by binding to His•Bind resin (Novagen) following manufacturer's instructions. Purified proteins were desalted by centrifugation with Amicon Ultra PL-10 Centrifugal Filter Devices (Millipore) and stored in 50 mM Tris-HCl, pH 8. Protein concentration was measured by using the Bio-Rad Protein Assay (Bio-Rad).
Electrophoretic mobility shift assay (EMSA)
Fluorescently labeled pMOS3 and pmos3-m probes that were used to test the interaction of AtVOZ2 with the MOS3/SAR3 promoter were prepared by mixing equal volumes of the sense oligonucleotide (0.1 nmol/µl in 10 mM Tris-HCl [pH 8.0]) and the Cy5-labeled antisense oligonucleotide (0.1 nmol/µl in 10 mM Tris-HCl [pH 8.0]) (Integrated DNA technologies), then heating the mixture at 65°C for 5 minutes and slow-cooling it to 30°C. The oligonucleotides used to prepare the pMOS3 probe were sense (5′-GACGTCCGGCGCAGCGTTTATCAGACGCTGGGATTAAAACA-3′) and the Cy5-labeled antisense oligonucleotide (5′-/Cy5/TGTTTTAATCCCAGCGTCTGATAAACGCTGCGCCGGACGTC-3′). To prepare the pmos3-m probe the mutated sense (5′-GACGTCCGGCGCATTTTTTATTTTTTTTTGGGATTAAAACA-3′) and mutated antisense (5′-/Cy5/TGTTTTAATCCCAAAAAAAAATAAAAAATGCGCCGGACGTC-3′) probes were used.
EMSA reaction mixtures (10 µl final volume) with purified AtVOZ2 and AtVOZ2m proteins and pMOS3 and pmos3-m promoter probes were combined in microfuge tubes on ice in the following order: 6 µl water, 1 µl 5× EMSA buffer (5× EMSA buffer: 20 mM HEPES [pH 7.9], 500 mM KCl, 40% glycerol [v/v], 25 mM EGTA [pH 8.0], 0.5 mM DTT, 1.25 mM ZnCl2. DTT and ZnCl2 were added to 5× buffer immediately before use), 2 µl purified protein (up to 2.4 µg), 1 µl fluorescently labeled probe (at 2 or 20 pmole/µl). Reactions were incubated at room temperature for 30 minutes. Loading dye (25 mM Tris-HCl [pH 7.5], 4% [v/v] glycerol, 0.02% [w/v] Bromophenol blue) was added (1 µl) and the samples were run immediately on 6% non-denaturing acrylamide gels for 1 hour at 100 V in TAE buffer. Fluorescent bands on gels were visualized using the Typhoon Trio imaging system.
Chromatin immunoprecipitation (ChIP)
ChIP analyses were performed as described previously with minor changes (Du et al., 2009). In short, protoplasts from ∼4-week-old WT plants were transfected with 40 µg of YFP control, AtVOZ1-YFP or AtVOZ2-YFP plasmids and cross-linked. Cross-linking and DNA isolation were done as before (Du et al., 2009). The sheared lysates were pre-cleared with agarose beads and incubated with 60 µl anti-GFP beads (D153-8, MBL) for 6 hours at 4°C. Beads were washed five times, resuspended in elution buffer and cross-linking was reversed by heating at 65°C for 12 hours. The immunoprecipitated DNA was purified using a DNA purification kit (Qiagen) and used as a template in PCR reactions (28 cycles) with MOS3 UE-F (5′-AGGAGGGAAAACGAATTGAGTC-3′) and MOS3 UE-R (5′-CCGAATTCCTTTCCAATTAAAGTCAAC-3′) that flanked the GCGTTTATCAGACGC sequence in the MOS3/SAR3 promoter. PCR amplification using primers in the actin-2 promoter served as a control for ChIP specificity; ACT2 UE-F (5′-GCCATCAAAGCAAAAGAACTAATC-3′) and ACT2 UE-R (5′-ATGAATTTATATAGGCGGGTTTATCTC-3′).
Results
AtVOZ transcription factors regulate transition to flowering
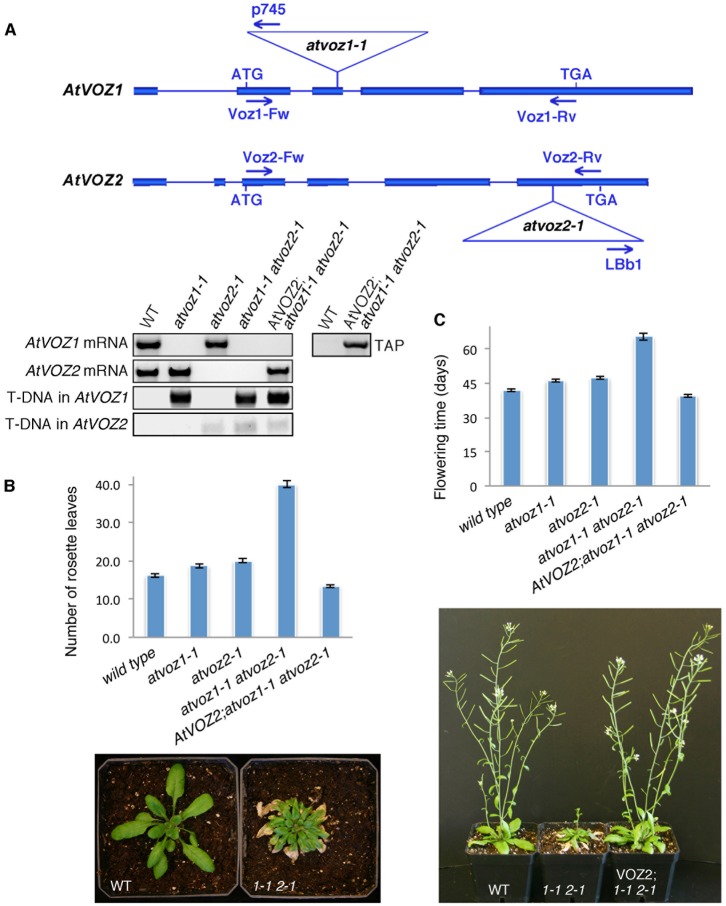
Arabidopsis Zn2+-finger transcription factors AtVOZ1 and AtVOZ2 share high sequence similarity and contain an N-terminal region with transactivating activity, and a C-terminal region that functions as the DNA-binding domain comprising a Zn2+-finger motif and a conserved basic region structurally similar to the NAC domain (Mitsuda et al., 2004; Jensen et al., 2010). To investigate the role of AtVOZs in plant growth and development, three Arabidopsis T-DNA insertion lines were examined: the single mutants atvoz1-1 (WiscDsLox489-492010) and atvoz2-1 (salk_115813c) obtained from ABRC and the double mutant atvoz1-1 atvoz2-1 generated by crossing single mutant lines (Fig. 1A). When plants were grown in long-day conditions (16 hours light, 8 hours dark), a severe delay in flowering time was observed for the double atvoz1-1 atvoz2-1 mutants compared to wild type (WT; ecotype Col-0) (Fig. 1B,C; supplementary material Table S1). In contrast, initiation of flowering differed only slightly, although significantly (P<0.05) for single mutants compared to WT, suggesting functional redundancy between the AtVOZ proteins (Fig. 1B,C, top panels; supplementary material Table S1). The number of rosette leaves at bolting was 2.5-fold higher (an average of 23.8 more leaves) for atvoz1-1 atvoz2-1 than for WT (Fig. 1B; supplementary material Table S1). Moreover, the flowering time measured as days to opening of the first flower was greatly delayed for the double mutant, which flowered at 65.5±1.5 days or 23.4 days later than WT (Fig. 1C; supplementary material Table S1). At that age, WT plants were fully mature with the mean height of 7.8±1.2 inches (n = 70) and producing seeds (Fig. 1C, bottom panel). In short days (8 hours light, 16 hours dark), the flowering time of atvoz1-1 atvoz2-1 double mutant (153.9±7.4 days) was also strongly delayed in comparison to WT (112.0±3.4 days) (supplementary material Table S1). Interestingly, in spite of the greatly delayed flowering time, the atvoz1-1 atvoz2-1 leaf count in SDs was similar to that of WT plants (supplementary material Table S1), suggesting that overall slower growth of the double mutant may have contributed to delayed flowering time in SD. Complementation of the double mutant line with AtVOZ2 (AtVOZ2; atvoz1-1 atvoz2-1) reversed all double mutant phenotypes in LDs and SDs (Fig. 1; supplementary material Table S1). In fact, in long days AtVOZ2 overexpression in atvoz1-1 atvoz2-1 resulted in slightly precocious flowering at fewer leaves than WT. These complementation results confirmed that the observed changes for atvoz1-1 atvoz2-1 were due to lack of AtVOZs and corroborated that these proteins overlap in function. Together, our results show that AtVOZs are important regulators of transition to flowering.
Fig. 1. AtVOZ proteins are activators of floral transition in Arabidopsis.
(A) Top: diagrammatic representation of AtVOZ1 and AtVOZ2 genes. Exons, blue rectangles; introns, blue lines. T-DNA insertion sites in atvoz mutants are indicated by triangles. Bottom: genotyping of atvoz1-1 and atvoz2-1 single mutants, atvoz1-1 atvoz2-1 double mutant, and AtVOZ2; atvoz1-1 atvoz2-1 complemented line with gene-specific or T-DNA-specific primers indicated in the above diagram. TAP-specific primer was used to detect expression of CTAP-AtVOZ2 in the complemented line. (B) Top: number of rosette leaves at bolting for WT, atvoz single and double mutants and complemented line under LD conditions. Mean values±s.e.m. are shown. Bottom: phenotypic comparison of WT and atvoz1-1 atvoz2-1 (1-1 2-1) plants at the beginning of bolting. Increased leaf number and extensive senescence is seen for atvoz1-1 atvoz2-1. (C) Top: flowering time, measured as number of days to opening of the first flower, for WT, atvoz single and double mutants and complemented line under LD conditions. Mean values±s.e.m. are shown. Bottom: phenotypic comparison of 9-week-old WT, atvoz1-1 atvoz2-1 (1-1 2-1) and complemented (VOZ2;1-1 2-1) plants at the time of opening of the first atvoz1-1 atvoz2-1 flower.
AtVOZ proteins function in early Arabidopsis development
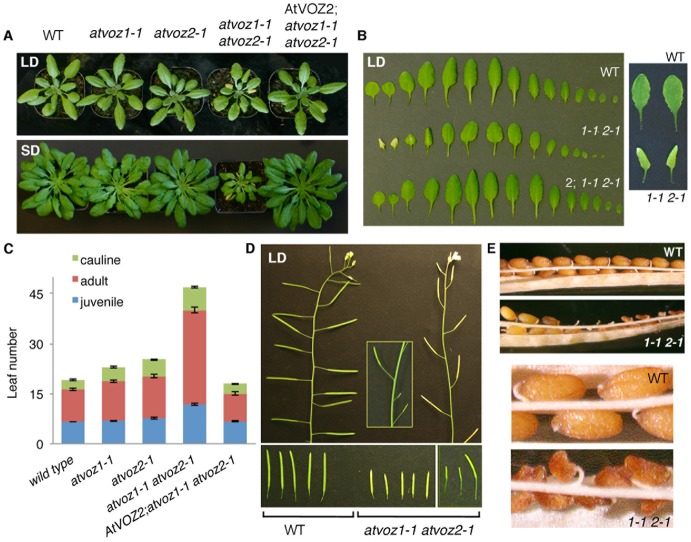
Aside from changes in flowering time, atvoz1-1 atvoz2-1 mutant plants displayed other pleiotropic phenotypes, indicating that AtVOZs perform additional functions in Arabidopsis growth. In early vegetative development, double atvoz1-1 atvoz2-1 mutants appeared smaller than WT, particularly under short-day conditions, with some of their leaves showing senescence (Fig. 2A). Senescence became more extensive and apparent as plants aged (Fig. 1B). Some of the leaves also curled into a distinctive funnel shape (Fig. 2B, right panel). atvoz1-1 atvoz2-1 leaves mostly had a round lamina and were smaller, which is characteristic of juvenile leaves (Fig. 2B, left panel). Indeed, analysis of leaf trichomes revealed a greater number of juvenile leaves (i.e. those without abaxial trichomes) in the double mutant compared to WT, as well as a large overall increase in the number of juvenile, adult (i.e. with abaxial trichomes) and cauline leaves at the time of flowering in LD conditions (Fig. 2C). The delay in transition from juvenile to adult leaves as compared to WT suggests that AtVOZs influence the vegetative phase change in Arabidopsis. Expectedly, single atvoz1-1 and atvoz2-1 mutants looked similar to WT and displayed only slight trichome and leaf count differences (Fig. 2A,C; supplementary material Fig. S1). AtVOZ2-complementation of the double mutant reversed all phenotypes (Fig. 2A–C).
Fig. 2. Pleiotropic phenotypes of atvoz1-1 atvoz2-1.
(A) Phenotypic comparison of WT, atvoz1-1 and atvoz2-1 single mutants, atvoz1-1 atvoz2-1 double mutant, and AtVOZ2; atvoz1-1 atvoz2-1 complemented line in LDs (4-week-old plants) and SDs (12-week-old plants). (B) Left: leaf spread of 4-week-old WT, atvoz1-1 atvoz2-1 (1-1 2-1) and AtVOZ2-complemented atvoz1-1 atvoz2-1 mutant (2; 1-1 2-1). Right: characteristic funnel shape of atvoz1-1 atvoz2-1 leaves. (C) atvoz1-1 atvoz2-1 displays increased number of juvenile, adult and cauline leaves in LDs. Mean values±s.e.m. are shown. (D) Top: shorter siliques of atvoz1-1 atvoz2-1 compared to WT. Inset: some atvoz1-1 atvoz2-1 plants display an alternating medium-size and very short siliques. Bottom: spread of WT (left) and atvoz1-1 atvoz2-1 siliques (middle, right). (E) Top: atvoz1-1 atvoz2-1 siliques contain both viable and abortive seeds. Bottom: close-up of WT seeds and atvoz1-1 atvoz2-1 abortive seeds.
AtVOZ proteins affect silique and seed development
Double atvoz1-1 atvoz2-1 mutants also exhibited changes in the reproductive phase following the flowering. atvoz1-1 atvoz2-1 siliques were smaller than WT siliques (9.7±1.7 mm compared to 14.5±1.2 mm, n>100) and consequently contained fewer seeds (Fig. 2D; supplementary material Table S2). A large fraction of double mutant seeds were non-viable (Fig. 2E). Individual siliques on a single plant contained variable numbers of seed abortants, ranging from siliques holding mostly or exclusively viable seeds to siliques containing only abortants or unfertilized ovules. The percentage of abortive seeds varied considerably from one plant to another and between experiments, but an increase in seed abortants was always observed in atvoz1-1 atvoz2-1 compared to WT plants. Measurements from one experiment are presented in supplementary material Table S2. In contrast to silique differences, inspection of flower architecture (i.e. number and arrangement of floral organs and flower size) under the dissecting microscope showed no dissimilarities between atvoz1-1 atvoz2-1 and WT.
AtVOZs control expression of FLC and FLC regulators
To investigate the molecular mechanisms behind the observed phenotypic changes of atvoz1-1 atvoz2-1 plants, we examined mRNA levels of several flowering-time genes in the atvoz1-1 atvoz2-1 mutant. Expression of the flowering repressor FLC was strongly increased in the double mutant, in agreement with the late flowering phenotype of atvoz1-1 atvoz2-1 (Fig. 3; supplementary material Fig. S2). Upregulation of FLC was reversed in double knockout plants complemented with AtVOZ2 (Fig. 3; supplementary material Fig. S2).
Fig. 3. AtVOZs control the FLC flowering pathway.
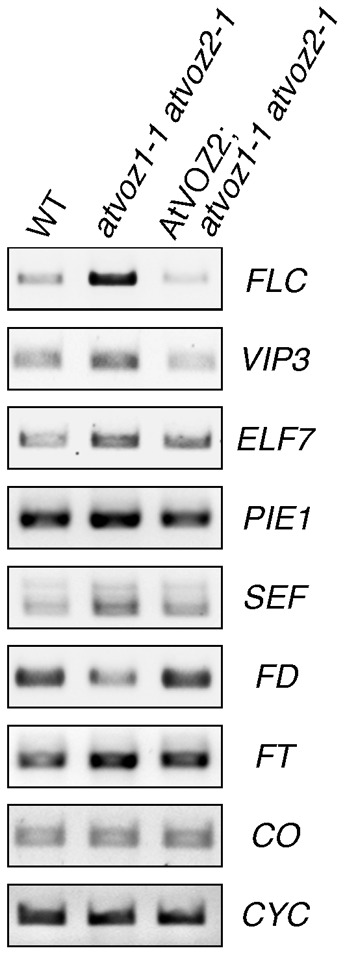
Expression of flowering genes in WT, atvoz1-1 atvoz2-1 and AtVOZ2; atvoz1-1 atvoz2-1 plants, measured by RT-PCR. Cyclophilin (CYC): template control.
A number of genes have been shown to control FLC expression. We examined transcript levels of several FLC regulators in atvoz1-1 atvoz2-1 (Fig. 3). Expression of VERNALIZATION INDEPENDENCE3 (VIP3) and EARLY FLOWERING7 (ELF7) was increased in the atvoz double mutant. VIP3 and ELF7 encode proteins that are related to components of the yeast PAF1-like complex, known to associate with RNA polymerase II and direct histone modifications by histone methyltransferases (He et al., 2004; Oh et al., 2004). Modifications of FLC by the Arabidopsis PAF1-like complex were postulated by Oh et al. to possibly involve recruitment of the FLC-chromatin remodeling factors such as PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 (PIE1) (Oh et al., 2004), a homologue of a member of the yeast SWR1 complex that is involved in histone H2A variant replacement (Deal et al., 2007). PIE1 exhibited only mild transcript level changes in atvoz1-1 atvoz2-1 (Fig. 3). However, a significant upregulation in the atvoz1-1 atvoz2-1 mutant was observed for another component of SWR1, the FLC activator AtSWC6/SERRATED LEAVES AND EARLY FLOWERING (SEF) (Lázaro et al., 2008) (Fig. 3). Together, our data show that AtVOZs regulate FLC levels, and that the observed FLC upregulation correlates with increased expression of genes involved in FLC chromatin modification. In contrast, CONSTANS (CO), the main photoperiod pathway gene, showed similar expression in WT and the double mutant (Fig. 3).
FLC encodes a MADS box protein that directly represses certain flowering-time genes. An important target of FLC is the FD gene, which encodes a bZIP transcription factor preferentially expressed in the shoot apex. FD associates with the flowering pathway integrator FLOWERING LOCUS T (FT) to activate downstream flowering activators (Abe et al., 2005). FT encodes a RAF kinase inhibitor-like protein and functions as a long distance signal between the leaves and the shoot meristem to promote flowering. In atvoz1-1 atvoz2-1, the increased FLC expression was correlated with decreased expression of FD, consistent with the late flowering of the mutant (Fig. 3). The transcript level of the flowering promoter FT showed a slight increase (Fig. 3). Since it is known that FD is required for FT to promote flowering, as an fd-1 mutation has been shown to suppress early flowering of 35S::FT overexpressing plants (Abe et al., 2005), the decrease in FD expression appears to be enough for delayed flowering in the atvoz1-1 atvoz2-1 mutant.
The atvoz1-1 atvoz2-1 mutant is responsive to vernalization
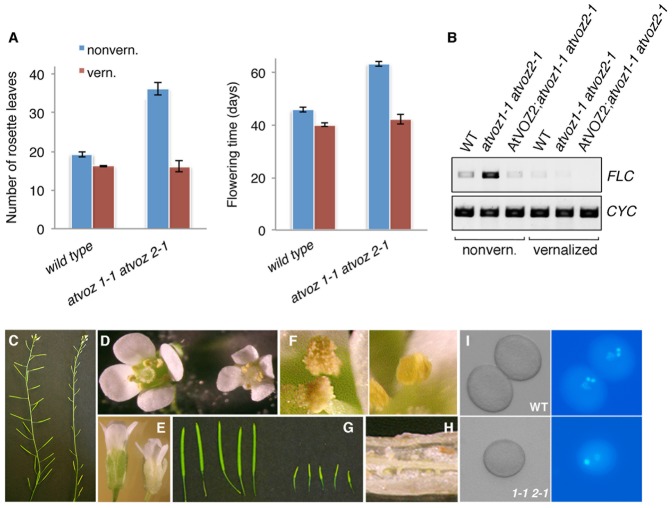
The delayed flowering of atvoz1-1 atvoz2-1 correlates with increased expression of the flowering gene FLC. FRI-containing lines and mutants in the autonomous pathway also have elevated FLC expression and delayed flowering, and show sensitivity to vernalization, which can abolish their late flowering through an epigenetic shut off of FLC expression. In contrast, photoperiod pathway mutants remain relatively uninfluenced by this treatment (Martinez-Zapater and Somerville, 1990; Koornneef et al., 1991; Mouradov et al., 2002; Moon et al., 2003; Lim et al., 2004). Similar to FRI-carrying lines and autonomous pathway mutants, atvoz1-1 atvoz2-1 plants responded to vernalization. Vernalized atvoz1-1 atvoz2-1 plants flowered much earlier than their non-vernalized counterparts. In fact, the flowering time was comparable to that of WT plants (Fig. 4A). Moreover, FLC transcript level was reduced to WT levels in the vernalized double mutant, in agreement with the complete recovery of flowering time (Fig. 4B).
Fig. 4. Vernalization reverses FLC-mediated delayed flowering of atvoz1-1 atvoz2-1 mutants.
(A) Rosette leaf number at bolting (left) and time to opening of the first flower (right) of non-vernalized (blue) and vernalized (red) WT and atvoz1-1 atvoz2-1 plants. Mean values±s.e.m. are shown. (B) FLC expression in non-vernalized and vernalized plants, measured by RT-PCR. Cyclophilin (CYC): template control. (C–I) Pleiotropic effects of vernalization on atvoz1-1 atvoz2-1. (C) Inflorescences of vernalized WT (left) and atvoz1-1 atvoz2-1 (right) plants. (D,E) Flower size of vernalized WT (left) and atvoz1-1 atvoz2-1 (right) plants. (F) WT anthers (left) and non-dehiscent atvoz1-1 atvoz2-1 anthers (right) from vernalized plants. (G) Silique sizes of vernalized WT (left) and atvoz1-1 atvoz2-1 (right) plants. (H) Silique from vernalized atvoz1-1 atvoz2-1 containing unfertilized ovules and abortive seeds. (I) Pollen from vernalized WT and atvoz1-1 atvoz2-1 flowers seen under bright field (left) and stained with DAPI (right).
However, in addition to reversing flowering time to WT levels, vernalization treatment caused several other changes in atvoz1-1 atvoz2-1 plants (Fig. 4C–I). The flowers of atvoz1-1 atvoz2-1 plants were mostly smaller and had non-dehiscent or shriveled anthers (Fig. 4D–F). Many had an aberrant number of stamens (∼33%). DAPI-staining of atvoz1-1 atvoz2-1 pollen revealed defects in pollen development in a small fraction (11%) of pollen grains; during normal development, pollen mitosis II of the generative cell creates two sperm cells that associate with the vegetative nucleus, but in abnormal atvoz1-1 atvoz2-1 pollen grains only one sperm cell was observed (Fig. 4I). Vernalized atvoz1-1 atvoz2-1 plants generated little or no seed, since they were mostly producing very small siliques that contained only aborted seeds or unfertilized ovules (Fig. 4C,G,H). It is unclear why vernalization treatment caused extensive pleiotropic effects in atvoz1-1 atvoz2-1 plants. Perhaps a vernalization-responsive gene or genes involved in reproductive organ and seed development is/are specifically deregulated in atvoz1-1 atvoz2-1.
AtVOZs directly regulate expression of MOS3, which encodes a nuclear pore constituent involved in flowering
To investigate whether AtVOZ transcription factors directly regulate the genes examined in Fig. 3, we searched for the consensus VOZ binding site (GCGTN×7ACGC) (Mitsuda et al., 2004) in their promoter sequences. Neither the FLC gene, nor the abovementioned FLC regulators and targets contained GCGTN×7ACGC in their promoter regions. In addition, none had either of the 2 suboptimal elements that have been reported to allow reduced AtVOZ binding (GCGTN×7ACGT and GCGTN×8ACGC). Considering this, AtVOZs might be regulating these flowering genes indirectly rather than by direct binding. Another possibility is that AtVOZs could recognize other, yet unidentified binding elements in promoter sequences of these genes.
To identify flowering genes that contain the VOZ binding element, we performed a genome-wide search for the presence of GCGTN×7ACGC in promoter sequences up to 1000 bp upstream from the translation initiation site. The search identified the gene MODIFIER OF SNC1,3 (MOS3)/SUPPRESSOR OF AUXIN RESISTANCE 3 (SAR3) whose promoter region contains the palindrome GCGTTTATCAGACGC. MOS3/SAR3 encodes a nuclear membrane protein with homology to human nucleoporin 96, a subunit of a nuclear pore complex involved in nucleocytoplasmic trafficking of macromolecules (Zhang and Li, 2005). MOS3/SAR3 regulates export of mRNAs, together with two components of the same nuclear pore subcomplex, Nup160/SAR1 and Seh1 (Parry et al., 2006; Wiermer et al., 2012). Mutants of MOS3/SAR3 are smaller and less robust than WT plants and flower significantly earlier and with fewer leaves (Zhang and Li, 2005; Parry et al., 2006). Nup160/sar1 are also early flowering (Dong et al., 2006) and double mutants mos3/sar3 nup160/sar1 exhibit severe developmental defects (Parry et al., 2006). The mos3/sar3 mutation strongly promotes flowering in the FRI-containing background, connecting MOS3/SAR3 with the FLC flowering pathway (Jacob et al., 2007). Another nuclear pore protein, AtTPR, has been shown to regulate flowering time and upregulate expression of FLC, further linking nuclear pore function with the flowering time (Jacob et al., 2007). Gene expression analysis in atvoz1-1 atvoz2-1 showed a substantial increase in MOS3/SAR3 expression, indicating that AtVOZs control this gene (Fig. 5A; supplementary material Fig. S2). The observed upregulation was restored to WT levels in the complemented line AtVOZ2; atvoz1-1 atvoz2-1.
Fig. 5. AtVOZs directly repress expression of the MOS3/SAR3 gene, which encodes a nuclear pore protein involved in regulation of flowering time.
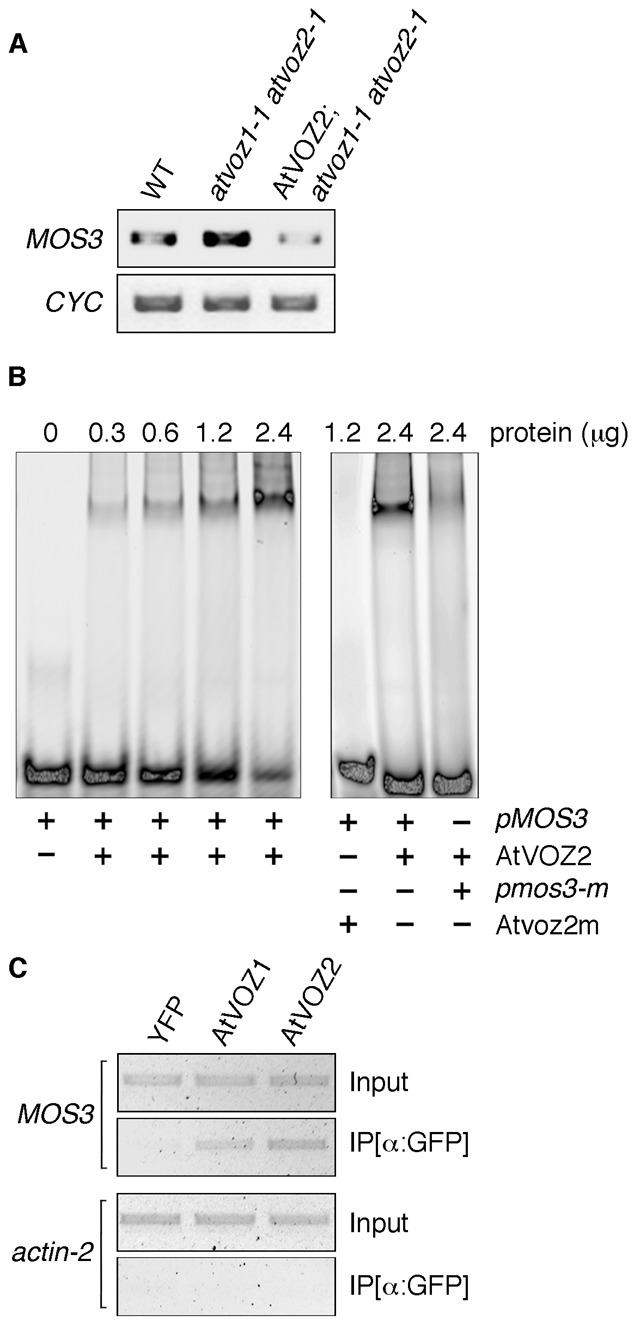
(A) MOS3/SAR3 expression measured by RT-PCR. Cyclophilin (CYC): template control. (B) Left: electrophoretic mobility shift assay (EMSA) showing dose-dependent binding of purified AtVOZ2 to MOS3 promoter probe (pMOS3; 2 pmoles). Right: mutated AtVOZ2 (Atvoz2m) does not bind to pMOS3 (20 pmoles). AtVOZ2 binding is reduced when the consensus VOZ binding element in MOS3 probe is mutated (pmos3-m; 20 pmoles). (C) Chromatin immunoprecipitation (ChIP) analysis showing in vivo binding of AtVOZs to MOS3 promoter. Complexes of AtVOZ1-YFP and AtVOZ2-YFP bound to DNA were precipitated with GFP-antibody beads (IP [α:GFP]). Precipitated sequences were PCR-amplified using MOS3-specific primers spanning the consensus VOZ binding element. YFP, negative control; actin-2, control for assay specificity. Input: PCR amplification of MOS3 and actin-2 promoter regions in extracts prior to immunoprecipitation.
To test whether AtVOZs regulate MOS3/SAR3 directly by binding to its promoter, we performed electrophoretic mobility shift assay (EMSA) using a fluorescently labeled 41-bp-long promoter probe encompassing the GCGTTTATCAGACGC element (Fig. 5B). EMSA revealed that purified AtVOZ2 bound to the probe in a dose-dependent manner. The binding was dependent on the zinc finger domain of AtVOZ2, as point mutations in two conserved amino acids (C249A/H253A) in that region completely abolished this interaction (AtVOZ2m, Fig. 5B). Furthermore, introducing mutations in the GCGTTTATCAGACGC sequence (pmos3-m, Fig. 5B) diminished AtVOZ2 binding, indicating that AtVOZ2 interacts with the consensus VOZ-binding element. Due to difficulties expressing AtVOZ1, this protein was not tested by EMSA. The binding of both proteins was confirmed in plant cells by chromatin immunoprecipitation (ChIP) analysis. PCR of immunoprecipitated DNA using primers flanking the AtVOZ binding site in the MOS3/SAR3 promoter revealed that AtVOZ1 and AtVOZ2 associate with the MOS3 promoter in vivo (Fig. 5C). Together, our results show that AtVOZs directly regulate MOS3/SAR3, which is required for proper timing of flowering, by binding to its promoter and repressing its expression, and suggest that at least part of AtVOZ activities are exerted through influencing the nuclear pore complex.
Discussion
Our data demonstrate an important role of AtVOZ1 and AtVOZ2 transcription factors in regulating the transition from vegetative growth to flowering. atvoz1-1 atvoz2-1 plants exhibit delayed flowering, which correlates with increased FLC expression and shows a robust response to vernalization. Vernalization causes a decrease in FLC transcript abundance and restores flowering time of atvoz1-1 atvoz2-1 to WT levels. Recently, Yasui et al. showed that phytochrome B interacts with AtVOZs (Yasui et al., 2012). They have also reported that atvoz1 atvoz2 double mutant is late flowering. In addition to flowering-time control, we observed other phenotypes, such as delayed juvenile phase and defects in seed production, in the atvoz1-1 atvoz2-1 double mutant, suggesting a broader role for AtVOZs in plant development. Functions additional to flowering-time control are often observed for genes in flowering pathways. For instance, MSI1, a flowering-time gene in the autonomous pathway, is also involved in gametophyte and seed development (Bouveret et al., 2006). Furthermore, due to additional functions, double mutants in some autonomous pathway genes display severe pleiotropic phenotypes or even lethality (Koornneef et al., 1998; Veley and Michaels, 2008). Homologues of AtVOZ1 and AtVOZ2 are found in various vascular plants as well as in the moss Physcomitrella patens (Mitsuda et al., 2004), indicating that AtVOZs are not specific to flowering plants. This is consistent with their additional role in growth and development.
Our experimental data indicate that AtVOZs directly regulate expression of the nuclear pore protein MOS3/SAR3, required for export of mRNA from the nucleus. Like AtVOZs, MOS3/SAR3 is required for proper plant growth and flowering time, further functionally linking these proteins (Zhang and Li, 2005; Dong et al., 2006; Parry et al., 2006). As AtVOZs directly regulate MOS3/SAR3, it is possible they exert at least part of their activities by controlling the nuclear pore function. It is conceivable that the effect of AtVOZs on flowering could result from altered nucleocytoplasmic localization and consequently activity of some flowering-time transcripts, a possibility that requires further functional analyses. A similar mechanism was proposed by Faria et al. for vertebrate Nup96 (Faria et al., 2006), which is required in mice immunity for nuclear mRNA export of specific interferon-regulated genes in response to viral infection. They observed that reduced levels of Nup96 resulted in nuclear retention of these transcripts and postulated that such retention likely contributes to the lower levels of the immune proteins encoded by these mRNAs at the plasma membrane. Nuclear pore proteins have also been directly linked to transcript abundance (Dong et al., 2006; Wiermer et al., 2012). It is conceivable that the observed changes in transcript accumulation in atvoz1-1 atvoz2-1 could partly be ascribed to that aspect of nuclear function. Additionally, the yeast nuclear pore Nup84 complex (equivalent to vertebrate Nup107-160 complex) is capable of activating transcription in vivo by tethering target genes to the nuclear pore and coupling transcription with mRNA export (Menon et al., 2005). Since none of the AtVOZ-controlled flowering genes mentioned in Fig. 3 carry the consensus VOZ binding site, it is intriguing to think that AtVOZs may control their expression indirectly via regulation of the nuclear pore complex.
Recently MdVOZ1, one of 5 predicted apple VOZ1 and VOZ2 genes, was implicated in reproduction in apple (Mimida et al., 2011). Like Arabidopsis VOZ transcripts, MdVOZ1 mRNA was detected in multiple tissues, with a strong accumulation in fruit, sepals and petals, and weak expression in stamens and carpels. In <10% of transgenic Arabidopsis plants constitutively expressing apple MdVOZ1, sterile flowers with enhanced elongation of flower stalks and abnormal inflorescences were observed. This suggested involvement of MdVOZ1 in the development of reproductive organs (Mimida et al., 2011). In contrast to morphological changes in Arabidopsis plants constitutively expressing MdVOZ1, overexpression of AtVOZ2 in atvoz1-1 atvoz2-1 double mutant resulted in plants that were visually indistinguishable from WT, suggesting that apple and Arabidopsis VOZ homologues diverged in their functions. However, lack of functional AtVOZs in Arabidopsis resulted in plants that produced smaller siliques and displayed substantial seed abortion, providing a correlation between the role of apple and Arabidopsis VOZ proteins in seed production.
Interestingly, MdVOZ1 was shown to interact with MdFT1 and MdFT2, apple orthologues of Arabidopsis FT. Based on this association and the expression patterns of apple VOZ1 and FTs, it was suggested that apple FTs might be involved in reproductive organ and fruit development in addition to flowering-time control. It is not known whether AtVOZ1 or AtVOZ2 also interact with FT in Arabidopsis. If so, such association could represent another level of flowering regulation, perhaps by: (i) targeting a specific subset of flowering genes, (ii) promoting modulation of AtVOZ and/or FT activities by their protein–protein interactions, or (iii) an intriguing idea is that AtVOZs could assist the FT florigen in leaf-to-meristem movement. In line with the third possibility is a recent report suggesting that Nicotiana benthamiana homologues of AtVOZ1, sc4i21 and Ni67, are utilized by plant-adapted rhabdoviruses for cell-to-cell movement (Min et al., 2010). Also, AtVOZ1 is primarily expressed in the phloem (Mitsuda et al., 2004), in accord with FT movement.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Science Foundation (MCB-5333470 to A.S.N.R). We are grateful to Amy Lowery and Sarah-Marie Lyons for help with maintaining plant lines and assistance in atvoz1-1 atvoz2-1 studies. We thank Drs Denghui Xing and Irene Day for helpful comments on this manuscript and the ABRC for providing T-DNA insertion lines of the AtVOZ single mutants. Plasmid constructs and mutants generated in this research are available upon request. We thank the Colorado State University Libraries Open Access Research and Scholarship Fund for paying the publication charges for this article.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Bouveret R., Schönrock N., Gruissem W., Hennig L. (2006). Regulation of flowering time by Arabidopsis MSI1. Development 133, 1693–1702 10.1242/dev.02340 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Deal R. B., Topp C. N., McKinney E. C., Meagher R. B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19, 74–83 10.1105/tpc.106.048447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. H., Hu X., Tang W., Zheng X., Kim Y. S., Lee B. H., Zhu J. K. (2006). A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26, 9533–9543 10.1128/MCB.01063-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., Poovaiah B. W. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Faria A. M., Levay A., Wang Y., Kamphorst A. O., Rosa M. L., Nussenzveig D. R., Balkan W., Chook Y. M., Levy D. E., Fontoura B. M. (2006). The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 24, 295–304 10.1016/j.immuni.2006.01.014 [DOI] [PubMed] [Google Scholar]
- Golovkin M., Reddy A. S. N. (2003). A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc. Natl. Acad. Sci. USA 100, 10558–10563 10.1073/pnas.1734110100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Doyle M. R., Amasino R. M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18, 2774–2784 10.1101/gad.1244504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Mongkolsiriwatana C., Veley K. M., Kim S. Y., Michaels S. D. (2007). The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 144, 1383–1390 10.1104/pp.107.100735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K., Kjaersgaard T., Nielsen M. M., Galberg P., Petersen K., O'Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196 10.1042/BJ20091234 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., van der Veen J. H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 10.1007/BF00264213 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Alonso–Blanco C., Blankestijn–de Vries H., Hanhart C. J., Peeters A. J. (1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro A., Gómez–Zambrano A., López–González L., Piñeiro M., Jarillo J. A. (2008). Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59, 653–666 10.1093/jxb/erm332 [DOI] [PubMed] [Google Scholar]
- Lee I., Amasino R. M. (1995). Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162 10.1104/pp.108.1.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M. H., Kim J., Kim Y. S., Chung K. S., Seo Y. H., Lee I., Kim J., Hong C. B., Kim H. J., Park C. M. (2004). A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16, 731–740 10.1105/tpc.019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez–Zapater J. M., Somerville C. R. (1990). Effect of light quality and vernalization on late-flowering mutants of Arabidopsis thaliana. Plant Physiol. 92, 770–776 10.1104/pp.92.3.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon B. B., Sarma N. J., Pasula S., Deminoff S. J., Willis K. A., Barbara K. E., Andrews B., Santangelo G. M. (2005). Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102, 5749–5754 10.1073/pnas.0501768102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS� C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941 10.1105/tpc.13.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida N., Kidou S., Iwanami H., Moriya S., Abe K., Voogd C., Varkonyi–Gasic E., Kotoda N. (2011). Apple FLOWERING LOCUS T proteins interact with transcription factors implicated in cell growth and organ development. Tree Physiol. 31, 555–566 10.1093/treephys/tpr028 [DOI] [PubMed] [Google Scholar]
- Min B. E., Martin K., Wang R., Tafelmeyer P., Bridges M., Goodin M. (2010). A host-factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Mol. Plant Microbe Interact. 23, 1420–1432 10.1094/MPMI-04-10-0097 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Hisabori T., Takeyasu K., Sato M. H. (2004). VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 45, 845–854 10.1093/pcp/pch101 [DOI] [PubMed] [Google Scholar]
- Moon J., Suh S. S., Lee H., Choi K. R., Hong C. B., Paek N. C., Kim S. G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623 10.1046/j.1365-313X.2003.01833.x [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 Suppl. 1, S111–S130 10.1105/tpc.001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Zhang H., Ludwig P., van Nocker S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16, 2940–2953 10.1105/tpc.104.026062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18, 1590–1603 10.1105/tpc.106.041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R. S. (2003). Phase change and the regulation of developmental timing in plants. Science 301, 334–336 10.1126/science.1085328 [DOI] [PubMed] [Google Scholar]
- Rédei G. P. (1962). Supervital mutants of Arabidopsis. Genetics 47, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh–Kumar S. P., Deng X. W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778 10.1111/j.1365-313X.2004.02328.x [DOI] [PubMed] [Google Scholar]
- Schmitz R. J., Amasino R. M. (2007). Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Biophys. Acta 1769, 269–275 10.1016/j.bbaexp.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., Peacock W. J., Dennis E. S. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 10.1105/tpc.11.3.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., Dean C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 10.1126/science.296.5566.285 [DOI] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veley K. M., Michaels S. D. (2008). Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiol. 147, 682–695 10.1104/pp.108.118927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Cheng Y. T., Imkampe J., Li M., Wang D., Lipka V., Li X. (2012). Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 70, 796–808 10.1111/j.1365-313X.2012.04928.x [DOI] [PubMed] [Google Scholar]
- Yasui Y., Mukougawa K., Uemoto M., Yokofuji A., Suzuri R., Nishitani A., Kohchi T. (2012). The phytochrome-interacting VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24, 3248–3263 10.1105/tpc.112.101915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X. (2005). A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17, 1306–1316 10.1105/tpc.104.029926 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.