Abstract
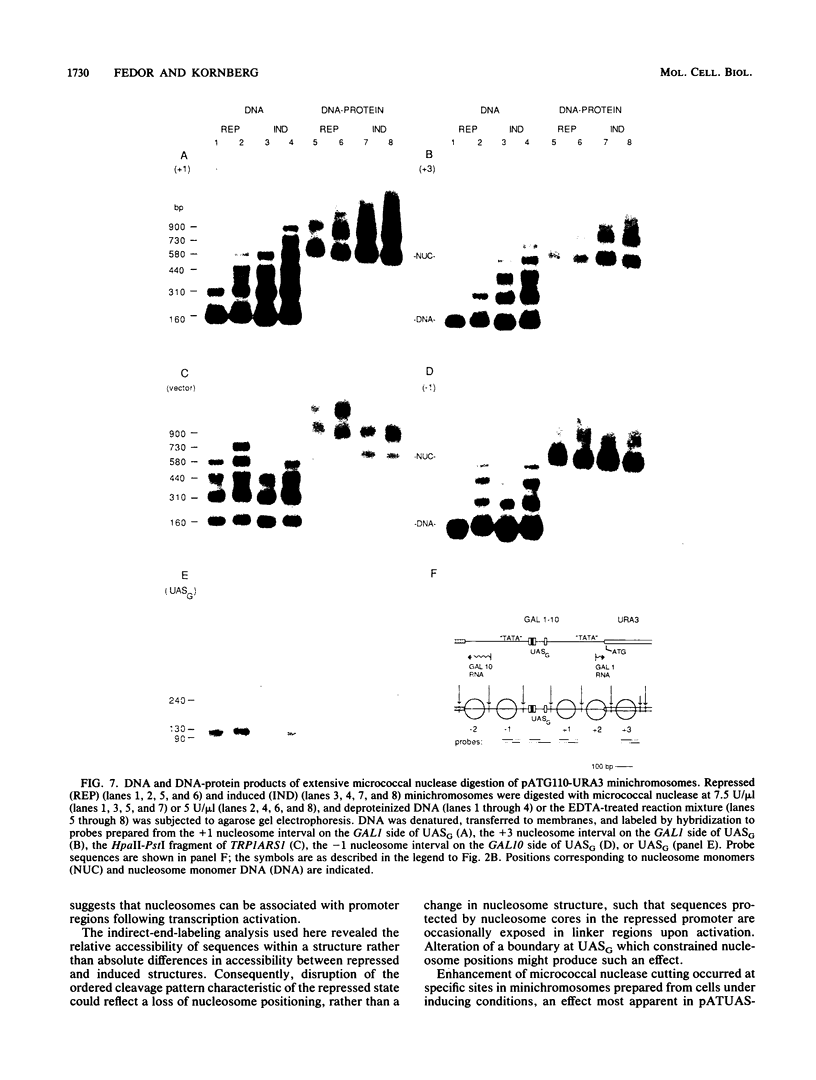
Conversion of the positioned nucleosome array characteristic of the repressed GAL1-GAL10 promoter region to the more accessible conformation of the induced state was found to depend on the upstream activation sequence, GAL4 protein, a positive regulator of transcription, and galactose, the inducing agent. The effect of the GAL4 protein-upstream activation sequence complex on the structure of adjacent chromatin required no other promoter sequences. Although sequences protected by histones in the repressed state became more accessible to micrococcal nuclease and (methidiumpropyl-EDTA)iron(II) cleavage following induction of transcription, DNA-protein particles containing these sequences retained the electrophoretic mobility of nucleosomes, indicating that the promoter region can be associated with nucleosomes under conditions of transcription activation.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allegra P., Sterner R., Clayton D. F., Allfrey V. G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J Mol Biol. 1987 Jul 20;196(2):379–388. doi: 10.1016/0022-2836(87)90698-x. [DOI] [PubMed] [Google Scholar]
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. M., Johnston S. A., Hopper J. E., Jaehning J. A. Transcription of multiple copies of the yeast GAL7 gene is limited by specific factors in addition to GAL4. Mol Gen Genet. 1987 Jun;208(1-2):127–134. doi: 10.1007/BF00330433. [DOI] [PubMed] [Google Scholar]
- Bergman L. W. A DNA fragment containing the upstream activator sequence determines nucleosome positioning of the transcriptionally repressed PHO5 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2298–2304. doi: 10.1128/mcb.6.7.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman L. W., Kramer R. A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Jun 10;258(11):7223–7227. [PubMed] [Google Scholar]
- Bergman L. W., Stranathan M. C., Preis L. H. Structure of the transcriptionally repressed phosphate-repressible acid phosphatase gene (PHO5) of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):38–46. doi: 10.1128/mcb.6.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Chemical footprinting of 5S RNA chromatin in embryos of Drosophila melanogaster. EMBO J. 1984 Dec 20;3(13):3101–3108. doi: 10.1002/j.1460-2075.1984.tb02265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Qasba P. K. Alkaline transfer of DNA to plastic membrane. Biochem Biophys Res Commun. 1984 Jul 18;122(1):340–344. doi: 10.1016/0006-291x(84)90480-7. [DOI] [PubMed] [Google Scholar]
- Cimino G. D., Gamper H. B., Isaacs S. T., Hearst J. E. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Dorbic T., Wittig B. Chromatin from transcribed genes contains HMG17 only downstream from the starting point of transcription. EMBO J. 1987 Aug;6(8):2393–2399. doi: 10.1002/j.1460-2075.1987.tb02517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Kikuchi Y., Nogi Y., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Isolation and characterization of the regulatory gene GAL4. Mol Gen Genet. 1983;191(1):31–38. doi: 10.1007/BF00330886. [DOI] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988 May;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Y., Barnard M. B., Xu M., Matsui S., Rose S. M., Garrard W. T. The active immunoglobulin kappa chain gene is packaged by non-ubiquitin-conjugated nucleosomes. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3738–3742. doi: 10.1073/pnas.83.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Razvi F., Worcel A. The role of DNA-mediated transfer of TFIIIA in the concerted gyration and differential activation of the Xenopus 5S RNA genes. Cell. 1986 Apr 25;45(2):209–218. doi: 10.1016/0092-8674(86)90385-5. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Ryoji M., Worcel A. Gyration is required for 5S RNA transcription from a chromatin template. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1305–1309. doi: 10.1073/pnas.83.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezetic J. A., Luse D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986 Apr 11;45(1):95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Lohr D., Hopper J. E. The relationship of regulatory proteins and DNase I hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res. 1985 Dec 9;13(23):8409–8423. doi: 10.1093/nar/13.23.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984 Nov 26;12(22):8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. The chromatin structure of an actively expressed, single copy yeast gene. Nucleic Acids Res. 1983 Oct 11;11(19):6755–6773. doi: 10.1093/nar/11.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Torchia T., Hopper J. The regulatory protein GAL80 is a determinant of the chromatin structure of the yeast GAL1-10 control region. J Biol Chem. 1987 Nov 15;262(32):15589–15597. [PubMed] [Google Scholar]
- Lorch Y., Kornberg R. D. A region flanking the GAL7 gene and a binding site for GAL4 protein as upstream activating sequences in yeast. J Mol Biol. 1985 Dec 20;186(4):821–824. doi: 10.1016/0022-2836(85)90400-0. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Matsui T. Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol Cell Biol. 1987 Apr;7(4):1401–1408. doi: 10.1128/mcb.7.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Petryniak B., Lutter L. C. Topological characterization of the simian virus 40 transcription complex. Cell. 1987 Jan 30;48(2):289–295. doi: 10.1016/0092-8674(87)90432-6. [DOI] [PubMed] [Google Scholar]
- Proffitt J. H. DNase I-hypersensitive sites in the galactose gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Jun;5(6):1522–1524. doi: 10.1128/mcb.5.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to DNA sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):103–108. doi: 10.1101/sqb.1978.042.01.011. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D. Structure and function of the yeast URA3 gene. Differentially regulated expression of hybrid beta-galactosidase from overlapping coding sequences in yeast. J Mol Biol. 1983 Nov 15;170(4):883–904. doi: 10.1016/s0022-2836(83)80193-4. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. E. In vivo DNA-binding properties of a yeast transcription activator protein. Mol Cell Biol. 1987 Sep;7(9):3260–3267. doi: 10.1128/mcb.7.9.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987 Jan 8;325(7000):173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Larsen P. L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988 Jun 17;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. L., Xu Y. Z., Bellard M., Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986 Feb;5(2):293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Martinson H. G. Gamma rays and bleomycin nick DNA and reverse the DNase I sensitivity of beta-globin gene chromatin in vivo. Mol Cell Biol. 1987 May;7(5):1917–1924. doi: 10.1128/mcb.7.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. Studies on the mechanism of transcription of nucleosomal complexes. Eur J Biochem. 1980 Jan;103(2):219–226. doi: 10.1111/j.1432-1033.1980.tb04306.x. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. Transcription by eukaryotic RNA polymerases A and B of chromatin assembled in vitro. Eur J Biochem. 1979 Aug 1;98(2):317–327. doi: 10.1111/j.1432-1033.1979.tb13191.x. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Thevenin G., Oudet P., Chambon P. Transcription of in vitro assembled chromatin by Escherichia coli RNA polymerase. J Mol Biol. 1979 Mar 5;128(3):411–440. doi: 10.1016/0022-2836(79)90095-0. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Williamson P., Felsenfeld G. Transcription of histone-covered T7 DNA by Escherichia coli RNA polymerase. Biochemistry. 1978 Dec 26;17(26):5695–5705. doi: 10.1021/bi00619a015. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984 Sep 6;311(5981):81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Yaniv M., Cereghini S. Structure of transcriptionally active chromatin. CRC Crit Rev Biochem. 1986;21(1):1–26. doi: 10.3109/10409238609113607. [DOI] [PubMed] [Google Scholar]
- Zakian V. A., Scott J. F. Construction, replication, and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982 Mar;2(3):221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]