Abstract
Meiotic crossover (CO) recombination involves a reciprocal exchange between homologous chromosomes. COs are often associated with gene conversion at the exchange site where genetic information is unidirectionally transferred from one chromosome to the other. COs and independent assortment of homologous chromosomes contribute significantly to the promotion of genomic diversity. What has not been appreciated is the contribution of another product of meiotic recombination, noncrossovers (NCOs), which result in gene conversion without exchange of flanking markers. Here, we review our comprehensive analysis of recombination at a highly polymorphic mouse hotspot. We found that NCOs make up ~90% of recombination events. Preferential recombination initiation on one chromosome allowed us to estimate the contribution of CO and NCO gene conversion to transmission distortion, a deviation from Mendelian inheritance in the population. While NCO gene conversion tracts are shorter, and thus have a more punctate effect, their higher frequency translates into an approximately two-fold greater contribution than COs to gene conversion–based allelic shuffling and transmission distortion. We discuss the potential impact of mammalian NCO characteristics on evolution and genomic diversity.
Keywords: noncrossover, gene conversion, recombination, meiosis, hotspot
Introduction
Meiosis is a specialized cell division program in which a diploid precursor cell undergoes one round of DNA replication followed by two consecutive rounds of DNA segregation and cellular division to generate haploid gametes for sexual reproduction. The first cellular division cycle, meiosis I, is a reductional division in that homologous chromosomes (homologs) segregate to daughter cells, resulting in half the chromosomal complement (e.g., in humans, 23 homolog pairs are reduced to 23 chromosomes), while during the second cellular division cycle, meiosis II, sister chromatids segregate.1 It can be argued that the primary mandate of meiosis I is to induce homologs to find each other, stably pair, and accurately segregate2 (see also Kauppi et al.3). Failures in this process lead to gamete aneuploidy, which is the leading cause of developmental disability and spontaneous miscarriage in humans.4 However, the DNA interactions ensuring the meiotic reductional division also have critically important consequences for evolution and genomic diversity.
Meiotic recombination: damaging the genome in order to propagate it
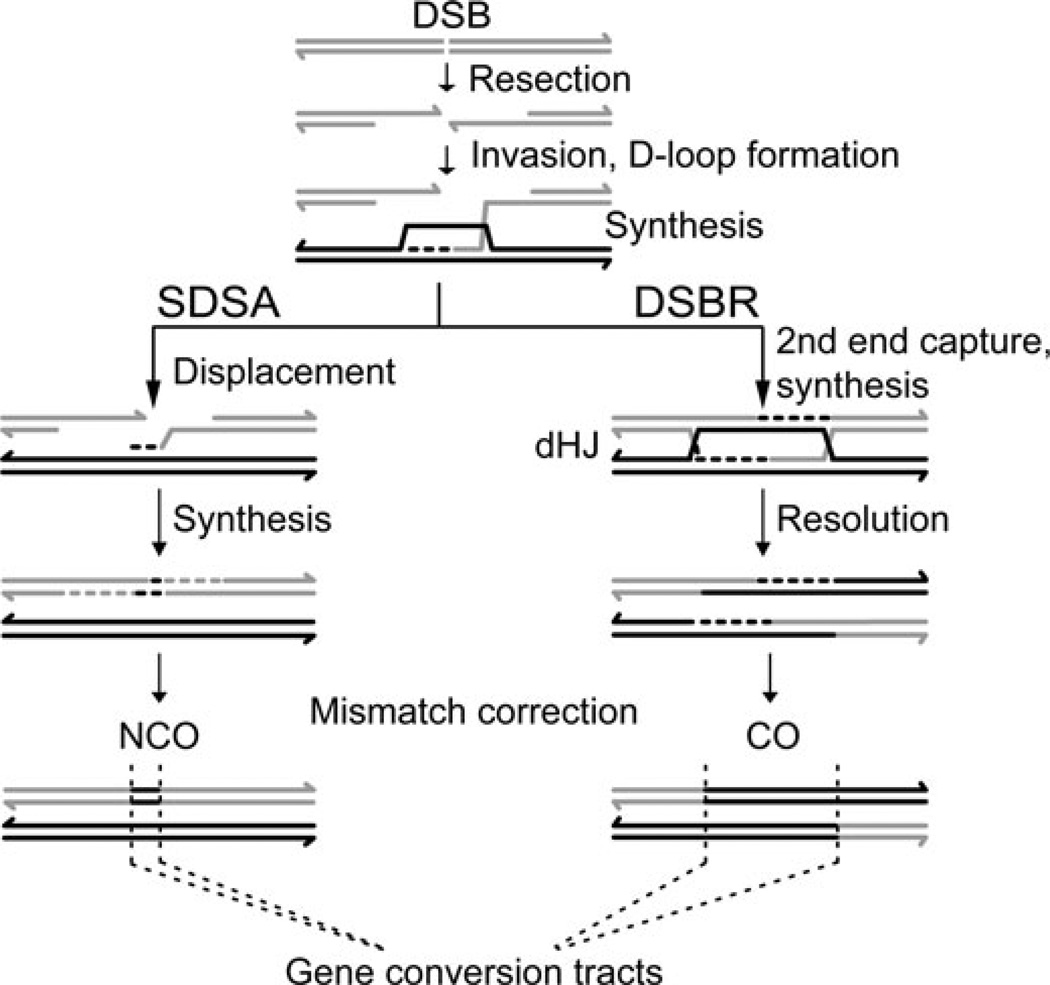
Meiosis has coopted ancient DNA repair mechanisms that predate sexual reproduction to provoke homologs to stably pair.5 To make use of these mechanisms, meiotic cells induce programmed DNA double-strand breaks (DSBs) at hotspots throughout the genome by expressing the SPO11 transesterase (Fig. 1).6 The location of meiotic hotspots is variable and will be discussed further below. DSB resection generates 3′ single-stranded tails that are bound by strand invasion proteins to catalyze D-loop formation with an intact homologous duplex that serves as a template for DNA repair synthesis. The homolog is the preferred repair template in meiosis rather than the identical sister chromatid, which is used in mitosis. Based upon studies primarily performed in budding yeast, there are two major homologous recombination pathways that bifurcate from the D-loop intermediate. In one pathway, known as synthesis-dependent strand annealing (SDSA), the extended 3′ end of the invading strand is displaced after repair synthesis from the homolog to anneal to the second 3′ end on the other side of the DSB.7 SDSA is thought to generate non-crossovers (NCOs) exclusively, which is a patch-like repair with no exchange of flanking markers. In the other pathway, known as double-strand break repair (DSBR), the second 3′ end is captured by the D-loop and a double Holliday junction (dHJ) forms.8 dHJs can be resolved by structure-specific endonucleases; most such resolution events are thought to generate a crossover (CO), which is a reciprocal exchange of flanking markers. Each homolog pair requires at least one CO to be physically linked as a bivalent, and these COs, in conjunction with sister chromatid cohesion established during DNA replication, tether the four chromatids (two from each homolog) as a tetrad.2 Thus, COs provide the connections necessary for accurate reductional segregation in meiosis I.
Figure 1.
Meiotic recombination pathways. DSBs are induced preferentially at hotspots located throughout the genome. Resection of the DSB from 5′ to 3′ generates 3′ single-stranded tails, which invade the intact homolog (black) creating a D-loop intermediate. DNA repair synthesis (dashed lines) is primed by the invading 3′ end and templated by the homolog. In synthesis-dependent strand annealing (SDSA), the newly synthesized 3′ tail is displaced from the homolog, whereupon it anneals to the second, homologous 3′ end of the DSB. Subsequent repair synthesis and ligation reseals the DSB and generates a noncrossover (NCO). In double-strand break repair (DSBR), the D-loop captures the second end of the DSB and a double Holliday junction (dHJ) is formed. Resolution of the dHJ can generate a crossover (CO). Both NCOs and COs can result in gene conversions, as indicated, where sequences of the homolog that receives a DSB (gray) are converted to the genotype of the intact homolog (black).
How meiosis contributes to genetic diversity
Meiosis generates genetic diversity through three principal mechanisms. First, pairs of homologous chromosomes are independently assorted from each other into haploid gametes, for 2n possible combinations, where n is the number of homolog pairs. Second, CO recombination reciprocally exchanges chromosome segments between homologs, altering the association of maternal and paternal alleles at the point of exchange. Finally, gene conversion during homologous recombination results in non-Mendelian transmission of genetic information at the site of DSB repair (Fig. 1). If the region around a DSB is polymorphic between homologs, heteroduplex DNA can form during homologous recombination and mismatch correction will convert those polymorphisms. Thus, within the meiotic tetrad, information is transferred unidirectionally from one parental chromatid to another, resulting in 3:1 transmission of alleles within the gene conversion tract. While the contribution of independent assortment and CO recombination to genomic diversity has long been appreciated, the contribution of gene conversion, particularly as a result of NCOs, has been difficult to assess.
Toward a comprehensive assessment of meiotic gene conversion
COs are a minor outcome of homologous recombination in mammalian meiosis. Based upon cytological markers, COs likely represent only ~10% of DSB repair products in mouse spermatocytes.9 The remaining DSBs had been inferred to be repaired as interhomolog NCOs; however, when analyzed, the proportion of NCOs at recombination hotspots was much lower in mammals than expected.10–12 This finding raised the possibility that many mammalian meiotic DSBs could be repaired by other mechanisms, such as repair between sister chromatids or even by nonhomologous processes. However, repair of this nature would serve no obvious purpose in promoting homolog recognition, pairing, and tethering. An important technical consideration is that COs can always be identified because they result in the exchange of flanking markers, but NCOs can only be detected if they generate a gene conversion incorporating a polymorphism. This raised the question as to whether the low frequency of NCOs identified in previous studies could be due to the low density of polymorphisms in the analyzed recombination hotspots.
To counter this problem, we sought to characterize meiotic recombination at a highly polymorphic mouse hotspot, hypothesizing that even if NCO gene conversion tracts are very short, we would capture a substantial fraction of events.13 We summarize our findings below and discuss some of the implications.
Frequency and distribution of COs and NCOs at a mouse hotspot: variations between strain backgrounds
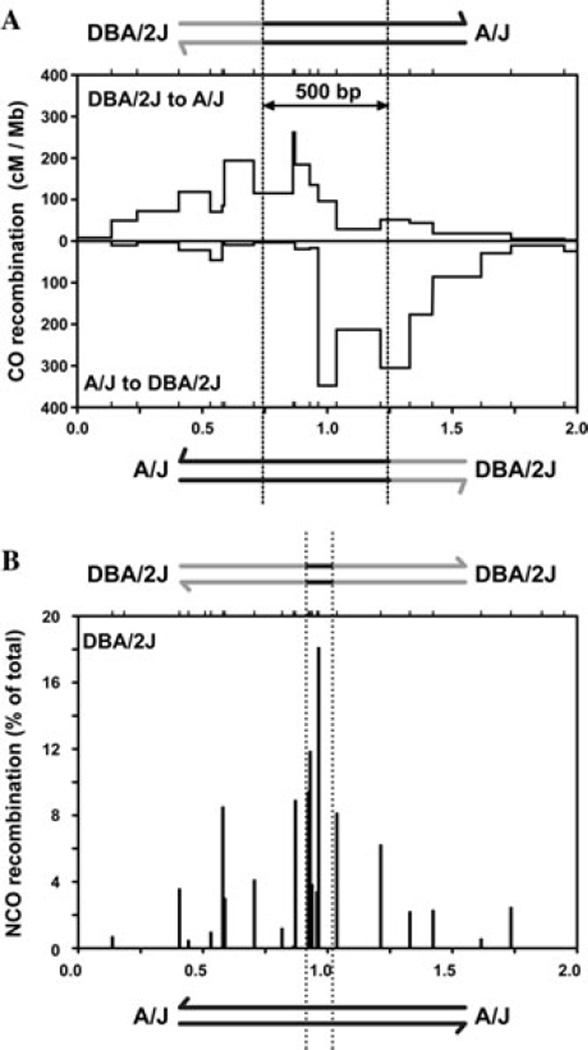
We determined that a previously identified mouse hotspot termed A3 (Ref. 14) had a high density of polymorphisms between inbred Mus musculus strains, ranging from 1.6% to 1.8%, with polymorphisms located approximately every 30 bp in the center of the hotspot. We compared the distribution of COs and NCOs in sperm from F1 hybrid mice from different strain combinations. Recombinant DNA molecules were amplified using allele-specific PCR, according to previously developed methods.15,16 The A3 hotspot was active for CO recombination in all F1 hybrids analyzed with a CO frequency of ~10−3 per sperm genome, ~200-fold greater than the genome average. In A/J × DBA/2J F1 hybrids, the observed distribution of CO exchange points differed depending on which orientation of CO product was amplified from sperm DNA: When amplifying molecules in the DBA/2J to A/J orientation, most CO exchange points (i.e., where the amplified DNA sequence switches from the DBA/2J to A/J genotype) clustered to the left (Fig. 2A, top), but when amplifying molecules in the A/J to DBA/2J orientation, most CO exchange points clustered to the right (Fig. 2A, bottom). This asymmetric distribution pattern of CO exchange points has been proposed by Jeffreys et al. to be caused by preferential formation of DSBs on one chromosome compared to the other,17 as the broken chromatid copies genetic information from the unbroken homolog (Fig. 1). Based on this model, we infer that DSBs at A3 are to be strongly biased in favor of the DBA/2J chromosome compared with the A/J chromosome. As a result, CO gene conversion strongly favors transmission of information from the unbroken A/J chromosome. By determining how offset the distribution of exchange points is between orientations, we estimated that the mean CO gene conversion tract length at A3 is ~500 bp (Fig. 2A). This estimate is similar to those at the handful of human and mouse hotspots that show similar asymmetric CO patterns.17–20 Consistent with preferential DSB formation on the DBA/2J chromosome leading to biased NCO gene conversion (Fig. 1), approximately nine-fold more NCOs were observed on the DBA/2J chromosome compared with the A/J chromosome.
Figure 2.
Recombination at the A3 hotspot. (A) CO activity in centimorgans (cM) per Mb in A/J × DBA/2J F1 hybrids. When amplifying COs in the DBA/2J to A/J orientation, exchange points cluster to the left (top), while in the A/J to DBA/2J orientation, exchange points cluster to the right (bottom). The offset in exchange points is due to preferential DSB formation on the DBA/2J chromosome. Dashed lines indicate the distribution center of each orientation, and the offset between them is used to estimate the mean CO gene conversion tract length of 500 bp. (B) NCOs (as % of total NCOs detected) in all F1 hybrids on the DBA/2J chromosome. Ticks at the top of graphs represent tested polymorphisms. The x-axis scale for (A) and (B) is in kilo bases.
In contrast to A/J × DBA/2J F1 hybrids, C57BL/6J × DBA/2J F1 hybrids showed a symmetric distribution pattern of CO exchange points at the A3 hotspot:13 COs clustered at the same point when amplified in either the C57BL/6J to DBA/2J or DBA/2J to C57BL/6J orientation. Moreover, NCOs were similar in frequency for both the C57BL/6J and DBA/2J chromosomes. Thus, DSB formation appears to be equally frequent on each chromosome in this F1 hybrid strain background.
High-resolution analysis of NCO gene conversions
NCO gene conversions peak at the same central polymorphisms that are at the center of CO exchange points (compare Fig. 2A and B). In comparison to COs, however, NCO gene-conversion tracts were universally short. We determined that the median of the maximal NCO gene-conversion tracts in the center of the hotspot was ~100 bp, or approximately five-fold shorter than the mean of CO gene conversion tracts. Most NCO gene conversions incorporated only a single polymorphism, validating our hypothesis that a high polymorphism density is required to capture a significant fraction of NCOs. The greater sensitivity of detection of NCOs compared with earlier studies allowed us to determine that there are substantially more NCOs than COs at A3. For example, in the A/J × DBA/2J F1 hybrid, the NCO frequency is ~2.3% on a per meiosis basis, which is 10-fold higher than the CO frequency (0.22%). Thus, for the first time, the ratio of NCOs to COs at a hotspot approximates the cytological estimate of the ratio of global DSBs to COs observed in mouse spermatocytes.9 The high NCO to CO ratio supports the hypothesis that most meiotic DSBs provoke interactions between homologs, which are critical for meiotic progression, in particular homolog pairing.
NCOs peaked in number at the center of the A3 hotspot; however, a substantial proportion of NCOs occurred in the flanking regions of the hotspot. Considering NCOs on the DBA/2J chromosome for all of the strain combinations analyzed, only about half of NCOs localized within the central 43 bp of the A3 hotspot (Fig. 2B). The remaining NCO gene conversions were located in the flanking ~750 bp on either side. As NCO gene conversions are short, we infer that the distribution of NCOs approximates the distribution of DSBs. Thus, DSBs do not form exclusively at the center of A3, but rather span a broad region encompassing ~1.5 kb. Direct analysis of meiotic DSBs in yeast21–23 demonstrates that DSBs are distributed throughout the width of hotspots, and also frequently spread into the flanking regions, as inferred for the A3 hotspot. If NCO gene conversion patterns at other mammalian hotspots are similar to what we observe at A3 and what has been shown in yeast, then meiotic DSBs are clearly not restricted to the center of hotspots, as is frequently modeled in mammals (e.g., Refs. 24 and 25); thus, while knowing the exact center of a hotspot is useful,26 it provides only part of the information regarding DSB distribution and the resultant gene conversion patterns.
Transmission distortion and allelic shuffling due to gene conversion: NCO supremacy?
Meiotic recombination can have a major impact on genomic architecture. COs break up allelic blocks on chromosome-wide scales, but gene conversion at hotspots from COs and NCOs also contribute to allelic shuffling, albeit over much smaller distances. Analyses of genetic maps in humans indicate that the major impact of meiotic recombination on genetic diversity occurs at the scale of hotspots, implicating gene conversion as a main driver of diversity.27 Although NCO gene conversion tracts are smaller than those of COs, the contribution of NCOs can be significant owing to their high frequency. While it is formally possible that A3 has an unusually high NCO to CO ratio, the concordance with the cytological ratio of DSBs to COs suggests that A3 may be typical of mammalian hotspots.
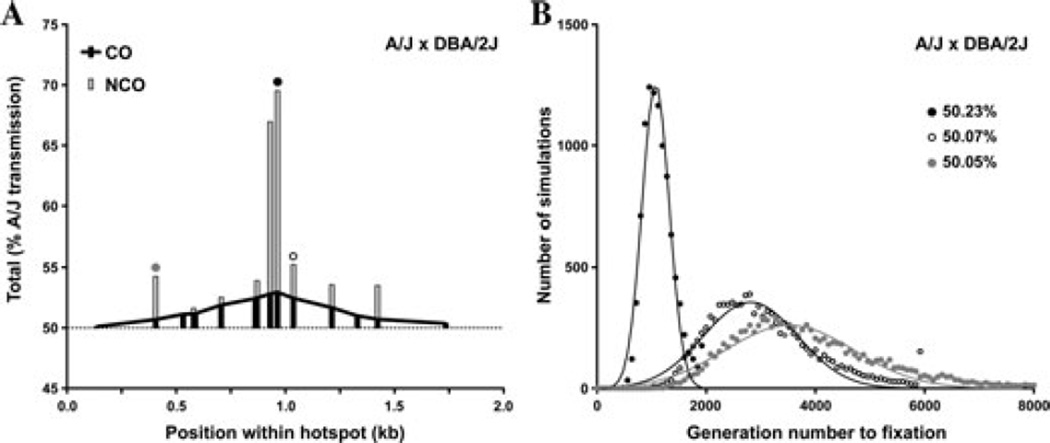
Importantly, gene conversion can also lead to transmission distortion, that is, deviation from the expected 50:50 gametic ratio of parental alleles. Transmission distortion in this case is a consequence of preferential DSB formation on one homolog leading to over transmission of alleles from the other, uncut homolog. The impact of gene conversion on transmission distortion depends on several variables: the absolute DSB frequency at the hotspot, the magnitude of DSB bias for one homolog over the other, and the length of the subsequent gene conversion tract. While transmission distortion due to CO gene conversion has been analyzed at several recombination hotspots in mouse and human,17–20 the ability to capture a significant fraction of NCOs at A3 provided an opportunity to assess the contribution of both CO and NCO gene conversion to transmission distortion. The percent transmission of the A/J allele peaks at ~70% for the polymorphism at the center of A3 when considering all recombination events (black circle, Fig. 3A). When considering all gametes, the transmission distortion from CO gene conversion translates into a gametic ratio of 50.044:49.956 for the A/J allele. However, because of the much higher frequency of NCOs in the center of the hotspot, adding the transmission distortion from NCO gene conversion increases this gametic ratio to 50.23:49.77. If the polymorphism at the center of the hotspot also causes the DSB bias between A/J and DBA/2J, population simulations predict that transmission distortion will lead to fixation of the A/J polymorphism and hotspot quiescence (i.e., a dampening of hotspot activity) in less than 1,200 generations (black circles, Fig. 3B).
Figure 3.
Transmission distortion and allelic fixation at the A3 hotspot. (A) Percent A/J transmission for A/J × DBA/2J F1 hybrids from COs (black bars and line) and NCOs (gray bars). The dotted line at 50% is Mendelian transmission. Circles above the bars demarcate polymorphisms analyzed in B. (B) Monte Carlo simulations (Wright-Fisher model) to determine the number of generations to fixation for the three indicated polymorphisms, with the simplified assumption that the polymorphism is causative for the DSB bias. Calculated gametic ratios are indicated.
Similar simulations predict that the high frequency of recombination and the strong DSB preference for DBA/2J would also lead to fixation of other nearby polymorphisms in the A3 hotspot. Polymorphisms just adjacent to the center of the hotspot may experience a more equitable contribution of NCOs and COs to transmission distortion (open circle, Fig. 3), whereas polymorphisms more distant from the center may undergo transmission distortion largely driven by NCOs (gray circle, Fig. 3). Thus, while short NCO gene conversion tracts have a more punctate effect on transmission distortion than COs, the substantially higher ratio of NCOs to COs at A3 results in a 1.9-fold greater contribution of NCOs to transmission distortion (Fig. 3A). We predict that this greater contribution of NCOs would be observed genome wide, as the approximately five-fold longer CO gene conversion tracts are counterbalanced by ~10-fold more NCOs. Thus, NCOs are predicted to have a significant impact on allele fixation, hotspot quiescence, and, ultimately, genomic diversity.
Short NCO gene conversion tracts in mammals: implications for hotspot longevity
The breadth of NCO gene conversions across the A3 hotspot also has consequences for the longevity of hotspots. Recent studies have determined that the location of DSB hotspots is largely controlled in mouse and humans by PRDM9, a meiosis-specific histone H3 methyltransferase.28 PRDM9 has an array of Zn-finger DNA-binding motifs that target sites for SPO11-dependent DSBs.29,30 The Zn-finger domain of PRDM9 is rapidly evolving,31,32 suggesting that PRDM9 has undergone positive selection to alter its DNA-binding residues. The rapidly evolving DNA binding domain of PRDM9 may function to counteract the consequence of gene conversion on hotspot loss,33 referred to as the hotspot paradox. This paradox posits that during meiosis any homolog that preferentially undergoes a DSB (the “hotter” allele) is under transmitted compared to the “colder” allele of the other homolog. In this manner, hotspots would rapidly extinguish themselves, prompting the question: how can hotspots exist? Rapid evolution of PRDM9 DNA-binding sites acts to alter hotspot locations genome wide, providing a potential mechanism to counter hotspot loss.
Features of NCOs themselves nonetheless act to mitigate the speed of hotspot extinction. Biochemical data indicate that PRDM9 preferentially binds to a DNA sequence at the center of hotspots,30 and genetic data suggest that a single nucleotide polymorphism can alter PRDM9 binding.34 The short conversion tracts of NCOs and the wide distribution of NCOs throughout the A3 hotspot make it likely that a significant fraction of NCO gene conversion tracts do not span the PRDM9-binding site. In contrast, we infer from the pattern of CO exchange points that the majority of CO gene conversions would involve the PRDM9-binding site. The combination of the high NCO to CO ratio, short gene conversion tracts of NCOs, and the wide distribution of NCOs implies that many DSBs can promote homolog pairing without contributing to hotspot quiescence. In this manner, even extremely active hotspots can have longevity over evolutionary time scales.
Mammalian homologous recombination mechanisms are analogous in many ways to those in budding yeast. For example, many of the enzymes and structural proteins that mediate meiotic recombination are conserved. However, clear mechanistic differences exist between mammals and yeast, which can have profound consequences. Yeast does not have a PRDM9-dependent mechanism to target sites for SPO11 cleavage; instead, hotspots are, for the most part, associated with promoters of genes (see Ref. 23 and references therein). Further, yeast DSB hotspots are smaller in width, with a median of less than 200 bp, while mammalian hotspots, like A3, are wider, spanning ~1.5–2 kb. In yeast, COs outnumber NCOs, and gene conversion tracts are substantially longer, averaging ~2 kb for COs and 1.8 kb for NCOs.35 If gene conversion mechanisms in mammals were identical to those in yeast, repair of most DSBs would convert the PRDM9-binding site, dramatically increasing the speed of hotspot quiescence.
The localization of hotspots in budding yeast to promoters means that hotspot sequences are evolutionarily constrained for reasons unrelated to meiotic recombination.36 Hotspots are short lived in mammals—for example, hotspots in chimps differ in location from those in humans31—so clearly their location is not highly constrained. However, prolonged longevity of sequences that are favorable for DSB formation in mammals may also be found advantageous for as yet undetermined reasons.
Conclusion
Our comprehensive analysis of the A3 hotspot has provoked new insights for how gene conversion can influence genomic diversity and evolution. Approximately 90% of meiotic recombination products at A3 are NCOs, approaching for the first time the frequency predicted based upon cytological determination of the numbers of DSBs versus COs and providing support for the model that frequent DNA repair interactions promote homolog pairing during meiosis. Despite the more punctate effect of NCO gene conversion upon transmission distortion, the high frequency of NCOs results in approximately two-fold greater contribution to transmission distortion than COs. While half of NCOs occur in the center of the hotspot, the remaining NCOs are dispersed across the width of the hotspot. Due to the short gene conversion tracts associated with NCOs, NCOs in the flanking regions can promote homolog pairing and result in allelic shuffling without causing hotspot quiescence. Thus, many of the features of mammalian recombination—a high ratio of NCOs to COs, short gene conversion tracts, and wide hotspots—promote increased longevity of mammalian hotspots over evolutionary time scales.
Acknowledgments
We thank Aaron Gabow and Alex Lash of the MSKCC Bioinformatics Core for calculations of fixation and Liisa Kauppi and Robert J. Klein for helpful discussions. This work was supported by NIH grants HD040916 and HD53855 (M.J. and S.K.).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes. Dev. 2010;24:1201–1207. doi: 10.1101/gad.1944710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 3.Kauppi L, Jasin M, Keeney S. The tricky path to recombining X and Y chromosomes in meiosis. Ann. N.Y. Acad. Sci. 2012;1267:18–23. doi: 10.1111/j.1749-6632.2012.06593.x. This volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 2007;16:R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 5.Hunter N, Aguilera A, Rothstein R. Meiotic Recombination, in Topics in Current Genetics, Molecular Genetics of Recombination. Vol. 17/2007. Heidelberg: Springer-Verlag; 2007. pp. 381–442. [Google Scholar]
- 6.Keeney S. In: Spo11 and the Formation of DNA Double- Strand Breaks in Meiosis, in Recombination and Meiosis. Egel R, Lankenau D-H, editors. Berlin, Heidelberg: Springer-Verlag; 2007. pp. 81–123 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 9.Cole F, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 2012;14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways inmouse meiosis. Mol. Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 11.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 12.Jeffreys AJ, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat. Genet. 2005;37:601–606. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- 13.Cole F, Keeney S, Jasin M. Comprehensive, finescale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol. Cell. 2010;39:700–710. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelmenson PM, et al. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics. 2005;169:833–841. doi: 10.1534/genetics.104.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauppi L, May CA, Jeffreys J. Analysis of meiotic recombination products from human sperm. Methods Mol. Biol. 2009;557:323–355. doi: 10.1007/978-1-59745-527-5_20. [DOI] [PubMed] [Google Scholar]
- 16.Cole F, Jasin M. Isolation of meiotic recombinants from mouse sperm. Methods Mol. Biol. 2011;745:251–282. doi: 10.1007/978-1-61779-129-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 2002;31:267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 18.Baudat F, de Massy B. Cis- and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS Genet. 2007;3:e100. doi: 10.1371/journal.pgen.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bois PR. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol. Cell Biol. 2007;27:7053–7062. doi: 10.1128/MCB.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb AJ, Berg IL, Jeffreys A. Sperm cross-over activity in regions of the human genome showing extreme breakdown ofmarker association. Proc. Natl. Acad. Sci. USA. 2008;105:10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Massy B, Rocco V, Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. Embo J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calabrese P. Apopulation genetics modelwith recombination hotspots that are heterogeneous across the population. Proc. Natl. Acad. Sci. USA. 2007;104:4748–4752. doi: 10.1073/pnas.0610195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pineda-Krch M, Redfield RJ. Persistence and loss of meiotic recombination hotspots. Genetics. 2005;169:2319–2333. doi: 10.1534/genetics.104.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer CC, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale MJ. PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11:104. doi: 10.1186/gb-2010-11-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segurel L, Leffler EM, Przeworski M. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 2011;9:e1001211. doi: 10.1371/journal.pbio.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grey C, et al. Mouse PRDM9DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diversemetazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coop G, Myers SR. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum. Mol. Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 35.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]