Summary
DNA methylation, mediated by the combined action of three DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B), is essential for mammalian development and is a major contributor to cellular transformation. To elucidate how DNA methylation is targeted, we mapped the genome-wide localization of all DNMTs and methylation, and examined relationships between these markers, histone modifications, and nucleosome structure in a pluripotent human tumor cell line in its undifferentiated and differentiated states. Our findings reveal a strong link between DNMTs and transcribed loci and that DNA methylation is not a simple sum of DNMT localization patterns. Comparing the epigenomes of normal and cancerous stem cells, and pluripotent and differentiated states, shows that the presence of at least two DNMTs is strongly associated with loci targeted for DNA hypermethylation. Taken together, this study sheds important new light on determinants of DNA methylation and how it may become disrupted in cancer cells.
Introduction
Methylation at the 5-position of cytosine within the CpG dinucleotide is an epigenetic modification of DNA linked to transcriptional repression when present within promoter regions, or to transcriptional activity when present within gene bodies (Jones, 1999; Maunakea et al., 2010). Global genomic DNA methylation patterns are established and maintained by the combined action of three enzymatically active DNA methyltransferases; DNMT1, DNMT3A, and DNMT3B. DNMT1 is traditionally referred to as the maintenance enzyme, copying methylation after replication, while DNMT3A/DNMT3B are de novo enzymes that establish new patterns of methylation during differentiation (Baylin and Jones, 2011; Goll and Bestor, 2005). While extensively studied, much remains unknown regarding the division of labor amongst DNMT family members in methylating the genome.
While there is evidence for redundancy among the DNMTs, there is also mounting evidence for functional specificity. During development for example, genetic inactivation of Dnmt3a results in mice that develop to term and are grossly normal at birth, but die by four weeks of age. In contrast, Dnmt3b null mice fail to develop normally past E9.5 and no viable offspring are produced (Okano et al., 1999). In disease, genetic reduction of Dnmt1 levels reduces polyp formation in the ApcMin murine colorectal cancer model (Laird et al., 1995), over-expression of Dnmt3b in the same system results in enhanced size and number of tumors (Linhart et al., 2007), and conditional knockout of Dnmt3a in a mutant K-ras lung cancer model promoted tumor growth and progression (Gao et al., 2011). In humans, mutations in DNMT3B are responsible for the majority of Immunodeficiency, Centromere Instability, and Facial Anomalies (ICF) syndrome cases (Ehrlich et al., 2006). Collectively, these studies emphasize that DNMT family members possess distinct non-overlapping functions, however the regions of the genome bound and regulated by each DNMT, and the mechanisms by which they are directed to these sites, remain largely unknown.
One process that plays a role in regulating methylation patterns is the extensive array of post-translational modifications of amino acid side chains on the core histones H2A, H2B, H3, and H4, which collectively form the nucleosome core particle and package ~150bp of DNA. Histone modifications, like DNA methylation, play key roles in transcriptional regulation, genome stability, and embryonic development (Berger, 2007). Histone acetylation is generally associated with transcriptional activation, while the relationship between histone lysine methylation and transcriptional activity is dependent on the position and state (mono-, di-, or trimethylation, or me1, me2, and me3) of the modification (Barski et al., 2007; Wang et al., 2008). H3K27me3 and H2AK119 monoubiquitination (Ub), transcriptionally repressive marks mediated by the PRC2 and PRC1 polycomb complexes, respectively, have been positively associated with DNA methylation (Kallin et al., 2009; Vire et al., 2006). Loci regulated by polycomb in normal cells display an increased propensity to acquire aberrant DNA hypermethylation in cancer (Jin et al., 2009; Ohm et al., 2007; Schlesinger et al., 2007). The underlying positioning of nucleosomes themselves also influences DNA methylation (Chodavarapu et al., 2010). Taken together, these results strongly suggest that histone modifications and nucleosomal structure play key roles in determining DNA methylation patterns, however exactly how these chromatin determinants differentially affect each DNMT remains to be elucidated.
In the present study we sought to gain a more comprehensive understanding of the unique, cooperative, and overlapping targets of each enzymatically active DNA methyltransferase and to define relationships between these patterns and DNA methylation, histone modifications, and nucleosomal structure in human cells. Using pluripotent human embryonic carcinoma (EC) cells in both the undifferentiated and differentiated states as a model system, we mapped the genome-wide localization of key histone modifications positively and negatively associated with DNA methylation using chromatin immunoprecipitation (ChIP)-sequencing. Global DNA methylation patterns were established using methyl CpG-binding domain protein-based enrichment coupled with next generation sequencing (MBD-seq), and DNMT1, DNMT3A, and DNMT3B localization was defined using ChIP-seq. Finally, micrococcal nuclease digestion-based whole genome sequencing (MNase-seq) was employed to establish nucleosome positioning. Global patterns of epigenetic marks and transcription were then used to examine features of regions bound by each DNMT before and after differentiation. This work establishes, for the first time, direct connections between the DNMTs and other facets of the epigenome, and provides important new information about how DNA methylation is targeted.
Results
Characterization of global histone modification patterns in human embryonic carcinoma (EC) cells
To characterize the EC epigenome and how differentiation alters epigenetic marks, NCCIT human EC cells were treated with 10 μM all-trans retinoic acid (RA), or left untreated, then formaldehyde-fixed chromatin was prepared. NCCIT cells are derived from a nonseminomatous germ cell tumor and possess features of both seminomas and embryonal carcinomas. They resemble embryonic stem (ES) cells in a number of ways, but are adapted to grow as tumors. Upon treatment with RA, NCCIT differentiate into derivatives of all three embryonic germ layers and extraembryonic lineages ((Sperger et al., 2003) and Supplemental Fig. S1A). Following RA treatment over 21 days showed that expression of DNMT3B decreased, while DNMT1 and DNMT3A levels remained relatively constant (Figs. S1B–C). Global expression profiling revealed a high degree of correlation between NCCIT and WA09 (H9) human ES cells (Fig. S1D) and ontology analysis showed marked over-representation of terms related to embryogenesis and development for RA-differentiated NCCIT cells (Fig. S1E).
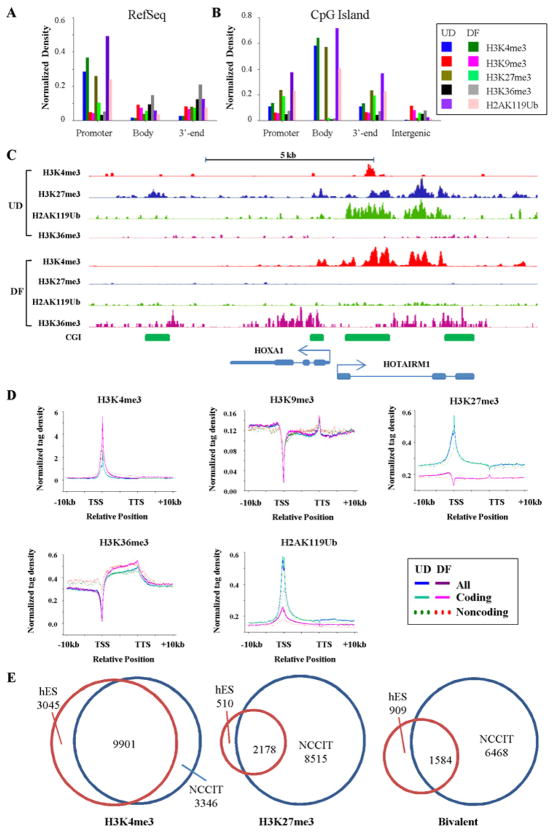
Based on these data, we focused subsequent RA treatments on day seven as a balance between inducing the differentiation program while retaining sufficient levels of DNMT3B to perform subsequent ChIP-seq experiments. Antibodies against the activating mark H3K4me3, the repressive marks H3K9me3, H3K27me3, and H2AK119Ub, and the gene body-associated mark H3K36me3, were used for ChIP-seq (summarized in Table S1). ChIP-seq data was annotated and the association of each histone modification with intragenic features was examined (summarized in Fig. 1A). H3K4me3 was most enriched in promoter regions; H3K27me3 and H2AK119Ub were also high at the TSS. H3K36me3, in contrast, was most highly enriched in gene bodies, while gene 3′-ends were mildly enriched for several histone marks (Fig. 1A). Given the importance of CpG islands (CGIs) to the biology of DNA methylation, we also stratified our ChIP-seq data to show marks associated with CGIs in different contexts. In general, CGIs associated with genes were marked by H3K4me3 and nearly devoid of H3K9me3 (Fig. 1B). Histone modifications within a portion of the HOXA locus are shown in Fig. 1C to illustrate a representative bivalent gene (H3K4me3, H3K27me3, and H2AK119Ub-positive) transitioning to a monovalent (H3K4me3 only) state following RA-induced differentiation. This figure also highlights the enrichment of H3K36me3 in the gene body. Confirmation of the presence of select histone modifications mapped by ChIP-seq at loci relevant to development and RA-induced differentiation of NCCIT cells, using ChIP-qPCR, is shown in Fig. S2.
Figure 1. Global analysis of histone modification patterns in EC cells.
(A) Relationships between genomic features and ChIP-seq peaks for five histone modifications in undifferentiated (UD) and seven day RA-differentiated (DF) NCCIT cells. The Y-axis represents the fraction (proportion) of the total length of the peaks in a genomic feature normalized to the total length of that feature present in the genome. Features were defined with hg19 RefSeq gene as: promoter (TSS-1kb), gene body (TSS to TTS), and 3′-end (TTS+1kb). (B) Enrichment of histone modifications in CpG islands (CGIs) based on context as in part A. (C) Representative ChIP-seq results at a locus which transitions from bivalent to monovalent during differentiation. Bent arrow: TSS (+1). (D) Normalized tag distribution profiles of five histone modifications across intragenic regions before and after differentiation. Each gene body is normalized to 0–100%, from the TSS to the TTS, with 10kb upstream (promoter) and 10 kb downstream of the gene body shown. Tag densities were normalized to total mapped read numbers in each sample. Different colors are used to represent UD or DF conditions and genes are further stratified by protein coding potential. (E) Venn diagrams summarizing overlap between histone modifications and bivalent loci in UD NCCIT compared to human ES cells.
We further investigated the localization of each histone mark by generating normalized tag density distribution plots across genes by type (protein coding vs non-coding, Fig. 1D), stratified by promoter CpG density (Fig. S3A), and across CGIs within different contexts (Fig. S3B). H3K4me3 localized to a sharp double peak spanning the TSS with a trailing shoulder extending into the gene body (Fig. 1D). H3K27me3 and H2AK119Ub profiles were similar in undifferentiated NCCIT cells. After differentiation, however, H3K27me3 patterns changed markedly, being almost a mirror image of their distribution in the UD state (Fig. 1D). H3K36me3 was depleted over the TSS but markedly and progressively enriched across gene bodies. We also examined the association of histone modifications with promoters of different CpG density. Stratifying genes in this way did not alter distribution profiles, but rather revealed that HCPs and LCPs were the most and least enriched, respectively, for most histone modifications (Fig. S3A). CGIs were markedly enriched for H3K4me3, particularly those associated with promoters (Fig. S3B). H3K27me3 and H2AK119Ub were similarly enriched at CGIs in the undifferentiated state, however both marks were reduced after differentiation and H3K27me3 became depleted (Fig. S3B), suggesting that many CGIs are bivalent in UD NCCIT cells but become monovalent after differentiation. Using recently published ChIP-seq data (Adamo et al., 2011), we compared histone marks in human ES and NCCIT cells. There was marked overlap between genes bound by H3K4me3 and H3K27me3 in hES and UD NCCIT cells (Fig. 1E), however NCCIT cells had more H3K27me3-containing loci. This result also likely accounts for NCCIT cells possessing a greater number of bivalent loci than hESs. Taken together, these data reveal similarities between hES and hEC cells, both in terms of expression and histone modification patterns, although notable differences also exist that may reflect their distinct cellular origins or state of transformation.
Mapping the localization of DNMT1, DNMT3A, DNMT3B, and DNA methylation genome-wide in NCCIT cells
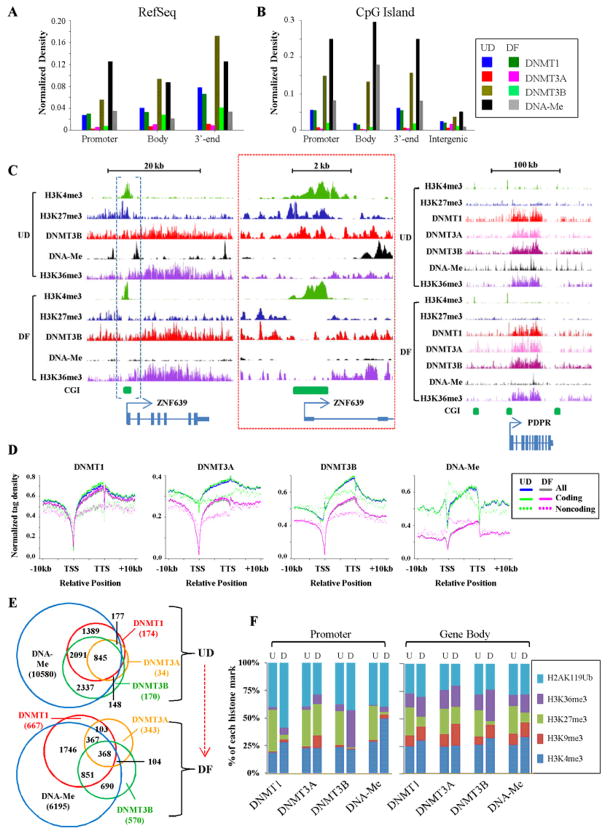
To examine how each DNA methyltransferase interfaces with DNA methylation and histone modifications, we used antibodies directed against DNMT1, DNMT3A, and DNMT3B (Fig. S1C) in ChIP-seq experiments, and determined global DNA methylation patterns using methyl-CpG binding domain protein-based enrichment coupled with next generation sequencing (MBD-seq), essentially as described in (Serre et al., 2009). Validation of the specificity of MBD-based enrichment for known methylated and unmethylated loci is shown in Fig. S4A. DNMT ChIP-seq and MBD-seq data is summarized in Table S1. DNMT3A was least enriched at intragenic loci, while DNMT1 and DNMT3B were present at higher levels, particularly DNMT3B in bodies and 3′-ends. After differentiation, DNMT3B levels generally declined in intragenic regions and CGIs (Figs. 2A–2B). Representative binding profiles are shown in Fig. 2C. Since repetitive regions are established targets of DNA methylation, we also stratified our localization data by repeat class, which showed that DNMT3B was enriched in SINEs and DNMT3A was enriched in satellite repeats (Fig. S4B). ChIP-qPCR and MBD pull-down coupled to qPCR were used to confirm DNMT binding and DNA methylation, respectively (Fig. S2), showing the validity of our whole-genome-based analyses.
Figure 2. DNMT1, DNMT3A, DNMT3B, and DNA methylation show distinct and overlapping distributions across the NCCIT genome.
(A) Enrichment of each enzymatically active DNA methyltransferase and DNA methylation within intragenic features of undifferentiated (UD) and RA-differentiated (DF) NCCIT cells as described in Fig. 1. (B) Enrichment of DNMTs and histone modifications within CpG islands (CGIs) based on context. (C) Representative ChIP-seq and MBD-seq results for the indicated DNMTs, DNA methylation, and histone modifications at two loci. In the left panel, the region enclosed by blue brackets is enlarged in the center (red boxed region), to highlight reduced DNMT3B binding at the TSS upon differentiation. (D) Normalized distribution profiles of DNMTs and DNA methylation across intragenic regions in NCCIT cells before and after differentiation as described in Fig. 1. (E) Venn diagrams summarizing overlap between genes marked by at least one DNMT and DNA methylation in UD and DF NCCIT cells. Further breakdown by promoter and body is shown in Fig. S5D. (F) Proportion of each histone mark overlaping with DNMT-bound or DNA methylated regions at promoters (left) or gene bodies (right). U-undifferentiated, D-RA-differentiated NCCIT.
To further examine how each DNA methyltransferase occupied intragenic regions, we created tag density plots across genes stratified by protein coding potential and CpG density, across CGIs, and across exons. DNMT1 and DNMT3B were most enriched in gene bodies, and, like H3K36me3, increased from 5′ to 3′ (Fig. 2D). DNMT1 was depleted at both the TSS and the TTS. DNMT3A and DNMT3B were depleted at the TSS in DF cells, but were either similar to gene body levels or slightly enriched at the TSS before differentiation. DNA methylation, like DNMT1, was depleted at the TSS and the TTS, but progressively elevated across the entire gene body (Fig. 2D). The depletion of DNA methylation and DNMTs coincided precisely with the TSS-associated peak of H3K4me3 (Fig. S5A), however H3K4me3 trails into and overlaps with gene body DNA methylation. Stratification by CpG density revealed that HCP genes generally had the highest levels of DNMT binding in the gene body and the largest depletion at the TSS. Interestingly, peaks of DNMT1, DNMT3B, and DNA methylation at the TSS distinguished the ICP/LCP from HCP genes (Fig. S5A). DNMT1 and DNMT3A were depleted from CGIs, however DNMT3B was enriched at CGIs before differentiation (Fig. S5B). DNA methylation was also slightly elevated at CGIs overall, although promoter-associated CGIs were always least methylated and absolute levels of H3K4me3 were much greater than DNA methylation at CGIs (Fig. S5B). This result suggests that DNMT3B is capable of binding and methylating a fraction of CGIs even though they contain H3K4me3. We plotted DNMT and DNA methylation patterns across exons as a function of coding potential and CpG density. With the exception of DNMT3A, all DNMTs and DNA methylation were at least mildly enriched in exons (Fig. S5C). Interestingly, non-coding RNA genes were distinct in their profiles for many epigenetic marks (Figs. 1D, 2D, and S5C), including DNMT localization. This finding could be due to non-coding transcripts generally being expressed at lower levels than coding transcripts (Birney et al., 2007), necessitating distinct epigenetic regulation. Taken together, our results show that DNMT1, DNMT3A, and DNMT3B have both unique and overlapping distributions across genomic features dependent on cellular differentiation status.
Interrelationships between DNA methyltransferases, DNA methylation, and histone modifications
The overlap among the epigenetic marks we then examined further. Venn diagrams focused on DNA methylation showed that while the majority of DNMT-bound regions are methylated, nearly half of all methylated loci do not appear to be stably occupied by a DNA methyltransferase (Fig. 2E). In addition, there are a small number of regions in UD cells occupied by at least one DNMT not positive for methylation, and this class of sequences increases more than four-fold after differentiation (Fig. 2E). Gene promoters and bodies were also separately compared, revealing even more DNMT-bound unmethylated loci within gene bodies (Fig. S5D). At promoters, DNMT1 overlapped most with DNMT3B and methylation and least with DNMT3A. After differentiation, methylated promoters were most highly associated with DNMT1. Finally, we examined co-occupancy of DNMT-bound loci with five different histone modifications. In undifferentiated NCCIT cells, promoter-associated DNMTs overlapped mostly with H3K27me3 and H2AK119Ub (Fig. 2F). After differentiation, DNMT1 overlap with H2AK119Ub increased and DNMT3B overlap with H3K36me3 increased. In gene bodies, overlap of DNMTs with H3K9me3 and H3K36me3 increased after differentiation. Overlap between DNA methylation and H3K4me3 is in large part due to the TSS-associated H3K4me3 peak trailing into the body (as illustrated in Fig. S5A). These results demonstrate that while most regions of the genome bound by DNMT are methylated, methylation is not a simple sum of DNMT binding patterns.
Differential enrichment of DNMTs and epigenetic marks at loci targeted for DNA hypermethylation – a comparison of two systems
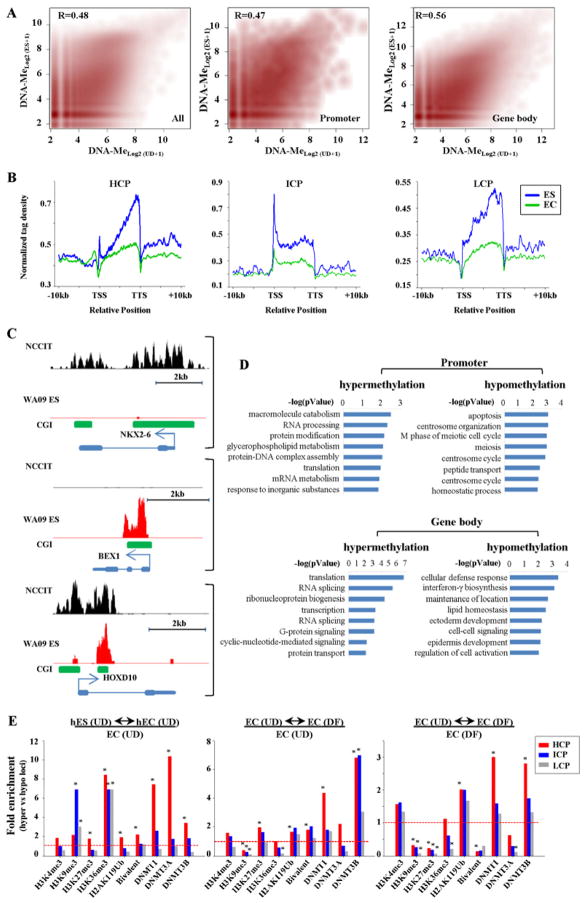
Given that both NCCIT EC and WA09 ES cells are pluripotent, but one is immortal (ES) and the other transformed (EC), we were interested in comparing their patterns of DNA methylation to define chromatin-associated features of loci that sustain aberrant DNA methylation changes linked to the cancer phenotype. To accomplish this, MBD-seq of WA09 human ES cells was performed to permit direct comparison of genome-wide methylation patterns using the same assay platform (Table S1). NCCIT and WA09 methylation patterns over the entire genome, at promoters, and in gene bodies were generally less well correlated with each other than gene expression patterns (Fig. 3A, compare to Fig. S1D). Tag density plots comparing hES and NCCIT intragenic DNA methylation reveal qualitatively similar patterns, although hES generally have higher levels of gene body methylation and differ to some degree at the TSS in all CpG density classes (Fig. 3B). Representative similarly and differentially methylated loci are shown in Fig. 3C.
Figure 3. DNA methylation and DNMT localization in normal versus malignant stem cells and features of loci targeted for methylation.
(A) MBD-seq data from pluripotent NCCIT and WA09 human ES cells compared over the entire genome (left), promoters (middle), and gene bodies (right). Pearson correlation coefficients are shown. (B) Normalized tag distribution profiles of DNA methylation across gene bodies in undifferentiated hES and NCCIT cells. (C) Representative MBD-seq results in UD NCCIT (black) compared to WA09 human ES cells (red) at loci hypomethylated (top, NKX2-6), hypermethylated (middle, BEX1), or similarly methylated (bottom, HOXD10) in WA09 relative to NCCIT. (D) Biological processes enriched in differentially methylated genes in NCCIT (UD) compared to WA09 using DAVID. (E) Left panel. Enrichment of DNMTs, histone modifications, and bivalent status at loci hypermethylated in EC (undifferentiated) relative to ES, compared to those loci that are hypomethylated in EC relative to ES cells. Middle-right panels. Enrichment of DNMTs and histone marks at loci that become hypermethylated upon RA-induced differentiation of NCCIT cells, relative to those loci that become hypomethylated upon differentiation (4X or greater change). Enrichments are relative to the histone modifications and DNMT binding patterns derived from NCCIT in the undifferentiated condition in the middle panel and the differentiated condition in the right panel. Significance was assessed using the chi-square test (*p<0.01). Dashed line – 1X enrichment level.
We hypothesized that loci differing in DNA methylation patterns between ES and EC represent the combined impact of adaptation to tumor growth and distinct cellular/developmental origin; cell culture-associated differences are unlikely to be a major contributor because both lines have been extensively cultured. Consistent with the hypothesis that some methylation differences between ES and EC are the result of differing developmental origin, ontology analysis showed enrichment of processes related to meiosis as hypomethylated in EC relative to ES (Fig. 3D). Since other methylation changes are likely linked to transformation, we examined the association of all differentially methylated loci with histone modifications, DNMT localization, and other characteristics of loci targeted for aberrant methylation, such as PcG regulation (Schlesinger et al., 2007). Furthermore, since many promoter CGIs are targeted for hypermethylation in adult cancers, we stratified all loci that become hypermethylated in undifferentiated NCCIT, compared to human ES, by their promoter region CpG density. This analysis revealed a modest but significant enrichment of PcG-mediated marks H3K27me3 and H2AK119Ub, individually and co-existent (bivalent), and H3K36me3 at HCP genes that were hypermethylated in UD NCCIT compared to hES cells (Fig. 3E, left graph). Others have shown that genes repressed by PcG in ES cells are 2–3X more likely to be targeted for aberrant DNA hypermethylation in cancers than non-PcG regulated loci (Schlesinger et al., 2007), consistent with our findings here. Genes with lower CpG density targeted for DNA hypermethylation showed marked enrichment for H3K9me3, consistent with a report showing that aberrantly hypermethylated loci in adult cancers display H3K9me3 enrichment in undifferentiated NTERA2 EC cells (Ohm et al., 2007). Interestingly, binding of all DNMTs, but particularly DNMT1 and DNMT3A, was highly enriched at HCP genes that were methylated in EC relative to ES (Fig. 3E), suggesting that their presence primes loci for de novo methylation.
While informative, the ES-EC comparison makes use of different cell lines and mapping of DNMT binding in ES is not currently available. Using loci that become hypermethylated in DF compared to UD NCCIT cells permits direct comparison of epigenetic marks involved in methylation targeting by comparing two states of the same cell line. To accomplish this, we correlated global DNA methylation patterns in UD and DF NCCIT to identify hyper- and hypomethylated loci (not shown). These genes were then examined for the presence of histone modifications and DNMTs that we mapped in both UD and DF states. As for the ES-UD comparison, loci that became hypermethylated upon NCCIT differentiation showed modest but significant associations with both PcG-mediated marks and bivalent status at HCP genes (Fig. 3E, middle graph). DNMT3B (at HCPs and ICPs), and to a lesser extent DNMT1 (at HCPs), were also highly enriched at loci in UD cells that sustained an increase in methylation upon RA treatment. In contrast to the ES-EC (UD) comparison, hypermethylated loci are not enriched for DNMT3A and are significantly depleted for H3K9me3 when comparing UD and DF NCCIT states. The former difference may be attributed to the high expression of DNMT3B in EC cells and the latter may be explained by H3K9me3 enrichment already being present on some genes in UD cells because they are transformed (Fig. 3E, left panel and (Ohm et al., 2007)). Interestingly, if we examine chromatin modifications associated with hypermethylated loci once they become methylated (in the DF condition), many of the same marks remain significantly enriched (e.g. DNMT1 and DNMT3B) or depleted (e.g. H3K9me3), however a significant depletion for both PcG regulated loci (defined by H3K27me3) and bivalency (Fig. 3E, right graph) is now observed. This result suggests that some loci targeted for DNA hypermethylation undergo an epigenetic switch in repressive marking (H3K27me3 to DNA methylation) and/or change from a poised to fully repressed state. These data taken together strongly suggest for the first time that DNMT binding does not necessarily equate to methylation of the bound region, however the presence of at least two DNMTs (DNMT1 and DNMT3A or DNMT3B), and the presence/absence of other histone modifications (that vary to some extent by cell type) render loci more susceptible to, or target, transformation and/or differentiation-associated DNA hypermethylation events.
Relationships between DNA methylation, DNMT occupancy, and transcription
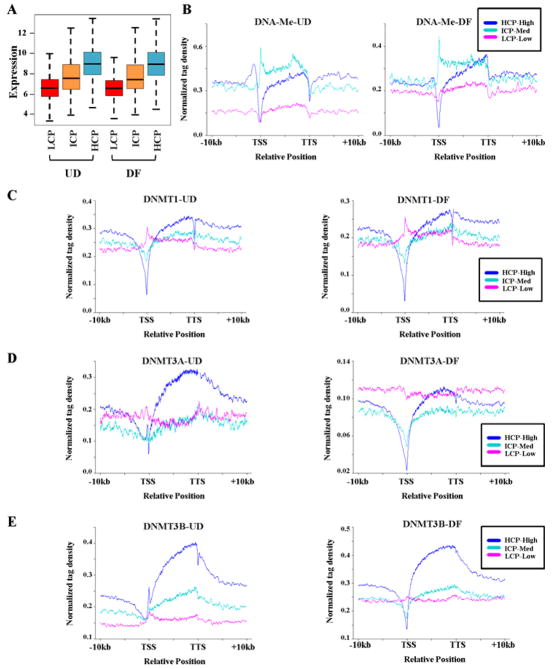
Since DNA methylation and each DNMT differentially associate with genes that undergo DNA methylation changes based on their promoter CpG density (Fig. 3), we first examined how CpG density relates to expression in NCCIT cells using our microarray our data (Fig. S1D). This analysis demonstrated a clear direct correlation between expression and promoter CpG density; with the majority of highly expressed genes being in the high CpG density class (HCP), the majority of moderately expressed genes falling into the ICP class, and most low expressed genes grouping into the LCP class (Fig. 4A). Given the associations between expression and promoter CpG density we observed, genes were stratified into the following combined classes: HCP-high expression, ICP-medium expression, and LCP-low expression to examine relationships between DNA methylation, DNMTs, and the three gene classes (Figs. 4B–4E). These plots show that DNA methylation at the HCP-high class of genes exhibits a peak of enrichment followed by a region of depletion flanking the TSS (hill-valley), followed by a gradual increase across the gene body and another sharp reduction at the TTS (Fig. 4B). For both UD and DF conditions, DNMT1 is markedly depleted at the TSS of HCP-high genes, elevated progressively across the gene body, followed by another depleted region at the TTS. Unlike DNA methylation, DNMT1 decreases only gradually in the 3′ region (Fig. 4C). DNMT3A showed the largest differentiation-specific differences among DNMTs, particularly at the TSS (Fig. 4D). Finally, DNMT3B showed the greatest gene body enrichment of all DNMTs, particular in the HCP-high class (Fig. 4E). A unique feature of DNMT3B in UD cells was a spike of enrichment just after the TSS in HCP-high genes. Tag density plots for these four marks stratified by expression level only are shown in Fig. S6 for comparison. Genes were further stratified based on whether they were up-regulated, down-regulated, or displayed no differential expression after RA treatment, revealing that up-regulated genes generally displayed lower levels of DNMTs and DNA methylation, particularly in the undifferentiated state, than genes that became down-regulated (Fig. S6). Taken together, these data reveal that DNMT1 and DNMT3B are most tightly linked to gene bodies and that their enrichment increases as a function of expression and CpG density, although the promoter region itself generally shows poor enrichment of these factors. These results also illustrate subtle but distinct localization patterns of each DNMT, particularly at the TSS and TTS, and that no single DNMT exactly parallels DNA methylation patterns.
Figure 4. DNA methyltransferases are differentially enriched in gene bodies as a function of expression and promoter CpG density.
(A) Relationship between promoter CpG density and expression in NCCIT cells. The box plot shows the relationship between log2 transformed gene expression level (stratified as: low expressed <7, medium expression 7–9, and high expression >9) and promoter CpG densities (HCP, ICP, and LCP). The bottom and top of each box represents the lower and upper quartiles respectively, the bar represents the median, and the whiskers represent the lowest and highest data within 1.5 interquartile range. (B–E) Tag density plots illustrating relationships between DNA methylation/DNMTs and expression stratified into: HCP-High (blue), ICP-Medium (green), and LCP-Low (red) classes. Normalized distribution profiles of (B) DNA methylation, (C) DNMT1, (D) DNMT3A, and (E) DNMT3B in NCCIT cells before (left) and after (right) differentiation.
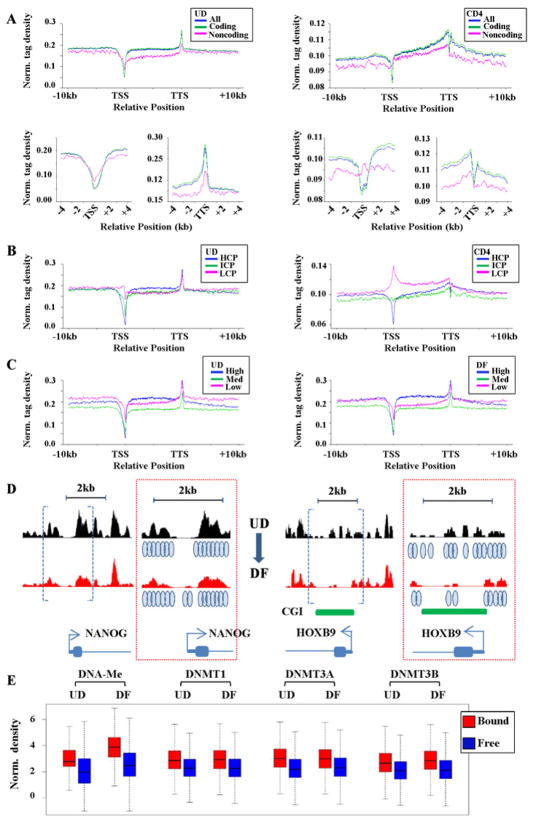
Global nucleosome positioning in NCCIT cells reveals links with DNA methyltransferase binding and DNA methylation, and distinct differences from normal human cells
Recent studies have suggested a positive link between DNA methylation and nucleosome occupancy (Chodavarapu et al., 2010), but global nucleosome positioning in transformed human cells has not, to our knowledge, been reported. To examine these associations, mononucleosomes from UD and DF NCCIT cells were isolated by micrococcal nuclease (MNase) digestion. Size fractionated gel-purified DNA of approximately 150bp was then subjected to massively parallel sequencing (MNase-seq, Fig. S7A), as described previously for human T cells (Schones et al., 2008). We obtained 359,468,369 and 328,468,369 reads from undifferentiated and RA-differentiated NCCIT cells, respectively, resulting in 16–18X genome coverage by 147bp nucleosome footprints (as calculated in (Valouev et al., 2011)), comparable in depth to a study using human T cells (Schones et al., 2008). The DANPOS algorithm was used to map nucleosomes, resulting in identification of 9,772,482 and 10,148,751 positioned nucleosomes in pluripotent and RA-differentiated NCCIT cells, respectively.
We then examined the relationship between nucleosome occupancy and gene features, promoter CpG density, and expression. To compare our data derived from a transformed cell line to nucleosome positions in normal human cells, the published CD4 T cell MNase-seq dataset was utilized (Schones et al., 2008). Tag density plots across intragenic loci show that nucleosome occupancy is relatively constant over genes in NCCIT cells, being marked by a depleted region at the TSS in both the undifferentiated (Fig. 5A) and differentiated states (Fig. S7B). Nucleosome positions in CD4 T cells showed notable similarities and differences with NCCIT cells. For example, nucleosome density increased more across the gene body in CD4 T cells than across gene bodies in NCCIT cells (Fig. 5A). Tag density plots centered on the TSS highlight the NDR present in both cell types. A TTS-centered plot emphasizes one of the most prominent differences; while CD4 T cells have another NDR at this location, NCCIT cells have a peak of nucleosome enrichment (Fig. 5A). Stratifying genes based on promoter CpG density shows that HCP and ICP genes are distinct from LCP genes in NCCIT cells; with the latter displaying a peak and the former two classes a depletion of nucleosome density at the TSS. HCPs in CD4 T cells have the deepest NDR at the TSS, like NCCIT cells, but both ICPs and LCPs show distinct nucleosome enrichments at the TSS (Figs. 5B, S7C). Moderate and highly expressed gene classes were most depleted of nucleosome density at the TSS (Fig. 5C). Representative nucleosome occupancy profiles at two loci displaying substantial differentiation-associated changes in expression in NCCIT cells are shown in Fig. 5D, and reveal subtle changes in nucleosome density at the TSS consistent with their change in expression. CGIs were uniformly depleted of nucleosomes (Fig. S7D), while exons were flanked by asymmetric peaks of nucleosome density (Figs. S7E–S7G). Taken together, these data provide not only the first comprehensive map of nucleosome positions in a human cancer cell line, but also begin to reveal differences between normal and transformed cells, which may collectively reflect the impact of cell of origin and transformation state on nucleosome positioning.
Figure 5. Relationships between global nucleosome positioning, genomic features, transcription, DNMTs, and DNA methylation.
(A) Normalized distribution profiles for nucleosome binding across intragenic regions for all, protein coding, and non-protein coding genes in pluripotent NCCIT cells. MNase-seq data from human CD4 T cells was used to create similar tag density plots. TSS and TTS (+/− 4kb)-centered plots are also shown to emphasize similarities and differences in these regions. (B) Tag density plots for nucleosome distributions across intragenic loci stratified according to promoter CpG density in UD NCCIT (left) and CD4 T cells (right). (C) Nucleosome distribution according to expression level in UD (left) and DF NCCIT (right) conditions. (D) Representative MNase-seq results at two loci that show among the largest change in expression upon differentiation of NCCIT cells. NANOG is down-regulated and this is accompanied by increased nucleosome density at the TSS. HOXB9 is up-regulated and shows the opposite trend in TSS nucleosome density. The browser data enclosed by blue brackets is enlarged at the right of each panel (red boxed region) with blue ovals indicating inferred positions of nucleosomes. (E) Analysis of relationships between DNMT occupancy, DNA methylation, and nucleosome bound versus free (linker) DNA. Using the method described in (Kaplan et al., 2009), nucleosome bound (red) and free (blue) DNA was defined and the correlation between these two states and regions enriched for DNMTs and DNA methylation was calculated. Enrichments are shown as box plots, with the black bar indicating the median, the box defining the first and third quartiles of the data, and whiskers indicating the range (excluding outliers). All bound versus free (linker) comparisons for a given mark are highly significant (P<2×10−16, Student’s two-sample t-test).
We focused subsequent analyses on relationships between nucleosome positioning, DNA methylation, and DNA methyltransferase localization. Using the method described by Kaplan et al. (Kaplan et al., 2009), we determined nucleosome bound and free (linker) DNA sequences, then compared enrichment of DNA methylation and each DNMT in both fractions. DNA methylation was clearly enriched in the nucleosome bound fraction. Not only did total DNA methylation within nucleosomal DNA increase after differentiation, but the difference between bound and unbound fractions also increased (Fig. 5E). All DNMTs also show significantly greater enrichment in the nucleosome-bound fraction before and after differentiation (Fig. 5E). These results therefore demonstrate that, despite the nucleosome generally being considered a barrier to DNA-based transactions, the DNMTs are preferentially targeted to this structure in vivo along with their mark, DNA methylation.
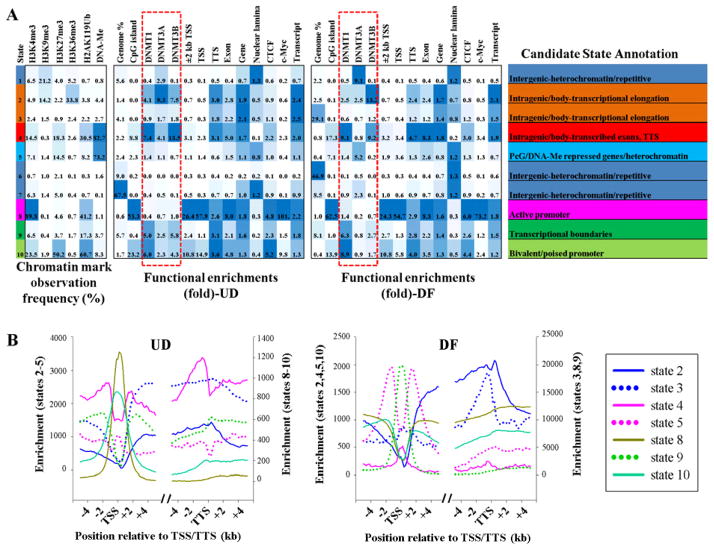
Defining chromatin states associated with DNA methyltransferase binding
To integrate all epigenetic mapping data across the two cell states and study relationships with DNMT localization, we applied a multivariate hidden Markov model (HMM) (Ernst et al., 2011), which makes use of combinatorial patterns of epigenetic modifications to distinguish chromatin states. Using all chromatin modifications except nucleosome positioning, chromatin states were learned. From this, ten states were selected that showed distinct biological enrichment (Fig. 6A). These states were distinctly associated with CGIs, gene features (TSS, TTS, etc.), transcription factor binding sites (c-Myc), the multifunctional insulator protein CTCF, and regions of heterochromatin associated with the nuclear lamina (Ernst et al., 2011) (Fig. 6A). Enrichment of states most associated with genes (states 2–5, 8–10) was plotted across the TSS and TTS (Fig. 6B), revealing distinct patterns that further reinforce the candidate state annotations. Most of the states changed relatively little upon differentiation, except 3, 6, and 7 (Fig. 6). DNMTs were most highly associated with states related to transcription, gene bodies, exons, and gene 3′-ends. DNMT3A also became more enriched in heterochromatin after differentiation. DNMT3B remained most highly associated with gene bodies while DNMT1 was linked more to promoter regions after differentiation (Fig. 6A). Our data clearly show a predominant interaction between DNMTs and DNA methylation and intragenic loci, and also that distinct classes of genes are differentially enriched for DNA methylation compared to the DNMTs (e.g. state 2 compared to state 4) further suggesting DNA methylation-dependent and -independent roles for the DNA methyltransferases.
Figure 6. Discovery and characterization of chromatin states in NCCIT cells.
(A) A multivariate hidden Markov model was used to learn chromatin states jointly across undifferentiated and differentiated NCCIT cells. In the left part of the table, emission parameters that were learned de novo from recurrent combinations of epigenetic marks over the entire genome are shown. Each value represents the frequency of the epigenetic mark at genomic positions corresponding to that particular state. Functional enrichment of different features and candidate state annotations for each state in UD and DF conditions are shown on the right. Blue shading – intensity scaled by column. (B) Comparison of enrichments for states associated with genes (states 2–5, 8–10 only) across the TSS and the TTS regions in UD and DF cells. The break indicates the remainder of the gene body. States are graphed with different scales (right and left y-axes) to highlight their patterns, relative to other states, rather than absolute enrichments.
Discussion
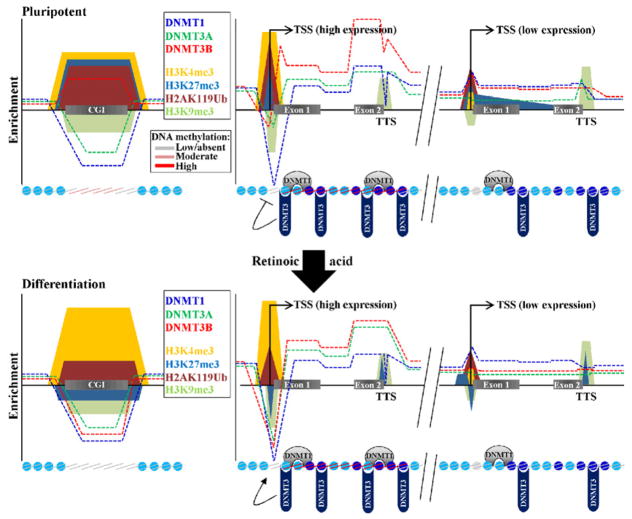
In this study we have investigated the relationships between a critical epigenetic mark, DNA methylation, the ‘writers’ of that mark (DNMT1, DNMT3A, and DNMT3B), histone modifications, and nucleosome structure in a cell type with relevance to both cancer and stem cell biology (summarized in Fig. 7). Genome-wide mapping of five histone modifications in EC cells reveals an epigenetic landscape displaying both similarities and differences with human ES cells. We also establish genome-wide binding profiles for all enzymatically active DNA methyltransferases and relate their binding patterns to the mark they directly mediate, DNA methylation, providing new insights into gene body methylation, possible methylation-independent functions of the DNMTs, and characteristics of loci targeted for DNA hypermethylation. Finally, we examine how the DNMTs and DNA methylation relate to the basic repeating unit of chromatin, the nucleosome, revealing an additional determinant of DNMT binding (the nucleosome itself), and distinct differences in nucleosome occupancy between normal and transformed cells. This study therefore sheds new light on how DNA methylation patterns are established and maintained in the context of chromatin that have relevance not only for better understanding the epigenetic underpinnings of cancer, but also for advancing our knowledge of how pluripotency, differentiation, and induced reprogramming are regulated.
Figure 7. Summary of key findings and model for targeting DNA methylation based on DNMT occupancy.
(A) The top portion of the figure represents summarized schematic enrichments of the indicated epigenetic marks, DNMTs, DNA methylation, and nucleosome positions in undifferentiated NCCIT cells based on our findings. Shown are three features, CGIs, highly expressed loci, and low expressed loci. DNMT enrichments are denoted with dashed lines, histone modifications by colored shapes, nucleosomes by blue circles (darker blue indicates more highly positioned or enriched), and DNA methylation by colored lines on the nucleosomes. Note that in general, DNMTs and DNA methylation are weakly associated with CGIs, except DNMT3B. DNMTs and DNA methylation are much more enriched at highly expressed (and CpG-rich) loci than at low expressed (CpG-poor) loci. The TSS is a nucleosome-free region regardless of expression level but is less pronounced in low expressed genes (gray circle). (B) Epigenetic marks after differentiation of NCCIT cells with retinoic acid. After differentiation of NCCIT, loci with DNMT1 and a DNMT3 tend to acquire DNA methylation and lose the H3K27me3 PcG marker and bivalency (shown by the colored shapes on nucleosomes, based on data in Fig. 3). The TSS and TTS therefore appear to set boundaries on methylation. Breakdown of these boundaries in cancer may permit aberrant methylation spreading into promoters and reduced gene body methylation.
A key realization not fully appreciated until the DNA methylome was examined on a genome-wide scale is that this mark is not strictly linked to transcriptional repression (Fig. 7). Rather, its function is dependent on genomic context; methylation in the promoter/5′-end is associated with repression, but methylation within the gene body is positively associated with expression (Maunakea et al., 2010). While the former has been intensively studied, the latter has not despite the fact that gene body methylation has been recognized for some time (Jones, 1999). Our ChIP-seq data reveal strong enrichment of DNMTs in gene bodies, especially DNMT3B and DNMT1. DNMT localization patterns are similar, but not identical to the distribution of DNA methylation. When expression level is factored in, DNMT3B is the most highly enriched DNMT as a function of expression. Further differences between DNA methylation and DNMT binding emerged, particularly at the TSS, suggesting that DNMTs are exerting methylation-independent regulatory roles within this region. Interestingly, Dhayalan et al. reported that the PWWP domain of DNMT3A binds to H3K36me3 and enhances DNMT3A activity (Dhayalan et al., 2010). It is unclear whether DNMT3B’s PWWP domain has a similar preference, although an analysis of methylation in ICF syndrome cells revealed substantial gene body hypomethylation (Aran et al., 2011). Further support for a direct link between DNMTs and transcribed loci comes from the recent report that DNMT3A and DNMT3B interact with RNA polymerase II subunits and the pol II elongation complex known as PAF1 (Rigbolt et al., 2011), which facilities transcriptional elongation and is also implicated in transcriptional initiation and mRNA processing (Kim et al., 2010). DNA methylation, like H3K36me, may repress spurious transcription within gene bodies that originate from repetitive elements, or regulate tissue-specific alternative promoter use or splicing. Elucidating the function of DNA methylation within the gene body and its role in disease represents a critical area of future research, which should be facilitated by the results presented here.
Data derived largely from gene knockout studies provides strong evidence for unique and overlapping roles within the DNMT family. Our results shed significant new light on this aspect of DNA methylation biology. Through integrative and comparative analysis we define how each DNMT interacts with various genomic features, with each other, and with other epigenetic marks. These analyses show that DNMTs possess both similarities and differences in their patterns of binding. For example, DNMT3A is least enriched within intragenic loci, while DNMT3B is most enriched within gene bodies. A recent study by Choi et al. also supports the division of labor hypothesis (Choi et al., 2011). These authors showed that genes targeted for de novo methylation by ectopic DNMT3A over-expression were associated with H3K4me3, while those targeted by DNMT3B were associated with H3K27me3 (Choi et al., 2011). Comparing DNMT localization to DNA methylation patterns reveals that, indeed, the majority of loci occupied by DNMTs are methylated, but also that nearly half of all methylated loci do not appear stably bound by a DNMT. The relatively small number of regions bound by at least one DNMT but not overlapping with methylation increases nearly four-fold after differentiation (sixfold in gene bodies). Hidden Markov modeling further supports a link between DNMTs and gene bodies, exons, and gene 3′-ends. The HMM model shows that some states have high DNA methylation and DNMT representation (e.g. state 4), while other states do not show equal representation (e.g. states 2, 9, and 10). Indeed, there is growing precedence for non-DNA methylation-related functions for DNMTs in gene regulation (Espada, 2012). Alternatively, loci occupied by DNMT, but not methylated, might be primed for later de novo methylation events, a notion supported by our comparison of the epigenomes of NCCIT cells in pluripotent and differentiated states and with hES cells (Fig. 7). It is also possible that certain biases inherent to all methylated DNA enrichment - next-gen sequencing methods and challenges associated with mapping reads within repetitive loci contribute to our finding of incomplete overlap between methylated regions, DNMT bound loci, and heterochromatic repetitive regions. Despite these potential issues, our data reveal the existence of consistent and novel links between DNMTs and transcribed loci, suggesting DNMTs and DNA methylation have important, but as yet unrecognized functions within intragenic regions and are not exclusively associated with heterochromatin or transcriptional repression.
In a recent study, approximately 20% of the genome exhibited significant nucleosome positioning preference, suggesting that the majority of nucleosomes are not subject to rigorous positioning, and that most promoters have a nucleosome depleted region (NDR) (Valouev et al., 2011). Our data confirm the presence of a genome-wide NDR flanking the TSS and we observed further depletion as a function of expression. By comparing NCCIT nucleosome positions to published nucleosome positioning data derived from CD4 T cells, we identified several differences. The TTS region in particular showed opposing trends in nucleosome density, and it will be of interest to determine the contribution of cellular transformation to this and other differences as more nucleosome profiles are generated. Our results show for the first time that DNMTs, like methylation (Chodavarapu et al., 2010), are more highly enriched in nucleosome-bound DNA compared to linker DNA, and methylation becomes further enriched in nucleosomes after differentiation (Fig. 7). Exactly how DNMTs are targeted to or access CpG sites within nucleosomal DNA is unknown. Chromatin remodelers that interact with DNMTs, such as SNF2H (Geiman et al., 2004), could facilitate this process. Alternatively, mechanisms may exist to couple DNA methylation to nucleosome reassembly following DNA replication, perhaps involving interactions between DNMTs, histone modifiers, and CAF1 (Sarraf and Stancheva, 2004).
Our results, and those of others (e.g. (Barski et al., 2007; Ernst et al., 2011)), reveal distinct boundaries at the TSS and TTS at the chromatin level. Data presented here show clear partitioning of DNMTs between promoters and gene bodies as well. It is intriguing to speculate that a defect in the TSS and/or TTS chromatin boundaries during tumorigenesis, or perhaps weaker boundaries in stem cells due to their more plastic nature, might permit DNA methylation/DNMTs to encroach into the promoter or permit activation of methylation at DNMT-bound but normally unmethylated loci (Fig. 7). Some of these methylation events may be part of normal cell-type specific epigenome changes associated with differentiation, leading defects in this process to cause methylation destined for one cell type to be aberrantly imposed on another (Irizarry et al., 2009). A boundary defect could account for gene body hypomethylation as well, either due to a breakdown in the targeting mechanisms or simple dilution of DNMTs. Several of the HMM states enriched for DNMTs, such as states 2 and 4, peak just inside the TSS and the TTS. By examining chromatin characteristics in two different systems, we showed for the first time that the presence of at least two DNMTs, along with other epigenetic marks such as those mediated by PcG complexes and H3K9me3, mark loci targeted for DNA hypermethylation in settings relevant to both adult cancers and differentiation programs (Fig. 7). Interestingly, both the maintenance DNMT, and at least one de novo DNMT, are enriched at methylated loci prior to the increase in methylation. DNMT3B represents a particularly strong candidate initiator of cancer-specific methylation changes because it is the only DNMT enriched at CGIs and highly over-represented at HCP genes that increase in methylation. Mapping DNMTs and chromatin marks in other cell types will be critical to refine models of methylation targeting, but have great potential to uncover previously unknown mechanisms and corrective measures for the methylation defects that typify cancer cells.
Experimental Procedures
Cell culture, treatments, RNA extraction, and quantitative PCR
NCCIT (from ATCC) and WA09 (H9, from WiCell) cells were grown as described in the Supplemental Experimental Procedures. All-trans retinoic acid (RA) (Sigma) was dissolved in DMSO. Cells were treated with 10 μM RA or an equivalent volume of DMSO and collected at day 0, 1, 4, 7, 14 or 21. Total RNA preparation and quantitative RT-PCR was performed as described in (Gopalakrishnan et al., 2009) and the Supplemental Experimental Procedures.
Expression microarrays
Gene expression profiling was performed using Affymetrix Human Gene 1.0 ST arrays. All samples were run in triplicate at the GHSU Cancer Center Genomics Core Facility and analyzed as described previously (Jin et al., 2009) and in the Supplemental Experimental Procedures.
Chromatin immunoprecipitation and western blotting
ChIP and western blotting were performed essentially as described (Jin et al., 2009). Detailed conditions and a listing of all antibodies used are provided in the Supplemental Experimental Procedures.
MBD methylated DNA enrichment
Enrichment of CpG methylated DNA for NGS is based on the method originally described in (Serre et al., 2009) with the modifications detailed in the Supplemental Experimental Procedures.
Nucleosome preparation
Genome-wide nucleosome positioning experiments (MNase-seq) were performed essentially as described in (Schones et al., 2008). Additional detail is provided in the Supplemental Experimental Procedures.
Library preparation and next generation sequencing
DNA fragments isolated by ChIP, MBD enrichment, or MNase digestion, were used to create single-end sequencing libraries using the Genomic DNA Sample Prep Kit (Illumina) with only minor modifications, as described in the Supplemental Experimental Procedures. Libraries were QC’d on a Bioanalyzer 2100 high sensitivity DNA chip and sequenced on an Illumina GAIIX (72 bp read) or a HiSeq2000 (100 bp read) at the Tufts University Genomics Core Facility or at BGI Americas.
Data analysis and HMM modeling
Raw sequence data was processed using the default settings for base calling to ensure read quality, with reads containing >two unidentifiable bases discarded, producing usable sequence data in standard FASTQ format. Reads were mapped against the human reference genome (hg19) using Novoalign software (http://www.novocraft.com) to produce alignment files. Alignment files were further analyzed by MACS1.4 (http://liulab.dfci.harvard.edu/MACS) for peak calling. Further processing and analysis of sequence data derived from all ChIP-seq, MBD-seq and MNase-seq experiments is described in the Supplemental Experimental Procedures. Hidden Markov modeling (HMM), using the histone modification and DNA methylation data, was performed as described in (Ernst et al., 2011) and the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
DNMT and DNA methylation maps are established in a pluripotent cancer cell line
DNMT binding is strongly linked to transcribed loci, exons, and nucleosomes
DNA methylation patterns are not simply the sum of DNMT binding patterns
Targeting of DNA hypermethylation is influenced by DNMT binding and histone marks
Acknowledgments
This work was supported by NIH grants R01CA114229, R01CA116028, R01AA019976 (KDR), NSF Postdoctoral Fellowship #0905968 (JE), RC1HG005334 (MK), and P01GM08535403 (SD and a Pilot Project grant to KDR). KDR is a Georgia Cancer Coalition Distinguished Cancer Scholar. We thank Kip Bodi and the Tufts University Genomics Core Facility for assistance with Illumina sequencing and Keji Zhao (NHLBI) for providing advice on nucleosome positioning experiments.
Footnotes
Accession Numbers
Sequencing and expression microarray data has been deposited into the NCBI Gene Expression Omnibus database under accession number GSE38938.
Supplemental information, including 7 figures, one table, and Supplemental Experimental Procedures, is located online at doi: xxx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature Rev Canc. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo K, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Heo K, Byun HM, An W, Lu W, Yang AS. Identification of preferential target sites for human DNA methyltransferases. Nucleic Acids Res. 2011;39:104–118. doi: 10.1093/nar/gkq774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Jackson K, Weemaes C. Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF) Orphanet J Rare Dis. 2006;1:2. doi: 10.1186/1750-1172-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J. Non-catalytic functions of DNMT1. Epigenetics. 2012;7:115–118. doi: 10.4161/epi.7.2.18756. [DOI] [PubMed] [Google Scholar]
- Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, Reddy S, Bell GW, Jaenisch R. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011;108:18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman TM, Sankpal UT, Robertson AK, Chen Y, Mazumdar M, Heale JT, Schmiesing JA, Kim W, Yokomori K, Zhao Y, Robertson KD. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic Acids Res. 2004;32:2716–2729. doi: 10.1093/nar/gkh589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009;18:3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Yao B, Li JL, Fields CR, Delmas AL, Liu C, Robertson KD. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- Kallin EM, Cao R, Jothi R, Xia K, Cui K, Zhao K, Zhang Y. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet. 2009;5:e1000506. doi: 10.1371/journal.pgen.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Le Proust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazell A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, Meissner A, Jaenisch R. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Rigbolt KTG, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Mecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D, Lee BH, Ting AH. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2009;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Delpus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Van der winden JM, et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.