Abstract
This article introduces a manually curated data collection for gene expression meta-analysis of patients with ovarian cancer and software for reproducible preparation of similar databases. This resource provides uniformly prepared microarray data for 2970 patients from 23 studies with curated and documented clinical metadata. It allows users to efficiently identify studies and patient subgroups of interest for analysis and to perform meta-analysis immediately without the challenges posed by harmonizing heterogeneous microarray technologies, study designs, expression data processing methods and clinical data formats. We confirm that the recently proposed biomarker CXCL12 is associated with patient survival, independently of stage and optimal surgical debulking, which was possible only through meta-analysis owing to insufficient sample sizes of the individual studies. The database is implemented as the curatedOvarianData Bioconductor package for the R statistical computing language, providing a comprehensive and flexible resource for clinically oriented investigation of the ovarian cancer transcriptome. The package and pipeline for producing it are available from http://bcb.dfci.harvard.edu/ovariancancer.
Database URL: http://bcb.dfci.harvard.edu/ovariancancer
Introduction
A wealth of genomic data, in particular microarray data, is publicly available through diverse online resources. Major databases of gene expression data, e.g. the Gene Expression Omnibus (GEO) (1) and ArrayExpress (2), offer the potential to identify sets of genes predictive of cancer survival and of patient resistance to chemotherapy using thousands of samples from multiple laboratories. Such high numbers of samples are needed to robustly identify and validate gene signatures for incorporation into routine clinical practice (3). However, inconsistent formatting among database interfaces, expression data storage and clinical metadata annotations present formidable obstacles to making efficient use of these resources.
Existing resources aiming to make large-scale high-dimensional analysis across multiple studies tend to serve only a few specifically targeted needs. To develop reproducible biomarker discovery methods appropriate for clinical translation, a data resource must be accurate and retain clinical variables of known importance as much as possible. The insilicoDB (4) project provides many curated gene expression data sets; however, it is not a focused resource in terms of retention or quality assurance of clinical annotations, or retention of all relevant data sets and clinical variables for any one cancer type. The other major database of curated gene expression studies, the Gene Expression Atlas (2), provides machine- rather than manually annotated data, resulting in reduced consistency of annotation across studies. These are among the only databases that offer basics such as uniform gene identifiers to enable cross-study analysis, and then for only the most common microarray technologies. Carey et al. (5) describe a framework for the curation, annotation and storage of microarray and high-throughput data in general. This framework allows, for example, institutions to provide researchers access to in-house and public data in a standardized and convenient fashion. However, there is no existing database that provides these resources for ovarian cancer.
Ovarian cancer is the fifth-leading cause of cancer deaths among women (6) and has been the focus of numerous clinical transcriptome investigations. The curatedOvarianData database is the result of a focused effort to enable meta-analysis of these studies and to provide the highest quality and most comprehensive gene expression data resource for any cancer. It provides standardized gene expression and clinical data for 2970 ovarian cancer patients from 23 studies spanning 11 gene expression measurement platforms, in the form of documented ExpressionSet objects for R/Bioconductor (7). Gene expression data were collected from public databases and author websites, processed in a consistent manner and mapped uniformly to official Human Gene Nomenclature Committee (HGNC) (8) gene symbols. Curation of clinical annotations was machine-checked for correctness of syntax and human-checked by two individuals to ensure accuracy. This data package is geared primarily towards bioinformatic and statistical researchers, providing an ideal resource for development and assessment of algorithms for high-dimensional classification, clustering and survival analysis. It will also be valuable to ovarian cancer researchers for biomarker identification and validation. In addition to providing all publicly available gene expression studies with patient survival in common forms of ovarian cancer, it includes tumours of rare histologies, normal tissues and uncommon early-stage tumours. Special effort is made to retain the most important clinical variables from author-provided metadata and from the original publications: overall survival, optimal debulking surgery and tumour stage, grade and histology.
We also developed a software pipeline for automated and reproducible production of this and comparable data libraries. The pipeline includes a controlled language for curation of clinical annotations, defined by a template, which is intuitive for non-programmers to create and edit, but which is also used directly for machine syntax checking of curated annotations. The pipeline handles all steps of the process including data download, microarray preprocessing, merging of duplicate probe sets and sample technical replicates, up-to-date probe-set to gene mapping and building of the R/Bioconductor objects and package.
One important application of the database is testing of hypothesized prognostic markers of ovarian cancer using multiple independent studies. We validated a recently proposed independent prognostic indicator of ovarian cancer, CXCL12 (9), using 13 published studies, demonstrating for this biomarker that numerous studies are needed to overcome the lack of power in individual studies of smaller sample size. We provide code in the documentation of the curatedOvarianData package demonstrating how this comprehensive analysis, which was previously impractical to achieve, is a straightforward application of the database.
Methods and implementation
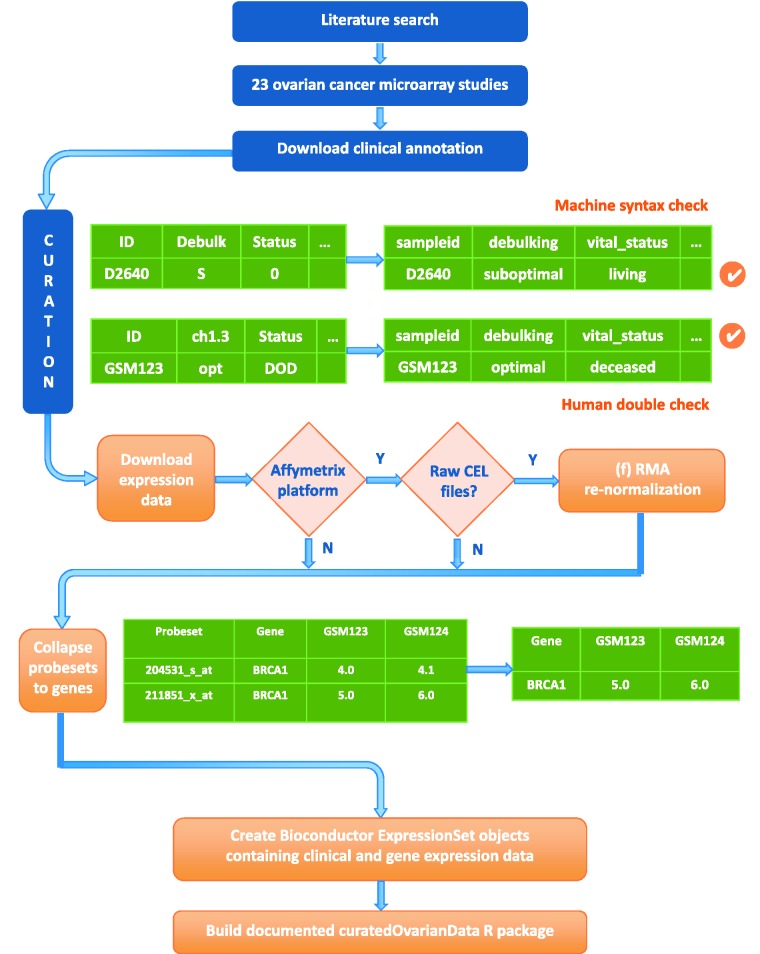
The pipeline for creating the data package from public databases (Table 1) is fully automated, with the exceptions of manual curation of clinical annotations (Figure 1). This manual curation was integrated in the pipeline with short R scripts that reformat user-provided annotations into a standardized template, which largely follows the format of The Cancer Genome Atlas (29). This template is provided in Table 2 and used as a unit test in the curatedOvarianData package, i.e. the curation is automatically checked for valid values in the package building process. Downloading phenotype data and expression data from GEO (1), syntax validation of curated clinical metadata, microarray data preprocessing, normalization, gene mapping and the creation of Bioconductor ‘ExpressionSet’ objects, which link gene expression data and phenotype annotations, were fully automated. The generation of the package is reproducible using the pipeline provided at https://bitbucket.org/lwaldron/curatedovariandata.
Table 1.
Data sets in the curatedOvarianData database
| Data set | Reference | Platform | Samples | Late Stagea (%) | Serous Subtype (%) | Median Survival (Months) | Median Follow-up (Months) | Censoring (%) |
|---|---|---|---|---|---|---|---|---|
| E.MTAB.386 | (10) | Ill. HumanRef-8 v2 | 129 | 99 | 100 | 42 | 55 | 43 |
| GSE12418 | (11) | SWEGENE v2.1.1_27k | 54 | 100 | 100 | N/A | N/A | N/A |
| GSE12470 | (12) | Agilent G4110b | 53 | 66 | 81 | N/A | N/A | N/A |
| GSE13876 | (13) | Operon Human v3 | 157 | 100 | 100 | 25 | 72 | 28 |
| GSE14764 | (14) | Affy U133a | 80 | 89 | 85 | 54 | 37 | 74 |
| GSE17260 | (15) | Agilent G4112a | 110 | 100 | 100 | 53 | 47 | 58 |
| GSE18520 | (16) | Affy U133 Plus 2.0 | 63 | 84 | 84 | 25 | 140 | 23 |
| GSE19829.GPL570 | (17) | Affy U133 Plus 2.0 | 28 | N/A | N/A | 47 | 62 | 39 |
| GSE19829.GPL8300 | (17) | Affy U95 v2 | 42 | N/A | N/A | 45 | 50 | 45 |
| GSE20565 | (18) | Affy U133 Plus 2.0 | 140 | 48 | 51 | N/A | N/A | N/A |
| GSE2109 | N/A | Affy U133 Plus 2.0 | 204 | 42 | 42 | N/A | N/A | N/A |
| GSE26712 | (19) | Affy U133a | 195 | 96 | 95 | 46 | 90 | 30 |
| GSE30009 | (20) | TaqMan qRT-PCR 380 | 103 | 100 | 99 | 41 | 53 | 45 |
| GSE30161 | (21) | Affy U133 Plus 2.0 | 58 | 100 | 81 | 50 | 83 | 38 |
| GSE32062.GPL6480 | (22) | Agilent G4112a | 260 | 100 | 100 | 59 | 56 | 53 |
| GSE32063 | (22) | Agilent G4112a | 40 | 100 | 100 | 53 | 81 | 45 |
| GSE6008 | (23) | Affy U133a | 99 | 54 | 41 | N/A | N/A | N/A |
| GSE6822 | (24) | Affy Hu6800 | 66 | N/A | 62 | N/A | N/A | N/A |
| GSE9891 | (25) | Affy U133 Plus 2.0 | 285 | 85 | 93 | 47 | 36 | 59 |
| PMID15897565b | (26) | Affy U133a | 63 | 83 | 100 | N/A | N/A | N/A |
| PMID17290060c | (27) | Affy U133a | 117 | 98 | 100 | 63 | 82 | 43 |
| PMID19318476 | (28) | Affy U133a | 42 | 93 | 100 | 34 | 89 | 48 |
| TCGA | (29) | Affy HT U133a | 578 | 90 | 98 | 45 | 52 | 48 |
These data sets provide curated gene expression and clinical data for a total of 2970 samples, including all publicly ovarian cancer gene expression experiments with individual patient survival information at the time of press.
aOnly FIGO Stages III and IV.
bData set is a subset of the samples from the retracted paper PMID17290060, Dressman et al. (27).
cPaper was retracted because of a misalignment of genomic and survival data (30); the corrected data are provided here.
N/A, not available.
Figure 1.
Flowchart of the data collection and curation pipeline. The software implementing this pipeline reproduces all steps from downloading of data to final packaging, requiring manual intervention only for identifying studies, curation of clinical metadata and documentation of the package.
Table 2.
Curated clinical annotations
| Characteristic | Allowed values | Description |
|---|---|---|
| sample_type | tumour, metastatic, cellline, healthy, adjacentnormal | Healthy, only from individuals without cancer; adjacentnormal, from individuals with cancer; |
| histological_type | ser, endo, clearcell, mucinous, other, mix, undifferentiated | ser, serous; endo, endometrioid; clearcell, mixture of ser + endo. Other includes sarcomatoid, endometroid, papillary serous, adenocarcinoma, dysgerminoma |
| primarysite | ov, ft, other | Ov, ovary; ft, fallopian tube |
| arrayedsite | ov, ft, other | ov, ovary; ft, fallopian tube |
| summarygradea | low, high | low, 1, 2, LMP (low malignant potential); high, 3, 2/3 |
| summarystage | early, late | early, FIGO I, II, I/II; late, FIGO III, IV, II/III, III/IV |
| tumourstage | 1, 2, 3, 4 | FIGO Stage (I–IV, translated to 1–4 for R usage) |
| substage | a, b, c, d | Substage (abcd) |
| gradea | 1, 2, 3 | Grade (1–3) |
| age_at_initial_pathologic_diagnosis | 1-99 | Age at initial pathologic diagnosis in years |
| pltx | y/n | Patient treated with Platin |
| tax | y/n | Patient treated with Taxol |
| neo | y/n | Neoadjuvant treatment |
| days_to_tumour_recurrence | decimal | Time to recurrence or last follow-up in days |
| recurrence_status | recurrence, no recurrence | Recurrence censoring variable |
| days_to_death | decimal | Time to death or last follow-up in days |
| vital_status | living, deceased | Overall survival censoring variable |
| os_binary | short, long | Dichotomized overall survival time; as defined by study |
| relapse_binary | short, long | Dichotomized relapse variable; as defined by the study |
| site_of_tumour_first_recurrence | metastasis, locoregional, etc. | Site of the first recurrence |
| primary_therapy_outcome_success | completeresponse, etc. | Response to any kind of therapy |
| bebulking | optimal, suboptimal | Amount of residual disease (optimal ≤ 1 cm) |
| percent_normal_cells | 0–100+/− | Estimated percentage of normal cells; 20− ≤ 20% |
| percent_stromal_cells | 0–100+/− | Estimated percentage of stromal cells |
| percent_tumour_cells | 0–100+/− | Estimated percentage of tumour cells; 80+ ≥ 80% |
| batch | character | Hybridization date or other available batch variable |
| uncurated_author_metadata | character | All original, uncurated metadata |
Data acquisition and curation
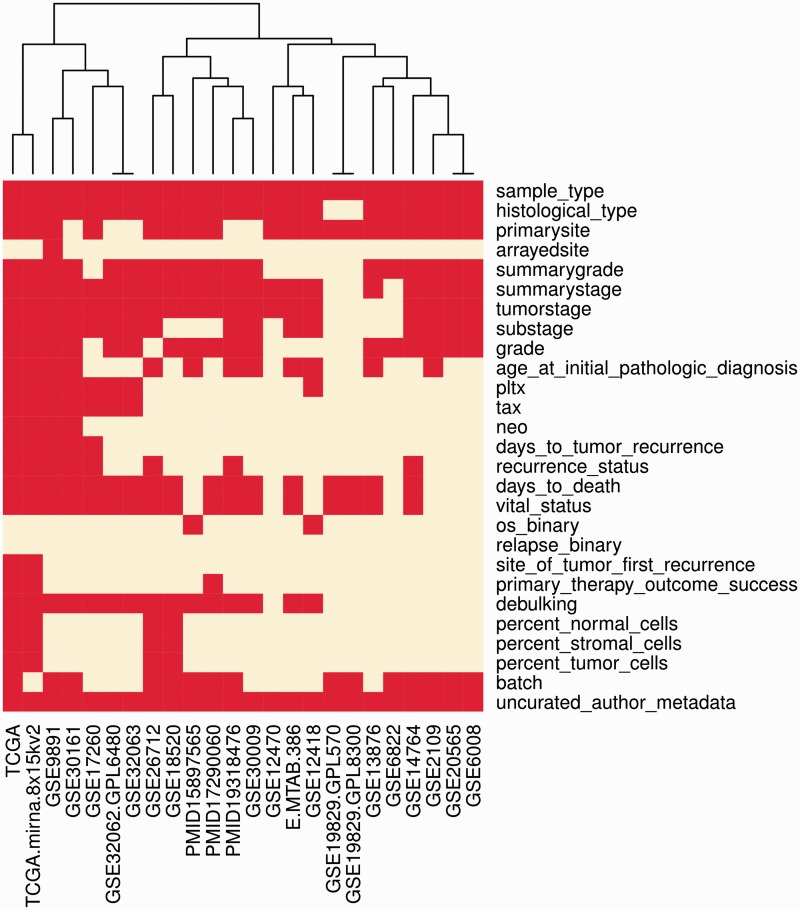
Our search for clinically annotated ovarian cancer microarray studies identified 21 published studies, which provided 23 publicly available data sets from various sources (Table 1). The search not only targeted studies of primary tumours annotated with patient survival but also included studies providing other potentially valuable clinical annotation. Other main factors of interest included drug resistance, outcome of the primary tumour debulking surgery, histology, stage and grade. We excluded studies not measuring gene expression (i.e. studies of genomic copy number), studies of cell lines, animal models, or non-primary tumours, and data sets not providing clinical information. Expression and clinical data were obtained from the two major public repositories GEO (i) and ArrayExpress (ii), otherwise from supplementary data of the original publications. Data from GEO were obtained using the GEOquery package (31). Clinical annotations were manually curated using one R script per data set, and original uncurated annotations were retained as a single field. Curated annotations were checked by syntax against a template, which standardized all the known clinically relevant indicators and allowable data values. Clinical data were twice independently curated (authors B.G. and T.R.), and all discrepancies were resolved for the final version. The availability of clinical data varied substantially across datasets (Figure 2).
Figure 2.
Available clinical annotation. This heatmap visualizes for each curated clinical characteristic (rows) the availability in each data set (columns). Red indicates that the corresponding characteristic is available for at least one sample in the data set. See Table 2 for descriptions of these characteristics.
Gene expression processing and gene mapping
Where raw data from Affymetrix U133a or U133 Plus 2.0 platforms were available, these were pre-processed by frozen Robust Multi-array Analysis (fRMA) (32), for other Affymetrix platforms by Robust Multi-array Average (RMA) (33), and otherwise we used pre-processed data as provided by the authors. Up-to-date maps from probe set IDs to gene symbols were obtained from BioMart (34). Where BioMart maps were not available but target sequences were provided for the microarray platforms, we used the BLAST algorithm (35) to map these sequences against the human genome (build GRCh37) and to identify the gene transcript targeted by each probe. Otherwise, the annotations provided with the platform on GEO were used. In the curatedOvarianData version of the package, genes with multiple probe sets were represented by the probe set with the highest mean across all data sets of the sample platform (36), and this original probe set identifier was also stored in the ExpressionSet object (7). We selected the same representative probe set for all studies of a common microarray platform. Finally, we provide two alternative versions of the package: NormalizerVcuratedOvarianData, where redundant probe sets are averaged after filtering probe sets with low correlation to their redundant probe sets, using the Normalizer function of the Sleipnir library for computational functional genomics (37), and FULLVcuratedOvarianData, which does not collapse redundant probe sets targeting the same gene transcript but instead provides a probe set to gene symbol map in the featureData slot of each ExpressionSet.
Final packaging
Technical replicate samples were merged by averaging. Microarray expression data and clinical metadata were then represented as ExpressionSet objects (7) for each study. The ExpressionSet objects were also populated with citations, platform identifiers and details, data preprocessing methods and warnings of retracted papers (27) and specimens also used in other studies (26, 28, 29, 38). ExpressionSets were packaged as the curatedOvarianData R library, which provides a reference manual including descriptions of the syntax template and summaries of the annotations, citation, microarray platform and other information for each study.
Discussion
We introduce a data package for the R/Bioconductor statistical programming environment that includes all current major ovarian cancer gene expression data sets (Table 1). The process of downloading clinically annotated public genomic data and proceeding to a final computational analysis is, despite recent efforts (4, 5), still long and prone to errors. This is particularly true when the various data sets need to be comparable for meta-analyses, which requires a fully standardized annotation. Our data resource provides a comprehensive and highly curated resource for efficient meta-analysis of the ovarian cancer transcriptome, for biological analysis and bioinformatic methods development. It additionally provides a complete computational pipeline to reproduce this process for other cancers or data sources.
Two common problems of publicly available genomic data are the scarcity of clinical annotation and inconsistent definitions of clinical characteristics across independent data sets (5). In our review of original papers and curation of clinical annotations, we were however able to retain, in most studies, the clinical variables of proven importance: overall survival, age, optimal debulking surgery, tumour histology, grade and stage (Figure 2). Other characteristics such as detailed treatment information or recurrence free survival times were rarely available; however, ovarian cancer has a relatively standard treatment regimen of platinum chemotherapy and no radiotherapy. The most important clinical variables were in general consistently defined between studies, with these definitions provided in Table 2. Notably, all studies used the Federation of Gynecology and Obstetrics (FIGO) staging system, and all but one study (11) defined suboptimal debulking surgery as residual tumour mass > 1 cm (Table 2). The relatively large number of well-annotated data sets in this database may allow interesting future work, addressing the problem of recovering missing annotations from genomic data only (40).
One important use of this database is the assessment of prognostic biomarkers. As a demonstration, we examined a recent study by Popple et al. (9), which analysed the expression of the chemokine protein CXCL12 using a tissue microarray of 289 primary ovarian cancers. CXCL12/CXCR4 is a chemokine/chemokine receptor axis that has previously been shown to be directly involved in cancer pathogenesis (41, 42). Ovarian cancer constitutively expresses CXCL12 and CXCR4, and both tumour CXCL12/CXCR4 expression and stroma-derived CXCL12 expression have been reported to be prognostic factors in human ovarian cancer (41). Popple et al. found that high levels of CXCL12 protein were associated with significantly poorer survival compared with patients whose tumours produce low amounts of this chemokine, independently of stage, residual disease (optimal debulking) and adjuvant chemotherapy. The patient cohort was heterogeneous, with various histologic types, grades and stages, leaving open the question of whether this biomarker would be generalizable to other patient populations. Furthermore, differences in protein abundance may not be associated with RNA abundance.
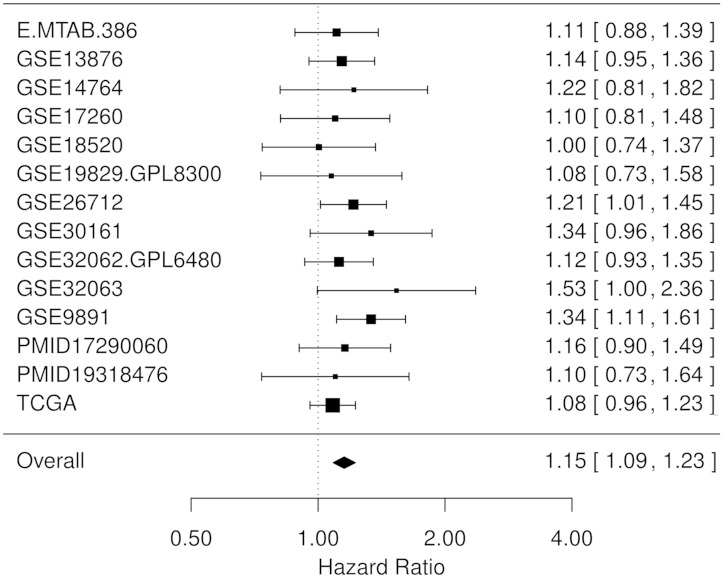
To investigate these questions, we analysed CXCL12 expression in all primary tumour samples included in curatedOvarianData for which overall survival information was available. To ensure that the expression values were on the same scale across studies, all data sets were centred by their means and scaled by their standard deviations. A population hazard ratio (HR) was then pooled with a fixed-effects model, in which the HR for each cohort was weighted with the inverse of the standard error. This is visualized as a forest plot in Figure 3. Although the effect is only significant (P < 0.05) in three cohorts individually, the pooled HR is significantly larger than 1 (HR = 1.15, 95% CI 1.09–1.23). HR refers to the HR between patients differing by one standard deviation in CXCL12 expression. This confirms the hypothesis that upregulation of CXCL12 is associated with poor outcome in 2108 patients from 13 independent studies with mixed stage, grade and histologies. The effect is thus small but consistently detected, emphasizing the importance of biomarker validation in sufficiently large data collections. To assess the independence of CXCL12 with stage and residual disease, we also analysed the 1776 patients from 10 studies where both FIGO tumour stage and success of debulking surgery were known. Adjustment for these two established predictors in multivariate analysis had little effect on the observed association between CXCL12 and overall survival (HR = 1.13, 95% CI 1.05–1.21). These HRs are comparable in magnitude to that reported by Popple et al. for ‘moderate’ CXCL12 staining (HR = 1.215, 95% CI 0.892–1.655), but lower than reported for ‘high’ staining (HR = 1.684, 95% CI 1.180–2.404). This potentially reflects that the function of this gene is at the protein level. Consistent with previous reports (9, 38), we found no significant association of the receptor CXCR4 with overall survival (HR = 0.95, 95% CI 0.9–1.01, P = 0.09). These analyses are straightforward and fully reproduced as examples in the package documentation. Additional analyses limited to more homogeneous patient subsets, e.g. limited to tumours of the same histology, are needed, but they are another straightforward application of the package.
Figure 3.
The database confirms CXCL12 as prognostic of overall survival in patients with ovarian cancer. Forest plot of the expression of the chemokine CXCL12 as a univariate predictor of overall survival, using all 14 data sets with applicable expression and survival information. HR indicates the factor by which overall risk of death increases with a one standard deviation increase in CXCL12 expression. A summary HR significantly larger than 1 indicates that patients with high CXCL12 levels had poor outcome and confirms in several lines of code the previously reported association between CXCL12 abundance and patient survival (9). Consideration of important clinicopathological features such as stage, grade, histology and residual disease (optimal surgical debulking) is also straightforward; examples are provided in the package vignette.
In constructing curatedOvarianData, we took several steps to minimize across-study batch effects. Where raw Affymetrix microarray data were available, we used a standardized pre-processing protocol. All data sets from the same platform were normalized with the same algorithms and parameters. For the Affymetrix U133A and U133 Plus 2.0, we chose the fRMA (32) normalizing algorithm, a variant of the standard RMA (33) algorithm that uses publicly available microarray databases to estimate probe-specific effects and variances, instead of using only the samples from the data set to be normalized. We provide example code in the database documentation for removing between-platform batch effects with the ComBat method (43). Such a batch effect removal is typically necessary when data sets are merged.
If different platforms are compared, then the mapping of probe sets to common identifiers such as gene symbols is a critical and error prone step. In particular when older platforms are considered, care must be taken to ensure that the probe sets target identical transcripts; gene identification is a persistent problem in genome-scale data integration. We used the BioMart database (34) to map stable manufacturer probe set identifiers or Genbank IDs to current standard gene symbols. For cases in which no stable identifiers were available, we used the BLAST algorithm (35) to identify gene symbols from the probe oligonucleotide sequences. When many genes are targeted by more than one probe set, several approaches of collapsing probe sets to single genes have been proposed (36, 44, 45). In the main version of the package, we selected the probe set with highest mean across all data sets from the same platform to represent each gene transcript, a method shown to perform well (36) and with the advantage of being traceable back to a single oligonucleotide probe sequence for each platform. We also provide two alternative packages with averaged and un-collapsed probe sets. The version with un-collapsed probe sets provides current HGNC symbols in the featureData slot of the ExpressionSet objects, which makes the application of alternative methods for collapsing probe sets to unique gene symbols straightforward, e.g. with the WGCNA R package (46).
We demonstrated meta-analytical use of the package by showing a survival association of the recently proposed prognostic biomarker CXCL12 (9). Other possible uses include the validation of multi-gene signatures, and identification of novel gene signatures and biomarkers for patient survival and response to chemotherapy. Finally, this package enables rigorous assessment of high-dimensional machine-learning algorithms in terms of their performance and computational requirements. We plan to continually include newly published ovarian cancer data sets in future versions of this package.
Conclusions
The curatedOvarianData package provides a comprehensive resource of curated gene expression and clinical data for the development and validation of ovarian cancer prognostic models, the investigation of ovarian cancer subtypes (10, 25, 29), and the comparative assessment of machine learning algorithms for gene expression data. This database greatly reduces the burden of time, expertise and error involved in assembling a compendium of curated gene expression data from tumours of known histopathology and from patients with known clinical progression. These advantages will be appealing to biostatisticians and bioinformaticians for development of analytical methods from high-dimensional genomic data, but the database will additionally provide a common, version-controlled and transparent platform for reproducible investigation of the ovarian cancer transcriptome. The pipeline for creating this database is published under an open license and will facilitate creating similar resources for other cancers. As such, we hope this database and pipeline will provide one part of the solution to reproducibility in high-dimensional genomic research.
Acknowledgements
The authors thank Shaina Andelman for her contributions to graphic design, and also Steve Skates, Jie Ding and Dave Zhao.
Funding
National Cancer Institute at the National Institutes of Health [1RC4CA156551-01 to G.P. and M.B.]; the National Science Foundation [CAREER DBI-1053486 to C.H.]. M.R. acknowledges support from the National Cancer Institute initiative to found Physical Science Oncology Centers [U54CA143798]. Funding for open access charge: National Science Foundation [CAREER DBI-1053486 to C.H.].
Conflict of interest. None declared.
References
- 1.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson H, Sarkans U, Kolesnikov N, et al. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N. Engl. J. Med. 2011;364:340–350. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 4.Taminau J, Steenhoff D, Coletta A, et al. inSilicoDb: an R/Bioconductor package for accessing human Affymetrix expert-curated datasets from GEO. Bioinformatics. 2011;27:3204–3205. doi: 10.1093/bioinformatics/btr529. [DOI] [PubMed] [Google Scholar]
- 5.Carey VJ, Gentry J, Sarkar R, et al. SGDI: system for genomic data integration. Pac. Symp. Biocomput. 2008:141–152. doi: 10.1142/9789812776136_0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 7.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seal RL, Gordon SM, Lush MJ, et al. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popple A, Durrant LG, Spendlove I, et al. The chemokine, CXCL12, is an independent predictor of poor survival in ovarian cancer. Br. J. Cancer. 2012;106:1306–1313. doi: 10.1038/bjc.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentink S, Haibe-Kains B, Risch T, et al. Angiogenic mRNA and microRNA gene expression signature predicts a novel subtype of serous ovarian cancer. PLoS One. 2012;7:e30269. doi: 10.1371/journal.pone.0030269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partheen K, Levan K, Osterberg L, et al. Expression analysis of stage III serous ovarian adenocarcinoma distinguishes a sub-group of survivors. Eur. J. Cancer. 2006;42:2846–2854. doi: 10.1016/j.ejca.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida S, Furukawa N, Haruta S, et al. Expression profiles of genes involved in poor prognosis of epithelial ovarian carcinoma: a review. Int. J. Gynecol. Cancer. 2009;19:992–997. doi: 10.1111/IGC.0b013e3181aaa93a. [DOI] [PubMed] [Google Scholar]
- 13.Crijns A, Fehrmann R, de Jong S, et al. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med. 2009;6:e24. doi: 10.1371/journal.pmed.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denkert C, Budczies J, Darb-Esfahani S, et al. A prognostic gene expression index in ovarian cancer - validation across different independent data sets. J. Pathol. 2009;218:273–280. doi: 10.1002/path.2547. [DOI] [PubMed] [Google Scholar]
- 15.Yoshihara K, Tajima A, Yahata T, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One. 2010;5:e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mok SC, Bonome T, Vathipadiekal V, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated Glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konstantinopoulos PA, Spentzos D, Karlan BY, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyniel JP, Cottu PH, Decraene C, et al. A genomic and transcriptomic approach for a differential diagnosis between primary and secondary ovarian carcinomas in patients with a previous history of breast cancer. BMC Cancer. 2010;10:222. doi: 10.1186/1471-2407-10-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonome T, Levine D, Shih J, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillet JP, Calcagno A, Varma S, et al. Multidrug resistance-linked gene signature predicts overall survival of patients with primary ovarian serous carcinoma. Clin. Cancer Res. 2012;18:3197–3206. doi: 10.1158/1078-0432.CCR-12-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferriss JS, Kim Y, Duska L, et al. Multi-gene expression predictors of single drug responses to adjuvant chemotherapy in ovarian carcinoma: predicting platinum resistance. PLoS One. 2012;7:e30550. doi: 10.1371/journal.pone.0030550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, Tsunoda T, Shigemizu D, et al. High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin. Cancer Res. 2012;18:1374–1385. doi: 10.1158/1078-0432.CCR-11-2725. [DOI] [PubMed] [Google Scholar]
- 23.Murph M, Liu W, Yu S, et al. Lysophosphatidic acid-induced transcriptional profile represents serous epithelial ovarian carcinoma and worsened prognosis. PLoS One. 2009;4:e5583. doi: 10.1371/journal.pone.0005583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouellet V, Provencher DM, Maugard CM, et al. Discrimination between serous low malignant potential and invasive epithelial ovarian tumors using molecular profiling. Oncogene. 2005;24:4672–4687. doi: 10.1038/sj.onc.1208214. [DOI] [PubMed] [Google Scholar]
- 25.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 26.Berchuck A, Iversen ES, Lancaster JM, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin. Cancer Res. 2005;11:3686–3696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 27.Dressman H, Berchuck A, Chan G, et al. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J. Clin. Oncol. 2007;25:517–525. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- 28.Berchuck A, Iversen ES, Luo J, et al. Microarray analysis of early stage serous ovarian cancers shows profiles predictive of favorable outcome. Clin. Cancer Res. 2009;15:2448–2455. doi: 10.1158/1078-0432.CCR-08-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dressman HK, Berchuck A, Chan G, et al. Retraction. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J. Clin. Oncol. 2012;30:678. doi: 10.1200/jco.2012.42.0331. [DOI] [PubMed] [Google Scholar]
- 31.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 32.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Durinck S, Moreau Y, Kasprzyk A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JA, Cai C, Langfelder P, et al. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huttenhower C, Schroeder M, Chikina MD, et al. The sleipnir library for computational functional genomics. Bioinformatics. 2008;24:1559–1561. doi: 10.1093/bioinformatics/btn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 39.Kauffmann A, Rayner TF, Parkinson H, et al. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics. 2009;25:2092–2094. doi: 10.1093/bioinformatics/btp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah NH, Jonquet C, Chiang AP, et al. Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinformatics. 2009;10(Suppl. 2):S1. doi: 10.1186/1471-2105-10-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kajiyama H, Shibata K, Terauchi M, et al. Involvement of SDF-1alpha/CXCR4 axis in the enhanced peritoneal metastasis of epithelial ovarian carcinoma. Int. J. Cancer. 2008;122:91–99. doi: 10.1002/ijc.23083. [DOI] [PubMed] [Google Scholar]
- 42.Kulbe H, Chakravarty P, Leinster DA, et al. A dynamic inflammatory cytokine network in the human ovarian cancer microenvironment. Cancer Res. 2012;72:66–75. doi: 10.1158/0008-5472.CAN-11-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 44.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Birkbak NJ, Gyorffy B, et al. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]