Abstract
The killer cell immunoglobulin-like receptors (KIR) play a fundamental role in the innate immune system, through their interactions with human leucocyte antigen (HLA) molecules, leading to the modulation of activity in natural killer (NK) cells, mainly related to killing pathogen-infected cells. KIR genes are hugely polymorphic both in the number of genes an individual carries and in the number of alleles identified. We have previously developed the Allele Frequency Net Database (AFND, http://www.allelefrequencies.net), which captures worldwide frequencies of alleles, genes and haplotypes for several immune genes, including KIR genes, in healthy populations, covering >4 million individuals. Here, we report the creation of a new database within AFND, named KIR and Diseases Database (KDDB), capturing a large quantity of data derived from publications in which KIR genes, alleles, genotypes and/or haplotypes have been associated with infectious diseases (e.g. hepatitis C, HIV, malaria), autoimmune disorders (e.g. type I diabetes, rheumatoid arthritis), cancer and pregnancy-related complications. KDDB has been created through an extensive manual curation effort, extracting data on more than a thousand KIR-disease records, comprising >50 000 individuals. KDDB thus provides a new community resource for understanding not only how KIR genes are associated with disease, but also, by working in tandem with the large data sets already present in AFND, where particular genes, genotypes or haplotypes are present in worldwide populations or different ethnic groups. We anticipate that KDDB will be an important resource for researchers working in immunogenetics.
Database URL: http://www.allelefrequencies.net/diseases/
Introduction
Natural killer (NK) cells are bone marrow-derived lymphocytes that play an active role in the innate immune system by interacting with human leucocyte antigen (HLA) class I molecules to kill pathogen-infected cells (1). Initially, NK cells were discovered as a result of their ability to target and kill tumour cell lines that expressed little or no HLA class I molecules (2). It is now known that the killing function in NK cells is dependent on a mixture of activating and inhibitory receptors present on the membrane and the interaction with their HLA ligand (3). Two main types of receptors are found in NK cells, C-type lectin-like (NKG2D, CD94/NKG2C, CD94/NKG2A) and the immunoglobulin-like superfamily (KIR, CD16, NKp30, NKp44, etc.). In the latter, the killer cell immunoglobulin-like receptors (KIR) that mostly bind Major histocompatibility complex (MHC) class I molecules have been shown to be the most polymorphic. Despite most of the NK cell receptors binding MHC class I-related molecules, several Ig-like receptors bind non-HLA ligands, for example, CD16 binds IgG, triggering an activating response, and NKp44, NKp30 and NKp46 are activating receptors that bind molecules expressed by pathogens and self-ligands (4–8).
The KIR gene cluster is located in the leucocyte receptor complex (LRC) at position 19q13.4 (4, 5). To date, 16 KIR genes have been identified, coding for receptors with activating (KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5A/B and KIR3DS1) or inhibitory (KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL5A, KIR2DL5B, KIR3DL1, KIR3DL2 and KIR3DL3) function, with KIR2DL4 appearing to have both functions. Two pseudogenes KIR2DP1 and KIR3DP1 have also been identified (9). Structurally, the activating and inhibitory functions of KIR are related to the length of their cytoplasmic tail that can be short (S) or long (L), distinguished in the nomenclature (9).
Variation in KIR can result from a different gene and/or allele content of an individual (10), giving rise to haplotype diversity and leading to a very large number of different genotypes that have been observed (presence/absence of KIR genes). The KIR genes KIR2DL4, KIR3DL2, KIR3DL3 and KIR3DP1 are present in nearly all individuals with a few exceptions (11), and are commonly known as ‘framework’ genes. The frequencies of inhibitory and activating genes vary in different populations, as reviewed in (11). A 24-kb band using HindIII digestion and Southern blot analysis distinguishes the haplotypes, termed A and B, that make up the genotype (12). The A haplotype is generally non-variable in its gene content—framework genes plus KIR2DL1, KIR2DL3, KIR2DS4 and KIR3DL1—although occasionally one of these genes may be missing (11). In contrast, the B haplotype contains one or more of the genes encoding activating KIRs—KIR2DS1/2/3/5 and KIR3DS1—and the genes encoding inhibitory KIRs—KIR2DL5A/B and KIR2DL2. In B haplotypes, variability is created by both the presence/absence of a gene and by allelic variation; in contrast, A haplotypes owe much of their variability to allele content (11). At the last release of IPD-KIR (Release 2.4.0), there were 601 KIR alleles reported (13). B haplotypes tend to be more prevalent in non-Caucasian populations, such as Australian Aborigines and Asian Indians, whereas in Caucasian populations, ∼55% will have one and 30% two A haplotypes (14, 15). It is thought that populations with higher frequencies of B haplotypes are those under strong pressure from infectious diseases. Such extensive diversity among modern populations may indicate that geographically distinct diseases have exerted recent or perhaps on-going selection on KIR repertoires. From a practical viewpoint, this makes the choice of controls very important for all disease association studies.
To collect allele, haplotype and genotype frequencies of several immune genes in different healthy human populations, the Allele Frequency Net Database (AFND) was developed (16). AFND stores large sets of data regarding HLA, KIR major histocompatibility complex class I chain related (MIC) and cytokine gene polymorphisms, and has shown to be frequently used in the immunogenetics field, receiving 200 hits per day on average. To date, 398 different KIR genotypes in 12 856 individuals from 109 populations have been reported to AFND.
Owing to its high level of polymorphism, many infectious and autoimmune diseases have been associated with KIR genes in different ways, e.g. associations with single genes (or single alleles) to associations with groups of genes and full genotypes (17–20). A disease association is defined as a statistically significant association between a genetic element (gene, allele, genotype, etc.) with a given disease outcome, either positive or negative i.e. the genetic profile makes the disease more likely/severe or less likely/severe than the control population. As such, the development of a database to store data regarding disease associations with those genes is a necessary step towards a more effective comprehension of such complex data. As KIR disease association studies are in its infancy compared with HLA, a decision was made to start collecting KIR disease associations, as a new module within AFND.
Materials and methods
Data curation
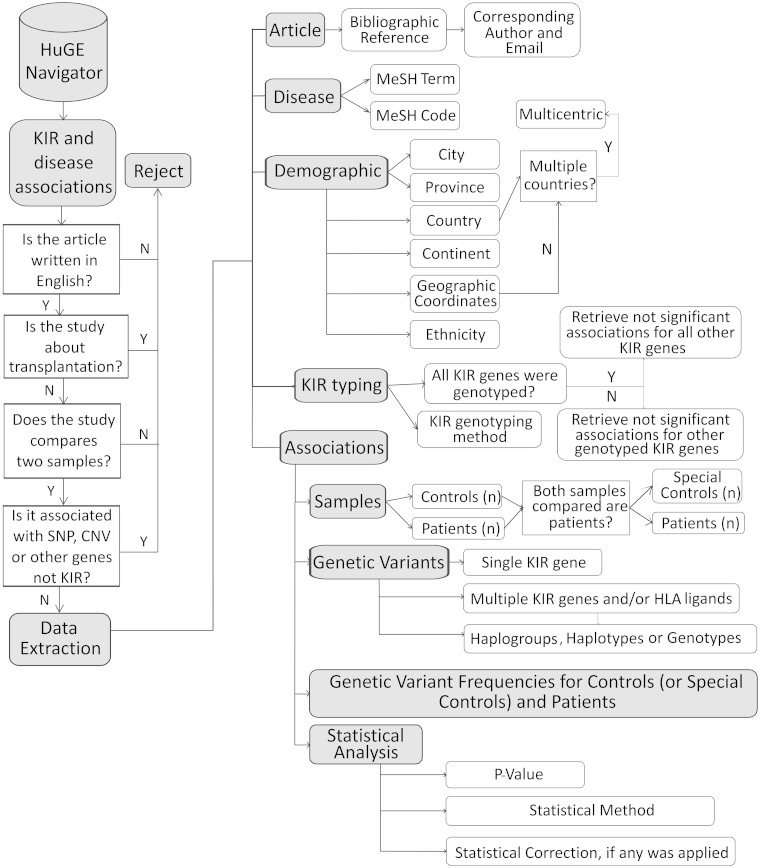
The first step towards creation of KDDB was the collection and extraction of data from peer-reviewed publications, following the workflow shown in Figure 1. Published KIR and disease association studies were extracted from the HuGE Navigator (version 2.0) (21), which is a web-based tool enabling searches of the scientific literature for studies on genetic associations with diseases. The HuGE Navigator makes use of the MeSH (Medical Subject Headings) terminology, which contains standardized keywords associated with clinically related published studies. In KDDB, we loaded MeSH terms that describe specific diseases with which associations have been found. Manual curation was performed to extract relevant data from retrieved studies. A set of consistent rules were applied to ensure that different curators extracted data in the same way (Figure 1). All studies identified based on the relevant MeSH terms were analysed and inserted into KDDB unless they did not pass one of the following criteria (as also shown on Figure 1): (i) the article was not written in English, as we do not have the capability to translate articles at present, (ii) the study design was not based on a gene frequency comparison between two samples with different clinical outcomes (future updates to KDDB will attempt to include more complex study designs), (iii) the article identified by the HuGE Navigator was not in fact related to KIR (i.e. misidentified), (iv) the study was not related to a disease specifically, but instead describe transplantation outcomes. Studies associating transplantation outcome and KIR have heterogeneous designs—some studies associate KIR–ligand matches/mismatches based on recipients and donors samples, and others correlate the risk of relapse with KIR combinations. These study designs are under evaluation to ascertain whether they can either fit the existing KDDB schema, or will be stored in a different database schema in the future. A data validation pipeline was created to ensure that quantitative data and metadata had been correctly extracted from each publication, involving two curators reviewing the same source publication to reduce the chance for misinterpretation or copy–paste errors.
Figure 1.
The data curation pipeline, the types of data that were extracted from each publication and the submission workflow developed within KDDB.
Implementation
The back end of the database was developed using a relational database schema. Users can connect to the database using the most common web browsers. The web interface of KDDB has been created to allow users to retrieve or query the database and to submit new data sets. For that purpose, interactive web pages for querying and submitting data were developed using the Active Server Pages (ASP) scripting environment and JavaScript language. The graphical display was designed using HyperText Markup Language (HTML) and Cascading Style Sheets (CSS), ensuring that the page will be viewable in most used web browsers. The data submission pipeline will be an important feature for future updates to KDDB, as we recognize the benefits of obtaining community input, including unpublished data sets (see Discussion).
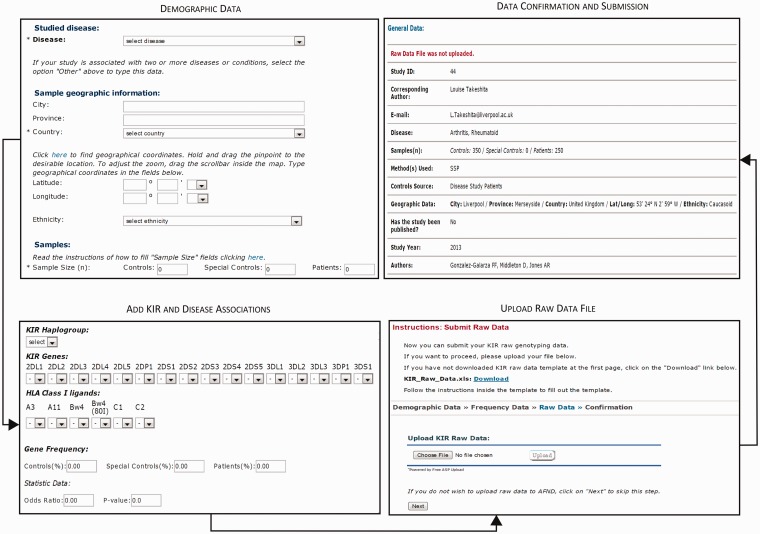
To submit studies to KDDB, a submission form pipeline was developed, which can be accessed through the AFND homepage by the menu ‘Submissions’ and the submenu ‘Add KIR and disease association study’ (Figure 2). This web form consists of four steps. The first step captures summary information about the study including the number of patients and controls. Information is also captured on the geographic location of the population, the ethnicity and the bibliographic reference. The second step captures the disease association data—the genes, alleles, haplotypes, KIR-HLA ligands, etc., the disease name, the frequency of patients and controls exhibiting the given genetic profile and the results of the statistical test. The third step (optional) allows users to upload anonymized raw data (the KIR genetic profile of every individual in the study). The fourth step allows users to review their data and submit. We anticipate that the submission pipeline will become an important tool for users to submit their own unpublished studies or studies missed in the curation process.
Figure 2.
Screenshots of the data submission pipeline within KDDB.
Results
Website organization and content
Using the HuGE Literature Finder tool, 159 articles remained after applying the exclusion criteria detailed in Figure 1. From all the articles, a total of 1027 KIR–disease associations were captured from 113 articles. A set of 46 articles was removed at this stage owing to studies lacking mandatory data/metadata or the numerical data were inaccessible, for example displayed only on charts. The genetic associations identified in this data compilation included those with single KIR genes, profiles of combined KIR genes and / or HLA class I ligands, and full KIR genotypes. In total, 70 unique MeSH terms have been associated with KIR across the studies in the present database. Classifying the studies by the main disease groups, 36 studies are related to infectious diseases, 61 studies are related to autoimmune or idiopathic diseases, 11 studies are related to pregnancy and 9 articles are related to cancer (Table 1). From these studies, a total of 1027 KIR records were inserted into KDDB, of which 496 are statistically significant KIR–disease associations.
Table 1.
Summary of data stored in the KDDB
| Disease type | No. of studies | No. of records (T/S) | No. of individuals |
|---|---|---|---|
| Infectious | 36 | 145/145 | 15 813 |
| Autoimmune or Idiopathic | 61 | 589/274 | 30 888 |
| Neoplasias | 9 | 135/47 | 5791 |
| Pregnancy related | 11 | 167/39 | 4879 |
| Total | 113a | 1027/496a | 56 214a |
aSome of the studies fall into more than one disease type category, e.g. tumours originated
from viral infections.
T, total records; S, significant associations.
The KIR and Diseases Database is part of Allele Frequencies Net Database, and can be accessed through Allele Frequencies Net homepage (http://www.allelefrequencies.net/) using the menu ‘KIR’ and the submenu ‘KIR and disease associations’ or via a direct URL access at http://www.allelefrequencies.net/diseases/. The website interface allows the user to retrieve and query KIR and disease associations applying a collection of filters. The user can restrict the search by gene or allele, country of origin of studied samples, continent of origin of studied samples or studied disease. Those filters can be applied alone or used in combination.
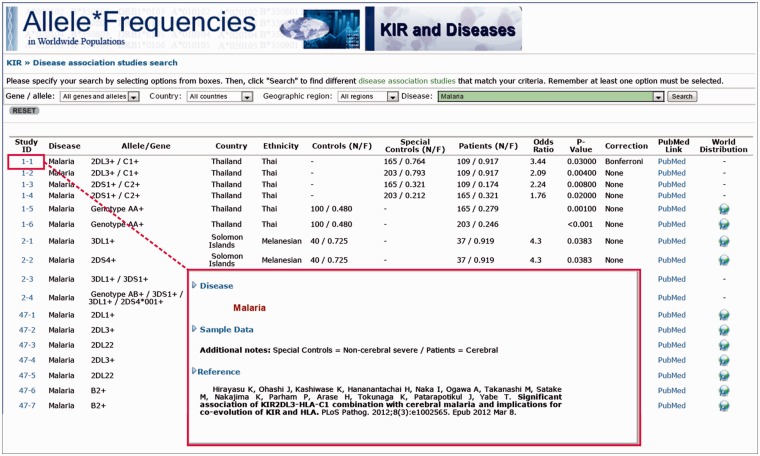
Results from a query are retrieved in a table format, with each row being a different disease association with KIR (Figure 3). In each row, the following information is displayed: (i) row number, (ii) the associated MeSH term, (iii) the country of origin of the sample, (iv) the associated KIR profile, (v) the sample size and gene frequencies for controls and patients, (vi) odds ratio value, (vii) P-value and (viii) statistical method used in comparisons. A link is provided, by clicking on the population name, to show the demographic information on the disease and corresponding control populations. As for normal populations in AFND, individual KIR gene frequencies or haplotype frequencies can be plotted on world maps. This enables a user to interpret disease association risks for KIR profiles in a geographic, ethnic group or individual population-based context. Additional functionality is under development for linking to external resources including to the IPD-KIR database (www.ebi.ac.uk/ipd/kir/), where the sequences and official nomenclature are maintained.
Figure 3.
The query interface within KDDB, showing the additional detail about a given association study retrieved by following the hyperlink.
Discussion
In our original search for frequency data in AFND in normal populations, we sourced publication data from >65 peer-reviewed journals—a complete list of data sets and journals may be consulted at http://www.allelefrequencies.net/datasets.asp. However, many disease studies, especially those that do not find statistically significant associations, are not published, and there is a risk that resources such as KDDB could suffer from publication bias. As such, we are contacting colleagues working in this field with a request to provide their data, even if it is unpublished or does not contain a statistically significant association. As unpublished studies are added to KDDB, we will add a filter to the query page allowing users to exclude these data sets if they wish to ensure quality control. We are also requesting users to upload anonymized raw data (individual KIR type and HLA ligands) to enable improved quality control measures (such as validation of frequency calculations) and to enable advanced analyses of the data. For example, having the individual data available will allow analyses such as looking at disease associations in the centromeric or the telomeric regions. It is known there is extensive linkage disequilibrium between KIR genes, but this exists separately in the centromeric half and the telomeric half (22). There is little linkage disequilibrium between the two halves, and the genes KIR3DP1 and KIR2DL4 are at the division between centromeric and telomeric sections.
We already have some associations in KDDB derived from the presence of the KIR gene and its HLA ligand, and it will be important to expand this collection and include raw data. Studies have shown that although KIR and HLA genes are coded on different chromosomes, there are correlations (both negative and positive) between the presence of the KIR gene and corresponding presence/absence of the ligand (23, 24). These correlations have been shown to be important in diseases. For example, a reciprocal relationship exists in populations between the frequencies of the KIR A haplotype and the HLA-C2 group. This is believed to be due to an increased risk of pre-eclampsia when the mother lacks the AA haplotype and the foetus carried the HLA-C2 group (25). Further, KIR2DL3 was found to be associated with the development of cerebral malaria when the HLA-C1 ligand is present (26).
The first release of KDDB reported here includes only data we have extracted and curated from the scientific literature, identified by the HuGE Navigator. We are aware that the HuGE Navigator does not retrieve all studies and as such we are using other search strategies, for example via Pubmed and Web of Knowledge to locate studies missed in the first pass curation process. We have currently excluded studies that do not fit into the simple model of a case–control disease association study. Capturing more complex stratification studies is possible in KDDB, but will necessitate either some loss of granularity of the data, or the development of a much more complex schema and display interface. KDDB also does not yet contain any raw data, although the schema and submission pipeline are developed and tested to receive such data. KDDB is going to be maintained through our own data mining and curation efforts and through the submission of data from contributing laboratories (with suitable quality control procedures, as currently used in AFND). We are also exploring holding community workshops in the future to collect and collate data sets not yet in the public domain.
At present, we are not aware of any other site designed for public deposition of the raw data associated with immunogenetic disease association studies, and thus, these are not available for public analysis. The release of KDDB provides a new home for this raw data, and we encourage research groups that have published studies in the past, or those in the process of publishing new studies, to deposit the raw data within KDDB. We also encourage feedback from the scientific community on the utility of the data submission and query interface and the general approach we have taken to curation.
Conclusions
Over the last 10 years of existence, AFND has provided the immunogenetics and histocompatibility community with an online repository for the examination of frequencies in different healthy populations. With the development of the KDDB, our aim is to cover disease studies that have been associated with KIR genes and to include studies in which no significant association has been found, to avoid publication bias. In the future, we will extend the alleles covered to include other loci and new data sets as they are published. We anticipate that KDDB will greatly facilitate meta-analyses and data re-use to understand the underlying function of KIR genes in a variety of disease processes.
Funding
PhD studentship from CNPq (National Council for Scientific and Technological Development—Brazil) (to L.Y.C.T.). Funding for open access charge: University of Liverpool Library.
Conflict of interest. None declared.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljunggren HG, Karre K. In search of the ‘missing self': MHC molecules and NK cell recognition. Immunol. Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Moretta L, Biassoni R, Bottino C, et al. Human NK-cell receptors. Immunol. Today. 2000;21:420–422. doi: 10.1016/s0167-5699(00)01673-x. [DOI] [PubMed] [Google Scholar]
- 4.Liu WR, Kim J, Nwankwo C, et al. Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on Chromosome 19q13.4. Immunogenetics. 2000;51:659–669. doi: 10.1007/s002510000183. [DOI] [PubMed] [Google Scholar]
- 5.Wende H, Colonna M, Ziegler A, et al. Organization of the leukocyte receptor cluster (LRC) on human Chromosome 19q13.4. Mamm. Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 6.Bashirova AA, Martin MP, McVicar DW, et al. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genomics Hum. Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 7.Brusilovsky M, Rosental B, Shemesh A, et al. Human NK cell recognition of target cells in the prism of natural cytotoxicity receptors and their ligands. J. Immunotoxicol. 2012;9:267–274. doi: 10.3109/1547691X.2012.675366. [DOI] [PubMed] [Google Scholar]
- 8.Moretta A, Marcenaro E, Parolini S, et al. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008;15:226–233. doi: 10.1038/sj.cdd.4402170. [DOI] [PubMed] [Google Scholar]
- 9.Marsh SG, Parham P, Dupont B, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55:220–226. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- 10.Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 13.Robinson J, Halliwell JA, McWilliam H, et al. IPD—the Immuno Polymorphism Database. Nucleic Acids Res. 2013;41:D1234–D1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajalingam R, Krausa P, Shilling HG, et al. Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics. 2002;53:1009–1019. doi: 10.1007/s00251-001-0425-5. [DOI] [PubMed] [Google Scholar]
- 15.Norman PJ, Carrington CV, Byng M, et al. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun. 2002;3:86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Galarza FF, Christmas S, Middleton D, et al. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–D919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamil KM, Khakoo SI. KIR/HLA interactions and pathogen immunity. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/298348. 298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol. Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 19.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu W, Gwinn M, Clyne M, et al. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 22.Gourraud PA, Meenagh A, Cambon-Thomsen A, et al. Linkage disequilibrium organization of the human KIR superlocus: implications for KIR data analyses. Immunogenetics. 2010;62:729–740. doi: 10.1007/s00251-010-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parham P, Norman PJ, Abi-Rached L, et al. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos. Trans. R Soc. Lond. B Biol. Sci. 2012;367:800–811. doi: 10.1098/rstb.2011.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guinan KJ, Cunningham RT, Meenagh A, et al. Signatures of natural selection and coevolution between killer cell immunoglobulin-like receptors (KIR) and HLA class I genes. Genes Immun. 2010;11:467–478. doi: 10.1038/gene.2010.9. [DOI] [PubMed] [Google Scholar]
- 25.Hiby SE, Apps R, Sharkey AM, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J. Clin. Invest. 2010;120:4102–4110. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirayasu K, Ohashi J, Kashiwase K, et al. Significant association of KIR2DL3-HLA-C1 combination with cerebral malaria and implications for co-evolution of KIR and HLA. PLoS Pathog. 2012;8:e1002565. doi: 10.1371/journal.ppat.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]