As indicated by leading healthcare agencies like the Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO), the rising antibiotic resistance of bacteria is one of the major concerns of modern healthcare, and the development of novel growth-based rapid diagnostics is a key in addressing this problem.[1,2] A common growth-based test used to help guide therapy is the antimicrobial susceptibility test (AST). Faster AST testing can lead to reduced inappropriate antibiotic use, decreased health care costs, and reduced prevalence of antimicrobial resistance.[3,4] To address this problem our group has been developing asynchronous magnetic bead rotation (AMBR) biosensors that have been used to detect the growth of individual bacterial cells.[5] Here, we introduce a radically new type of AMBR sensor that detects bacterial growth-based on the rotation of a cluster of magnetic microparticles formed through self-assembly. Using these self-assembled AMBR biosensors, the minimum inhibitory concentration (MIC) value of a uropathogenic Escherichia coli isolate was measured. While detection of the presence of pathogens is a widely covered topic in research and assay development, here we focus on demonstrating that AMBR can be used to perform AST testing. A new method for detecting the rotation of AMBR biosensors is also described, which hass enabled validation of a multi-well prototype for rapid observation of bacterial growth.
Magnetic microparticle self-assembly in the presence of a magnetic field has been investigated by other groups, showing many interesting structures and dynamics. In fact, magnetic microparticles can self-assemble into various structures in an external magnetic field, including rods and disk-like clusters.[6-13] In a rotating magnetic field, we show that these clusters exhibit asynchronous rotation, similar to the behavior of single particles, and this allows for their use as AMBR biosensors. We also experimentally demonstrate that the rotation rate of these self-assembled clusters—made of antibody-coated magnetic beads—can be used to measure bacterial growth and their response to antimicrobials. When bacteria grow, they alter the drag of the rotating magnetic bead cluster. This is a key feature of the AMBR sensor, as changes in the drag can be due to changes in viscosity, volume and/or shape, see Figure 1.
Figure 1.
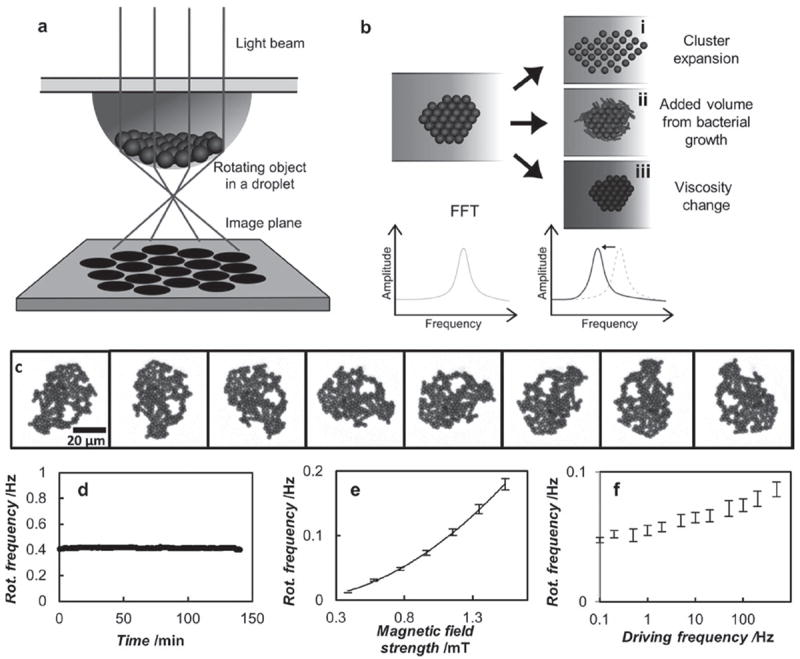
Overview of the AMBR biosensor technology. a) A schematic illustration of the droplet lensing effect used to amplify the rotational signal or the AMBR biosensor cluster: an LED or laser light is lensed by the droplet curvature, magnifying the shadow image of the particle cluster by 100-fold. After the magnification, the rotational period can be observed using a photodetector, which observes a periodic signal corresponding to the rotational period of the cluster. b) The rotational period of the cluster changes accordingly when i) the cluster expands, ii) bacteria etc. attach to the cluster, or iii) the viscosity of the surrounding fluid changes. c) An optical microscopy image sequence of a self-assembled AMBR biosensor rotating asynchronously in a rotating magnetic field. Frames are taken every 400 ms. d) The rotational frequency of a typical self-assembled AMBR biosensor as a function of time, where a few obvious outliers have been removed. The standard deviation of the frequency is 0.7%. e) The rotational frequency of a magnetic particle cluster as a function of the magnetic field strength. The data is fitted to a squared relationship. f) The rotational frequency as a function of the driving frequency, on a semi logarithmic axis. The error bars represent the deviations among four similar sized clusters (roughly 800 particles per cluster).
The rotational rate of magnetic particles in a rotating magnetic field is inversely proportional to the drag experienced by the particles when they are driven in the so-called asynchronous regime. This dependence enables their use as biosensors.[5,14-20] These AMBR biosensors are extremely sensitive to volumetric changes and they have been used to detect and monitor the growth of bacteria at the single cell level.[5,15] However, observing the AMBR biosensor signal of individual particles without a microscope can be challenging, due to their microscopic size and spherical form. Self-assembled AMBR biosensors were developed to address this challenge. Using a small self-assembled group of magnetic beads, instead of individual magnetic beads, allows for a more straight-forward implementation of AMBR biosensors on a multi-well prototype for bacterial growth studies.
Self-assembled systems can form in numerous ways and in countless shapes. Disk-like clusters of magnetic particles—which are of interest for the purposes of this paper—can be formed in aqueous solutions such as water or PBS-buffer. When formed in this fluidic environment, the clusters do not maintain their shape and, in our observations, change their shape nearly continuously. Due to the constantly changing drag, groups formed in this manner are difficult to use as AMBR biosensors. A similar shape-changing behavior was reported by Nagaoka et al.[6] To enable clusters to be used as AMBR biosensors, a curved interface was chosen, in combination with a modification of the surrounding media, which stabilized the cluster’s shape and location.
Cluster formation and dynamics are governed by the individual beads that make up the cluster. Here, for simplicity, we examine single bead behavior, which share the same dynamics as magnetic bead clusters. A single superparamagnetic microparticle placed in a rotating magnetic field (at a sufficiently high frequency) experiences a torque, τmag, in the direction of the rotating field:
| (1) |
where χ″ is the imaginary part of the magnetic susceptibility, Vm is the magnetic content volume, B is the magnetic field strength, μ0 is the permeability of free space, and ê is a unit vector pointing towards the direction of the magnetic field rotation. Neglecting inertial forces (which are minute compared to the drag forces) and Brownian rotation forces (which are minute compared to the magnetic torque) the opposing torque due to drag can be expressed by:
| (2) |
where κ is the Einstein shape factor (6 for a sphere), η is the dynamic viscosity of the surrounding fluid, V is the total volume of the rotating body, and θ is the angular orientation (θ̇ is the rotational rate of the object, in radians s−1). The rotational rate of the object can be solved by setting the magnetic torque equal to the fluidic drag and combining Equations 1 and 2, yielding:
| (3) |
At a constant temperature and a constant rotating magnetic field, the imaginary susceptibility χ″, magnetic content volume Vm, and the magnetic field strength B, all remain constant. Under these conditions, the rotation rate of the particle is primarily a function of the rotational drag:
| (4) |
where the drag (γ) is the product of the shape factor, viscosity, and volume: γ = κηV. The rotational period of the particle, T, can be written in terms of the rotational rate, θ̇, as T = 2π/θ̇. This can be combined with equation (4) to finally arrive at the equation relating the rotational period of the single bead superparamagnetic AMBR biosensor to the effective volume:
| (5) |
Therefore any changes in the drag of the cluster can be observed by measuring its rotational period in time.
Microscopy experiments were performed on a hanging droplet sample, where the self-assembled AMBR biosensor was formed on the air-water interface of an inverted 2 μL Müller-Hinton growth media droplet, see Figure 1a. The reason for using an inverted droplet sample was three-fold:
to minimize surface interactions between the interface and rotating magnetic beads,
to use gravity to help form the clusters in a reproducible location at the bottom of the droplet and to maintain the position during rotation, therefore “self-aligning”, and
to use the inverted droplet as a lens for magnification purposes, which is particularly useful for off-the-microscope measurements.
Magnifications can be 100-fold and higher, depending on the curvature of the drop and the distance away from the curved interface. These hanging-drop properties thus enabled the design of a simple and robust 16-well prototype that is shown in Figure 3b and discussed below.
Figure 3.
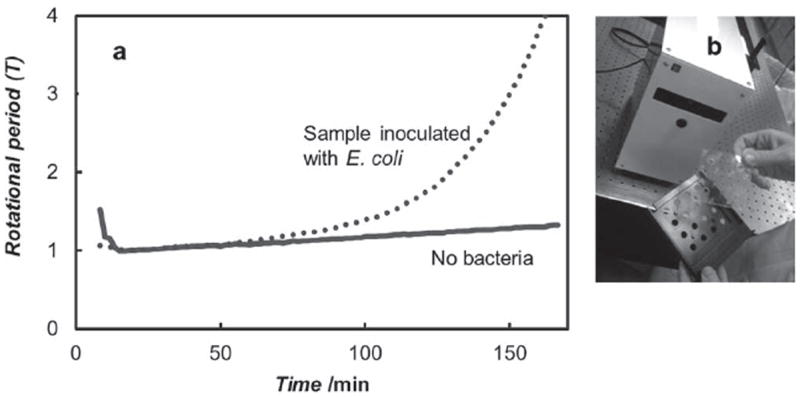
A Prototype device for rapid AST testing of bacteria using selfassembled AMBR biosensors. a) Data from the prototype showing the difference between a sample inoculated with E. coli bacteria compared to a sample with no bacteria. The rotational periods are normalized to one after initial stabilization. b) An image of the prototype device, including the transparent sample card holder and black drawer. The prototype houses sample cards with 16-wells, which are loaded with self-assembled AMBR biosensor to observe bacterial growth. The prototype is connected to a computer and controlled with a custom computer program written in NI LabView.
Once a cluster was formed and its location maintained at the bottom of a hanging drop, several properties of the fluidic sample can be measured using the cluster. Figure 1b shows how a change in sample properties can cause a change in the drag that the cluster experiences. Specifically, (i) cluster expansion, (ii) added volume from bacterial growth, or (iii) viscosity changes can be monitored by measuring a change in the drag and thus, the rotatioal period. Rotational periods are determined by standard FFT or autocorrelation analysis. One key aspect of being able to use the self-assembled cluster as an AMBR biosensor is that the cluster does not spontaneously break apart. Figure 1c shows a typical rotating cluster after being formed. Notice that while it has an amorphous shape, it maintains this shape throughout its rotation. This is especially important because change in shape will affect the rotation rate, as can be seen from Equation 5. To accomplish this stability in shape, a casein hydrolysate environment was used. In our studies, we found that groups could spontaneously rearrange if they were in water or PBS. However, upon addition of low amounts of casein hydrolysate (~1%), the group shape was stable throughout its rotation. It turns out that Müller–Hinton broth contains 1.75% of casein hydrolysate, which means that MH is well suited for self-assembled AMBR applications. Media that do not have casein hydrolysate, such as water and PBS, can be spiked with casein hydrolysate. Alternatively, different and more “sticky” beads can be used. Furthermore, MH is the standard broth used in growing non-fastidious bacteria and in performing AST testing in a clinical microbiology laboratory setting,[21] which is why it was used in this study.
Once a stable group is formed, its magnetic and rotational properties can be analyzed. While single particles, chains, and magnetic fluids have been magnetically rotated and characterized, a cluster, such as described here, has not been characterized in the literature, especially with respect to use as an AMBR sensor. Because the stabililty of the cluster is critical when used as an AMBR sensor, the first property analyzed was the rotational stability of the cluster. The rotational frequency (reciprocal of the rotational period) of an unperturbed AMBR cluster was found to be stable over the time period of hours, showing only a 0.7% variation over 2 hours (Figure 1d). This stability of the rotation rate forms the basis for the AMBR sensor’s sensitivity.[17] Furthermore, the rotational frequency of the AMBR cluster has a quadratic dependence on the driving magnetic field amplitude (at 100 Hz), which is expected from the literature for superparamagnetic rotators.[5,22] The rotational frequency is also expected to have only a weak dependence on driving frequency, which indeed is the case (Figure 1e). Notably, ferromagnetic rotators, which can also be used as AMBR biosensors, have a different dependency.[14,15]
Using the optical microscope setup to simultaneously measure the rotation rate of the clusters and optically monitor bacterial growth, we measured the MICs of two antibiotics for a uropathogenic E. coli strain (see Figure 2a–c). Unaffected cell growth can be observed in approximately one hour after the beads are washed and placed on the microscope. The self-assembled AMBR biosensors enabled MIC measurements and the determined values agreed with the traditional micro-dilution method. The clinical MIC of the uropathogenic E. coli isolate, as determined with an FDA approved automated system for susceptibility testing, the Vitek 2 (bioMerieux) was 16 μg mL−1 for streptomycin, and 2 μg mL−1 for gentamicin. The MIC values that we measured with AMBR were respectively 8 and 2 μg mL−1 (see Figure 2a,b), which are consistent with the reference MIC, within the accepted tolerance.[21] The growth and absence of growth measured with the AMBR sensors, see Figures 2a,b, also agree with what was observed microscopically. Figure 2c shows the microscopic images of the magnetic beads plus E. coli, in several antibiotic concentrations that were above and below the streptomycin MIC. With 0 μg mL−1 and 2 μg mL−1 concentrations, the magnetic beads (black) appear to be “pushed” apart and separated by a large number of E. coli cells (false colored red). In comparison, the 4 μg mL−1 and 8 μg mL−1 samples showed little or no apparent growth and the original structure of the cluster was maintained. When the AMBR biosensor would be used for identification purposes rather than here-described antimicrobial susceptibility testing, one would have to control for false positives, see Supporting Information for additional information. The authors note that the gold standard method for AST testing in a clinical laboratory is currently performed on pure cultures (in addition to being separate from ID), therefore specificity of the assay is not an issue for the current AST applications.
Figure 2.
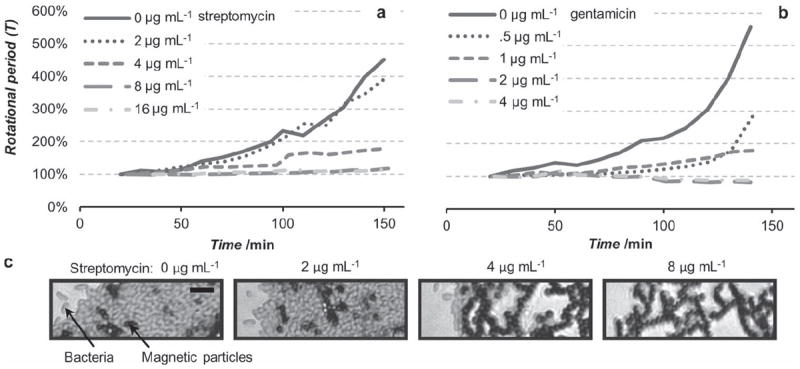
The rotational period of self-assembled AMBR biosensors used to observe the growth of uropathogenic E. coli with different concentrations of antibiotics of a) streptomycin and b) gentamicin. When the antibiotic is ineffective the bacteria keep growing and slow down the rotation of the cluster, and when it is effective the growth is inhibited keeping the rotational period constant. The MIC can be measured within 2 hours of inoculation. c) False colored optical microscopy images of a segment of the self-assembled AMBR biosensors in different concentrations of streptomycin (at 160 minutes), highlighting the presence or absence of the bacteria. Bacteria are false colored (red), and magnetic particles are black. Scale bar is 10 μm.
To further establish the feasibility of using the AMBR biosensor for clinical MIC testing, a 16-well prototype was built and tested, Figure 3. The prototype was built using off-the-shelf electronic components, including inductors for magnetic field generation, and laser diodes and photodiodes for observation, mirroring the schematic shown in Figure 1a. Data acquired with the prototype is shown in Figure 3a, demonstrating that the rotational signal time dependence for self-assembled AMBR biosensors inoculated with bacteria, compared to the biosensors with no bacteria present. A difference can be seen within two hours from sample introduction to the prototype. These results demonstrate that using a self-assembled cluster and standard off-the-shelf components, high-throughput off-the-microscope measurements can be achieved. Ongoing development has led to higher well count prototypes, lower protocol time (15 min), reduced starting concentration (1 × 104 CFU mL−1), and reduced time to results.
To summarize, we have reported on the development of a novel type of self-assembled AMBR biosensor, and its use for antimicrobial susceptibility testing (AST). Using the self-assembled AMBR biosensors, streptomycin and gentamicin minimum inhibitory concentrations against a uropathogenic bacteria, E. coli, were measured (8 and 2 μg mL−1 respectively), and found to agree with clinical reference methods. A high throughput prototype was constructed using exclusively off-the-shelf components for operating the biosensors. Therefore, self-assembled AMBR biosensors offer a high throughput method for measuring bacterial growth and its inhibition to chemical agents, with a potential to improve clinical AST applications.
Experimental Section
Magnetic Particles Functionalization
Streptavidin functionalized 2.8 μm diameter superparamagnetic particles (Invitrogen, Dynal M-280) were further functionalized with biotinylated anti E. coli antibodies (Abcam, ab20640-1), using a modified version of the manufacturers adsorption protocol.[23] Specifically, M-280 bead stock solution (100 μL), containing 6–7 × 108 beads mL−1 was combined with an equal volume of Phosphate Buffered Saline (PBS) with 0.1% Bovine Serum Albumin (BSA) and 0.1% Tween-20 solution, referred to as PBS-TB. Beads were magnetically separated and resuspended in PBS-TB solution (200 μL). This solution was pipetted into 4 mg mL−1 antibody solution (30 μL) and mixture was rotated in a 1.5 μL vial at room temperature for 24 hours, end-over-end at 60 rpm, and then allowed to sit at room temperature for 1.5 hours. Particles were magnetically separated and washed three times in PBS and resuspended in PBS (1 mL) resulting in a concentration of 6–7 × 107 beads mL−1.
Bacterial Growth Condition
A uropathogenic Escherichia coli isolate (obtained from Clinical Microbiology Laboratory, University of Michigan Hospital) was grown on Müller–Hinton (MH) agar media in 37 °C for 18–20 hours, suspended in Müller-Hinton broth and diluted to 0.5 McFarland standard, which corresponds roughly to 1.5 × 108 CFU mL−1.
Binding Protocol
To bind magnetic particles to the bacteria, the bacteria solution (12.5 μL) was combined with anti-E. coli-coated magnetic beads (10 μL) and MH broth (77.5 μL). The vial containing this mixture was shaken in a dish on a rocking platform shaker at ~180 rpm at 37 °C for 1.5–2 hours, to bind the bacteria to the magnetic beads. In our more recent experiments, this incubation time has been reduced to 10 minutes for bacterial concentration of 104 CFU mL−1. Bacteria-coated beads were then removed from solution using a handheld magnetic separator (Bio-Nobile, PickPen 1-M), temporarily released into MH (300 μL), dipped, but not released, in a second vial of MH and finally resuspended in MH (250 μL), resulting in a 3 ± 0.5 × 106 beads mL−1.
Samples of bacteria-coated beads were then combined with equal volumes of MH containing varying concentrations of antibiotics. For microscope experiments, two droplets (1.55 μL) of each antibiotic concentration were deposited onto PTFE-coated slides (Tekdon inc., 244-041-120) and inverted to create hanging droplets. Samples were taped and sealed using a greased (Apiezon, L grease) custom-cut (1.6 mm thick) rubber spacer and a glass slide. Sample preparation varied slightly when using the PDMS sample card holder for prototype measurements: final aliquots (7 μL) were put into the custom sample card holder and sealed with a second PDMS layer.
Self-assembly
Beads were pulled to the bottom of each droplet by holding the sample above a cone-shaped permanent magnet for 20 seconds. For measurements performed on a microscope, samples were then placed within a pair of Helmholtz coils (four coils total) on Olympus IX71 inverted microscope, in a custom built on-stage incubator that was held at 37 ± 1.5 °C. One of the two sets of coils was turned on creating a lateral oscillating magnetic field (100 Hz, 1 mT) to align particles. After 30 seconds of the one dimensional field, the second set of coils was turned on, creating an equivalent, orthogonal oscillating field 90° out of phase, resulting in a rotating magnetic field in the imaging plane (100 Hz, 1 mT). This rotating field formed and rotated clusters of beads at the bottom of the hanging droplets. The authors note that in some cases the 1D field could be used to eliminate the step of pulling the beads to the bottom of the drop using a cone magnet.
Observation
One droplet of each concentration was selected for observation. The only selection criterion was that the cluster not be visibly contaminated with foreign contaminants, such as fiber. Videos of each cluster rotation were taken at ten-minute intervals, at 50 fps, using a digital camera (Basler, piA640-210gm) with a 20× objective of the inverted microscope. Rotation rates were determined using a LabView program called StaT tracker (by the University of St Andrews Optical Trapping Group), which was modified to observe angular changes.
Prototype
The prototype used in Figure 3 was built by an engineering firm (Insight Product Development LLC, Chicago, IL) for the purpose of performing AST testing with the self-assembled AMBR biosensors. The design of the prototype was enabled by the self-assembled AMBR biosensors and self-aligning and lensing properties of the hanging droplet samples.
The prototype was designed to work with off-the-shelf electronic components; the rotating magnetic field is generated by passing sinusoidal currents (at 100 Hz) through standard inductors and the rotation rate of the AMBR biosensors is observed by aligning a collimated laser through each of the 16 sample holding droplets and monitoring the laser intensity with photodetectors. The sample holder card holds 16 hanging droplets and is made out of silicone rubber (polydimethylsiloxane, PDMS).
Supplementary Material
Acknowledgments
Funding was provided by the National Science Foundation (DMR 0455330, RK), the National Institute of Health (R21EB009550, RK and UL1RR024986, BHM, Postdoctoral Translational Scholar Program), Michigan Universities Commercialization Initiative (MUCI), and by a translational grant from the Wallace H. Coulter Foundation. Authors would like to thank Tomas Matusaitis of Insight Product Development, LLC for his expertise with prototype construction. Life Magnetics, Inc. has licensed this technology and now employs P.K. and B.H.M.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Paivo Kinnunen, Email: pkkinn@umich.edu, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA.
Dr. Brandon H. McNaughton, Email: bmcnaugh@umich.edu, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA; University of Michigan, Biomedical Engineering, 2200 Bonisteel Blvd., Ann Arbor, MI 48109, USA.
Theodore Albertson, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA.
Dr. Irene Sinn, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA
Sima Mofakham, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA.
Remy Elbez, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA.
Prof. Duane W. Newton, University of Michigan, Clinical Microbiology Laboratory, 1500 E. Medical Center Drive, Ann Arbor, MI 48109, USA
Prof. Alan Hunt, University of Michigan, Biomedical Engineering, 2200 Bonisteel Blvd., Ann Arbor, MI 48109, USA
Prof. Raoul Kopelman, Email: kopelman@umich.edu, University of Michigan, Department of Chemistry, 930 N. University, Ann Arbor, MI 48109-1055, USA.
References
- 1.Jorgensen JH, Ferraro MJ. Clin Infect Dis. 2009;49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 2.CDC - Get Smart: Antibiotic Resistance Questions and Answers. 2010 can be found under http://www.cdc.gov/getsmart/antibioticuse/anitbiotic-resistance-faqs.html.
- 3.Kerremans JJ, Verboom P, Stijnen T, Hakkaart-van Roijen L, Goessens W, Verbrugh HA, Vos MC. J Antimicrob Chemother. 2008;61:428–435. doi: 10.1093/jac/dkm497. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove SE. Clin Infect Dis. 2006;42:S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 5.Kinnunen P, Sinn I, McNaughton BH, Newton DW, Burns MA, Kopelman R. Biosensors Bioelectron. 2011;26:2751–2755. doi: 10.1016/j.bios.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagaoka Y, Morimoto H, Maekawa T. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.032502. [DOI] [PubMed] [Google Scholar]
- 7.Grzybowski BA, Stone HA, Whitesides GM. Nature. 2000;405:1033–1036. doi: 10.1038/35016528. [DOI] [PubMed] [Google Scholar]
- 8.Melle S, Calderón OG, Rubio MA, Fuller GG. Phys Rev E. 2003;68:041503. doi: 10.1103/PhysRevE.68.041503. [DOI] [PubMed] [Google Scholar]
- 9.Chen Q, Bae SC, Granick S. Nature. 2011;469:381–384. doi: 10.1038/nature09713. [DOI] [PubMed] [Google Scholar]
- 10.Climent E, Yeo K, Maxey MR, Karniadakis GE. J Fluids Eng. 2007;129:379–387. [Google Scholar]
- 11.Bacri JC, Cēbers A, Lācis S, Perzynski R. J Magn Magn Mater. 1995;149:143–147. [Google Scholar]
- 12.Whitesides GM, Grzybowski B. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 13.Grzybowski BA, Campbell CJ. Chem Eng Sci. 2004;59:1667–1676. [Google Scholar]
- 14.Behrend CJ, Anker JN, McNaughton BH, Kopelman R. J Magn Magn Mater. 2005;293:663–670. [Google Scholar]
- 15.McNaughton BH, Agayan RR, Clarke R, Smith RG, Kopelman R. Appl Phys Lett. 2007;91:224105. [Google Scholar]
- 16.McNaughton BH, Kinnunen P, Smith RG, Pei SN, Torres-Isea R, Kopelman R, Clarke R. J Magn Magn Mater. 2009;321:1648–1652. [Google Scholar]
- 17.Kinnunen P, Sinn I, McNaughton BH, Kopelman R. Appl Phys Lett. 2010;97:223701. doi: 10.1063/1.3505492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinn I, Kinnunen P, Albertson T, McNaughton BH, Newton DW, Burns MA, Kopelman R. Lab Chip. 2011 doi: 10.1039/C0LC00734J. [DOI] [PubMed] [Google Scholar]
- 19.Sinn I, Albertson T, Kinnunen P, McNaughton BH, Burns MA, Kopelman R. Anal Chem. 2012 doi: 10.1021/ac300128p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helgesen G, Pieranski P, Skjeltorp AT. Phys Rev A. 1990;42:7271–7280. doi: 10.1103/physreva.42.7271. [DOI] [PubMed] [Google Scholar]
- 21.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard–Eight Edition. Vol. 29. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. CLSI document M07-A8. [Google Scholar]
- 22.Janssen XJA, Schellekens AJ, van Ommering K, van Ijzendoorn LJ, Prins MWJ. Biosensors Bioelectron. 2009;24:1937–1941. doi: 10.1016/j.bios.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Bangs Laboratories, Inc. TechNote 204: Adsorption to Micro-spheres. 2008 can be found under http://www.bangslabs.com/files/bangs/docs/pdf/204.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


