In a previous publication of ours1 we showed the involvement of the myeloid translocation gene-related protein 2 gene (CBFA2T3) in a case of acute erythroid leukemia with the translocation t(1;16)(p31;q24). Because of lack of material available for analysis, we could not with certainty determine the leukemogenic mechanism, whether it be generation of a fusion gene with CBFA2T3 as one of the partners or loss of tumor suppressor activity, in which case genes KANK1and L1TD1, both homozygously lost, might be of the essence.
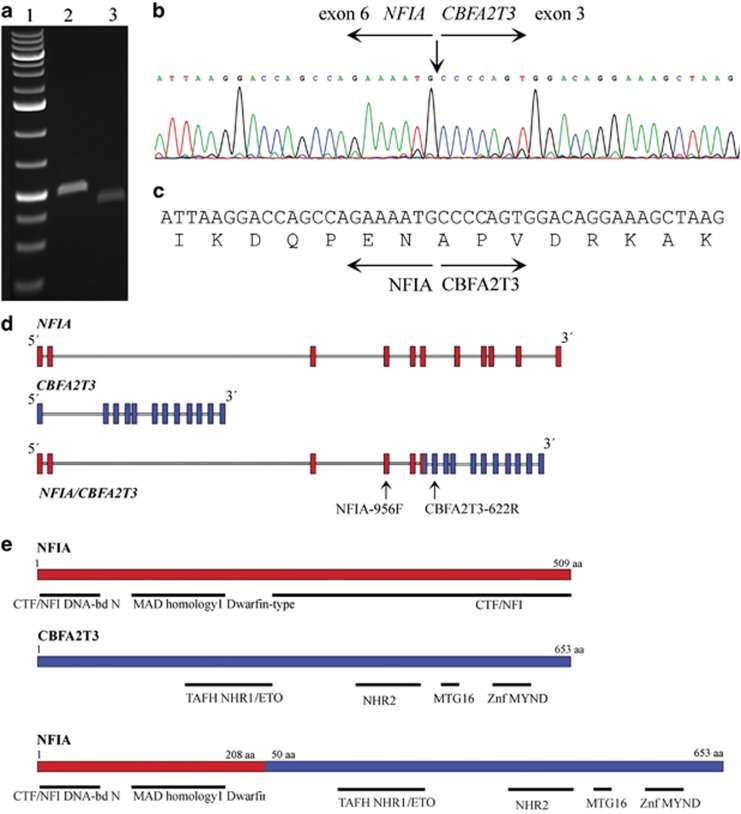
Some months after our article was published we got hold of 2 ml coagulated blood with 3% of abnormal cells from the same patient. As sequencing technology has also developed very fast lately, we decided to extract RNA from the sample and try to use it for high-throughput sequence analysis. The RNA was extracted and its quality checked by the Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA). A total of 3 μg of RNA was sent for high-throughput pair-end RNA-sequencing to the Norwegian Sequencing Center at the Ullevål Hospital (http://www.sequencing.uio.no/). The Illumina software pipeline was used to process image data into raw sequencing data and only sequence reads marked as ‘passed filtering' were used in the downstream data analysis. A total of 107 million reads were obtained. The FASTQC software was used for quality control of the raw sequence data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We used the fusion discovery software FusionMap (release date 16-04-2012)2 and the pre-built Human B37 and RefGene from the FusionMap website (http://www.omicsoft.com/fusionmap/). A list of over 500 possible fusion genes was obtained. A specific fusion involving the CBFA2T3 gene, which maps to chromosome band 16q24, and the nuclear factor I/A (NFIA) gene, which maps to chromosome band 1p31, was identified as number 10 in the list (seed count-rank 48; Supplementary Table 1). The involvement of the NFIA gene fits well with the fluorescence in situ hybridization (FISH) data on chromosome 1 previously obtained and published.1 The presence of the NFIA/CBFA2T3 fusion was verified by PCR using the NFIA-956 F and CBFA2T3–622 R primer combination (Supplementary Table 2). A specific PCR product of about 500 bp was identified and directly sequenced (Figure 1a). The specific fusion occurs between exon 6 of the NFIA gene (accession number NM_001134673.3) and exon 3 of the CBFA2T3 gene (accession number NM_005187.5; Figure 1b).
Figure 1.
Detection of the NFIA/CBFA2T3 fusion. (a) Gel picture showing the amplified fragment. Lane 1: ladder, lane 2: PCR product obtained with primer combination NFIA-924 F and CBFA2T3–622 R, lane 3: product of the NESTED-PCR obtained with primer combination NFIA-956 F and CBFA2T3–598 R. (b) Partial chromatogram showing the junction of the NFIA and CBFA2T3 genes. (c) Deduced amino acid sequence of the fusion transcript. (d) Schematic overview of the breakpoint region of the NFIA and CBFA2T3 genes. The exons are not in scale. Arrows point to primer positions. (e) Schematic overview of the position of the different domains of the NFIA and CBFA2T3 proteins and the NFIA/CBFA2T3 chimeric protein, according to ensembl (http://www.ensembl.org/index.html).
The NFIA/CBFA2T3 fusion gave an open reading frame and is expected to lead to a chimeric protein containing 208 amino-acid residues from NFIA (according to NP_001128145.1) and 603 residues from CBFA2T3 (according to NP_005178.4). The predicted fusion protein should thus consist of 811 amino acids (Figure 1c).
The NFIA gene encodes a member of the NFI family of transcription factors (http://genome.ucsc.edu). Interestingly, it has been found that NFIA exhibits a marked lineage-specific expression pattern in normal human hematopoiesis; it is upregulated in the erythroid lineage but fully suppressed in granulocytopoiesis.3 It has been shown that in early hematopoiesis, the NFIA expression level acts as a factor channeling hematopoietic progenitor cells into either the erythroid or granulocytopoietic lineage.3 The NFI proteins have a DNA-binding and dimerization domain in their N-terminal half, which contains four cysteine residues, and a transactivation and repression domain in their C-terminal half.4 The NFIA gene was found involved in an NFIA/EHF chimeric fusion in one breast cancer cell line out of 24 breast tumors analyzed (nine cell lines and 15 primary tumors).5 However, its role as either a passenger event or a direct, albeit infrequent, contributor to breast cancer development remains uncertain.
CBFA2T3 encodes an ETO myeloid translocation gene family protein, which interacts with DNA-bound transcription factors and recruits a variety of corepressors to facilitate transcriptional repression.6, 7, 8 The t(16;21)(q24;q22) translocation is one of the less common karyotypic abnormalities specifically associated with acute myeloid leukemia (AML). The translocation produces a chimeric gene made up of the 5'-region of the runt-related transcription factor 1 (RUNX1) gene fused to the 3′-region of CBFA2T3 (Figure 1d). In AMLs with either t(8;21) or t(16;21), the transcription factor RUNX1 is juxtaposed to one of the zinc finger nuclear proteins CBFA2T1 and CBFA2T3, respectively, resulting in transcriptional repression of RUNX1 target genes.6 Lately, its involvement as a partner in fusion genes was underlined by the identification of a IGH/CBFA2T3 fusion in a case of Burkitt lymphoma and a diffuse large B-cell lymphoma.9 This gene is also a putative breast tumor suppressor.10, 11 Interestingly, CBFA2T3 is downregulated during erythroid differentiation, and it has been suggested to have a repressive role in early, as well as late human erythroid differentiation.12 Hildebrand et al.6 demonstrated that the nuclear protein ETO (eight-twenty-one, a family to which also CBFA2T1, CBFA2T2 and CBFA2T3 belong) does not show reduced repressor activity even if it lacks the first 236 amino acids. As in the present fusion the altered CBFA2T3 protein lacks only the first 50 amino acids, we assume that its repressor activity is still retained (Figure 1e). More specifically, we hypothesize a pathogenetic parallel between AML showing a t(8;21) or t(16;21) and the present erythroleukemia with the 1;16-translocation with transcriptional repression of the NFIA target genes in the present case.
As the karyotype was described as 46,XY,der(1)t(1;1)(p31;q21),del(1)(p11p31),der(16)t(1;16)(p31;q24), that is, presented additional rearrangement besides the 1;16-translocation, we decided to screen the list of possible fusion genes in search of genes located in karyotypic breakpoints to see if those were involved in fusions as well. We identified four possible fusions (seed count-rank >12) where one of the genes mapped to a breakpoint position on chromosome 1. An analysis of the hypothetical fusions using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed in one of the two genes high-sequence identity with several genes and/or numerous repetitive sequences (for example, SINE). Hence, the reality of the putative fusions was seriously called into question and no further investigations were undertaken.
In addition to the present case, two more cases of erythroleukemia showing a t(1;16)(p31;q24) in their karyotype13, 14 can be found in the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer.15 All three patients (including ours) were very young children, and clinical outcome was poor. We assume that a NFIA/CBFA2T3 fusion existed also in these leukemias, but no evidence is at hand to corroborate or falsify this assumption.
In summary, we describe the first fusion gene identified in acute erythroleukemia. Knowledge of its specific functions in the neoplastic context is still incomplete, but pathogenetic similarities with other leukemic fusion genes are readily discernible. As for other leukemias characterized genetically by fusion genes, one may assume that the detailed pathogenetic knowledge now emerging may eventually form a starting point from which therapeutic attempts may begin.
Acknowledgments
This study was approved by the Regional Ethics Committee (REK number: S-07474a) and the institutional review board. The study was supported by grants from the Norwegian Cancer Society and the South-Eastern Norway Regional Health Authority. The sequencing service was provided by the Norwegian High-Throughput Sequencing Center, a national technology platform supported by the ‘Functional Genomics' and ‘Infrastructure' programs of the Research Council of Norway and the South-Eastern Regional Health Authorities.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Micci F, Thorsen J, Haugom L, Zeller B, Tierens A, Heim S. Translocation t(1;16)(p31;q24) rearranging CBFA2T3 is specific for acute erythroid leukemia. Leukemia. 2011;25:1510–1512. doi: 10.1038/leu.2011.100. [DOI] [PubMed] [Google Scholar]
- Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- Starnes LM, Sorrentino A, Pelosi E, Ballarino M, Morsilli O, Biffoni M, et al. NFI-A directs the fate of hematopoietic progenitors to the erythroid or granulocytic lineage and controls beta-globin and G-CSF receptor expression. Blood. 2009;114:1753–1763. doi: 10.1182/blood-2008-12-196196. [DOI] [PubMed] [Google Scholar]
- Bernard F, Gelsi-Boyer V, Murati A, Giraudier S, Trouplin V, Adelaide J, et al. Alterations of NFIA in chronic malignant myeloid diseases. Leukemia. 2009;23:583–585. doi: 10.1038/leu.2008.228. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand D, Tiefenbach J, Heinzel T, Grez M, Maurer AB. Multiple regions of ETO cooperate in transcriptional repression. J Biol Chem. 2001;276:9889–9895. doi: 10.1074/jbc.M010582200. [DOI] [PubMed] [Google Scholar]
- Kumar R, Cheney KM, Neilsen PM, Schulz RB, Callen DF. CBFA2T3-ZNF651, like CBFA2T3-ZNF652, functions as a transcriptional corepressor complex. FEBS Lett. 2010;584:859–864. doi: 10.1016/j.febslet.2010.01.047. [DOI] [PubMed] [Google Scholar]
- Kumar R, Selth LA, Schulz RB, Tay BS, Neilsen PM, Callen DF. Genome-wide mapping of ZNF652 promoter binding sites in breast cancer cells. J Cell Biochem. 2011;112:2742–2747. doi: 10.1002/jcb.23214. [DOI] [PubMed] [Google Scholar]
- Salaverria I, Akasaka T, Gesk S, Szczepanowski M, Burkhardt B, Harder L, et al. The CBFA2T3/ACSF3 locus is recurrently involved in IGH chromosomal translocation t(14;16)(q32;q24) in pediatric B-cell lymphoma with germinal center phenotype. Genes Chromosomes Cancer. 2012;51:338–343. doi: 10.1002/gcc.21919. [DOI] [PubMed] [Google Scholar]
- Kochetkova M, McKenzie OL, Bais AJ, Martin JM, Secker GA, Seshadri R, et al. CBFA2T3 (MTG16) is a putative breast tumor suppressor gene from the breast cancer loss of heterozygosity region at 16q24.3. Cancer Res. 2002;62:4599–4604. [PubMed] [Google Scholar]
- Kumar R, Manning J, Spendlove HE, Kremmidiotis G, McKirdy R, Lee J, et al. ZNF652, a novel zinc finger protein, interacts with the putative breast tumor suppressor CBFA2T3 to repress transcription. Mol Cancer Res. 2006;4:655–665. doi: 10.1158/1541-7786.MCR-05-0249. [DOI] [PubMed] [Google Scholar]
- Kiefer CM, Lee J, Hou C, Dale RK, Lee YT, Meier ER, et al. Distinct Ldb1/NLI complexes orchestrate gamma-globin repression and reactivation through ETO2 in human adult erythroid cells. Blood. 2011;118:6200–6208. doi: 10.1182/blood-2011-06-363101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller U, Haas OA, Ludwig WD, Bartram CR, Harbott J, Panzer-Grumayer R, et al. Phenotypic and genotypic heterogeneity in infant acute leukemia. II. Acute nonlymphoblastic leukemia. Leukemia. 1989;3:708–714. [PubMed] [Google Scholar]
- Castaneda VL, Parmley RT, Saldivar VA, Cheah MS. Childhood undifferentiated leukemia with early erythroid markers and c-myb duplication. Leukemia. 1991;5:142–149. [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F.Mitelman database of chromosome aberrations and gene fusions in cancerhttp://cgap nci nih gov/Chromosomes/Mitelman 2012. Available from: URL http://cgap.nci.nih.gov/Chromosomes/Mitelman .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.